Abstract
The evolutionarily conserved Mre11/Rad50/Nbs1 (MRN) complex is involved in various aspects of meiosis. Whereas available evidence suggests that the Mre11 nuclease activity might be responsible for Spo11 removal in Saccharomyces cerevisiae, this has not been confirmed experimentally. This study demonstrates for the first time that Mre11 (Schizosaccharomyces pombe Rad32Mre11) nuclease activity is required for the removal of Rec12Spo11. Furthermore, we show that the CtIP homologue Ctp1 is required for Rec12Spo11 removal, confirming functional conservation between Ctp1CtIP and the more distantly related Sae2 protein from Saccharomyces cerevisiae. Finally, we show that the MRN complex is required for meiotic recombination, chromatin remodeling at the ade6-M26 recombination hot spot, and formation of linear elements (which are the equivalent of the synaptonemal complex found in other eukaryotes) but that all of these functions are proficient in a rad50S mutant, which is deficient for Rec12Spo11 removal. These observations suggest that the conserved role of the MRN complex in these meiotic functions is independent of Rec12Spo11 removal.
In meiosis, one round of DNA replication is followed by two nuclear divisions that divide the genetic material equally over four haploid daughter cells. Meiotic recombination contributes to genetic diversity and is essential for correct disjunction of the homologous chromosomes in the first meiotic division. In meiotic prophase, after meiosis-specific DNA replication, the homologous chromosomes pair and recombine. In the following two nuclear divisions, the homologous chromosomes (meiosis I) and the sister chromatids (meiosis II) are segregated. The study of meiosis in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe has greatly contributed to our understanding of various meiotic processes. Because these model organisms are as distantly related to each other as to animals (35), detailed studies of similarities and differences between meiotic mechanisms in these yeasts are informative as to which mechanisms are conserved in higher eukaryotes.
The evolutionarily conserved Mre11/Rad50/Nbs1 (MRN) protein complex is involved in a wide range of early responses to DNA damage. Mutations in Nbs1 and Mre11 are responsible for the cancer-prone human disorders Nijmegen breakage syndrome and ataxia telangiectasia-like disorder. Central in this complex is the Mre11 nuclease, which is thought to be involved in double-strand break (DSB) end resection and DSB signaling (reviewed in reference 40).
The MRN complex is also involved in multiple aspects of meiosis. In S. cerevisiae, meiotic recombination is initiated by the topoisomerase-like protein Spo11 (17), which creates a DSB in the DNA. Spo11 remains covalently bound to the 5′ ends of the break (17) and is removed by endonucleolytic cleavage (28) to initiate subsequent DSB end resection and meiotic recombination. Meiotic DSB formation is abolished in S. cerevisiae MRN null mutants (6, 16). In an S. cerevisiae rad50S point mutant (a separation-of-function mutant with severe defects in meiosis but only mild defects in mitotic DNA repair) (2), meiotic DSBs are formed, but this mutant is unable to remove Spo11 from meiotic DSB ends (17). This observation has implicated the MRN complex in Spo11 removal. Since a nuclease-dead mre11-D56N mutant is defective in resecting meiotic DSBs (27), it has been proposed that the Mre11 nuclease activity is responsible for Spo11 removal, but this has not been confirmed experimentally. Also, in S. pombe, meiotic DSBs are formed by the Spo11 homologue, called Rec12 (7). An S. pombe rad50S mutant is defective in meiotic DSB repair, and this feature has been instrumental in the study of meiotic DSB formation in this organism (42). Although this phenotype is compatible with an involvement of the MRN complex in Rec12Spo11 removal, this has not been demonstrated experimentally.
Meiotic DSB formation in S. cerevisiae (30) is accompanied by an increase of micrococcal nuclease (MNase) sensitivity, suggesting that a more open chromatin structure could facilitate DSB repair. S. cerevisiae Mre11 is required for meiosis-specific chromatin remodeling, whereas Rad50 and Xrs2 (the S. cerevisiae Nbs1 homologue) are dispensable (29). Also in S. pombe, meiotic recombination hot spot activity at ade6-M26 has been associated with increased MNase sensitivity (23), but a role of MRN in this process has not been reported.
In the great majority of sexually reproducing eukaryotes, a meiosis-specific tripartite structure is formed during meiotic prophase; this is called the synaptonemal complex (SC) and is thought to be involved in various processes associated with chromosome pairing and recombination. Early in meiotic prophase, axial elements are formed along the sister chromatids of the individual chromosomes. Pairing and connection of the axial elements by transverse filaments lead to formation of the tripartite SC (in which the axial elements are now called lateral elements) (31). In S. cerevisiae rad50Δ and rad50S mutants, SC precursors are formed, but formation of a complete SC is blocked (2). In S. pombe wild-type (WT) cells, no fully formed SC is found, but instead linear elements (LEs) appear during meiotic prophase that show similarity to SC precursors in other organisms (4). It remains unknown if the MRN complex is involved in LE formation in S. pombe.
Mutants of sae2 in S. cerevisiae have a rad50S-like phenotype in meiosis as well as in mitotic cells (32), and it has been shown that Sae2 is required for Spo11 removal in meiosis (28). Recently, a novel gene, ctp1, was identified in S. pombe (1, 21), and its product shows homology to the mammalian tumor suppressor CtIP (33) and the more distantly related Sae2 protein in S. cerevisiae. CtIP/Ctp1 has been shown to interact with the MRN complex (33) and is involved in DSB end resection (21, 33), but a role for Ctp1CtIP in Rec12Spo11 removal in S. pombe has not been confirmed.
Whereas meiotic phenotypes of MRN and sae2 mutants have been studied extensively in S. cerevisiae, much remains unknown about the role of the MRN complex and Ctp1CtIP in S. pombe meiosis. In this study, we characterize meiotic phenotypes of S. pombe rad50 and rad32mre11 null and separation-of-function mutants. First, we demonstrate, for the first time in any organism, that the nuclease activity of Rad32Mre11 is required for Rec12Spo11 removal. Second, we demonstrate that Ctp1CtIP is required for Rec12Spo11 removal, confirming functional conservation between the distantly related S. pombe Ctp1CtIP and S. cerevisiae Sae2 proteins. Finally, we show that an S. pombe rad50S mutant has a defect in removing Rec12Spo11 but is proficient for meiotic recombination (measured in surviving spores), chromatin remodeling at the ade6-M26 recombination hot spot, and LE formation, whereas all of these functions are defective in the rad50Δ mutant.
MATERIALS AND METHODS
Yeast strains and techniques.
For strain construction and propagation, standard genetic methods and media were used (11). Strains used and constructed in this study are listed in Table 1.
TABLE 1.
S. pombe strains used in this study
| Straina | Genotype |
|---|---|
| 265 | h−smt0 leu1-32 ura4-D18 |
| 251 | h+ ura4-D18 leu1-32 |
| 260 | h−smt0 rad50::kanMX6 leu1-32 ura4-D18 |
| 284 | h+ rad50::kanMX6 leu1-32 ura4-D18 |
| 258 | h+ rad50-K81I leu1-32 ura4-D18 |
| 259 | h−smt0 rad50-K81I leu1-32 ura4-D18 |
| 263 | h+ rec12-152::LEU2 leu1-32 ura4-D18 |
| 264 | h−smt0 rec12-152::LEU2 leu1-32 ura4-D18 |
| 261 | h+ rad50::kanMX6 leu1-32 ura4-D18 rec12-152::LEU2 |
| 262 | h−smt0 rad50::kanMX6 leu1-32 ura4-D18 rec12-152::LEU2 |
| 266 | h+ leu1-32 ura4-D18 rec12-152::LEU2 rad50-K81I |
| 267 | h− smt0 leu1-32 ura4-D18 rec12-152::LEU2 rad50-K81I |
| 805 | h− smt0 ura4-D18 rad32-D65N |
| 814 | h+ rad32-D65N ura4-D18 |
| 810 | h−smt0 ura4-D18 rad32-D65N rad50::kan |
| 811 | h+ ura4-D18 rad32-D65N rad50::kan |
| 812 | h−smt0 ura4-D18 rad32-D65N rad50S |
| 813 | h+ ura4-D18 rad32-D65N rad50S |
| 352/354 | h−/h+ ade6-M216/ade6-M210 ura4-aim/ura4-aim ura4-D18/ura4-D18 rad50-K81I/rad50-K81I |
| 611 | h−smt0 ade6-M26 pat1-114 rec12-6HA:kanMX6† |
| 617 | h−smt0 pat1-114 ade6-M26 rec12-6HA:kanMX6 rad50-K81I |
| 649 | h−smt0 ade6-M26 rec12-6HA:kanMX6 rad50::kanMX6 pat1-114 |
| 418 | h+ mat1PD17::LEU2 leu1-32 arg6-1 |
| 417 | h−smt0 ade7-152 |
| 411 | h+ mat1PD17::LEU2 leu1-32 rad50-K81I arg6-1 |
| 412 | h−smt0 ade7-152 rad50-K81I |
| 407 | h+ mat1P::LEU2 leu1-32 rad50::kanMX6 arg6-1 |
| 408 | h−smt0 rad50::kanMX6 ade7-152 |
| 421 | h+ smt0 lys7-1 leu1-32 |
| 422 | h+ mat1PD17::LEU2 leu1-32 ade2-17 ura2-10 |
| 405 | h−smt0 lys7-1 leu1-32 rad50::kanMX6 |
| 406 | h+ mat1PD17::LEU2 ade2-17 ura2-10 leu1-32 rad50::kanMX6 |
| 415 | h−smt0 rad50-K81I lys7-1 leu1-32 |
| 416 | h+ mat1PD17::LEU2 rad50-K81I leu1-32 ade2-17 ura2-10 |
| 157 | h−smt0 ade6-M26 pat1-114 |
| h+ ade6-M26 pat1-114 leu1-32 | |
| h+ ade6-M26 rad50-kanMX6 pat1-114 leu1-32 | |
| h+ ade6-M26 rad32::ura4 pat1-114 leu1-32 | |
| h+ ade6-M26 rad50-K81I pat1-114 leu1-32 | |
| 207/209 | h−/h−smt0/smt0 pat1-114/pat1-114 ade6-M210/ade6-M216 ura4-aim/ura4-aim ura4-D18/ura4-D18 |
| 203/205 | h−/h−smt0/smt0 pat1-114/pat1-114 ade6-M210/ade6-M210 ura4-aim/ura4-aim ura4-D18/ura4-D18 rad50::kanMX6/rad50::kanMX6 |
| 148/152 | h−/h−pat1-114/pat1-114 ade6-M210/ade6-M216 ura4-aim/ura4-aim ura4-D18/ura4-D18 rad50-K81I rad50-K81I |
| 808 | h−smt0 pat1-114 rad32-D65N rec12-6HA:kan ade6-M26 |
| 834 | h−smt0 pat1-114 rad32-D65N rec12-6HA:kan ade6-M26 rad50::kan |
| 832 | h−smt0 pat1-114 rad32-D65N rec12-6HA:kan ade6-M26 rad50S |
| 824 | h−smt0 ctp1::kan pat1-114 rec12-6HA:kan |
Numbers are from the strain collection of E. Hartsuiker. The rec12-6HA-kanMX strains were created in the lab of K. Ohta. Other unpublished strains/constructs for this study were created by E. Hartsuiker.
Previously published procedures.
Measurement of meiotic spore viability and recombination (11), synchronization of meiotic cultures (4, 7), pulsed-field gel analysis (7), preparation of chromosome spreads and electron microscopy (4), and analysis of meiotic nucleosome remodeling at ade6-M26 (23) were described previously.
DNA-linked protein detection assay.
We developed a DNA-linked protein detection assay based on previously published procedures (17, 34). Premeiotic or meiotic cells (25 ml) were washed in 1 ml lysis buffer (8 M guanidine HCl, 30 mM Tris, 10 mM EDTA, 1% Sarkosyl, pH 7.5), resuspended in 750 μl lysis buffer, and lysed using glass beads (±0.8 g). The cell extract was incubated at 70°C for 15 min; these strongly denaturing conditions remove noncovalently bound proteins from the DNA. After clarification (15 min at 13,000 rpm in an Eppendorf centrifuge), one aliquot of extract was set aside for DNA quantification (see below), while the rest was loaded on a CsCl gradient consisting of 1-ml layers with densities of 1.82, 1.72, 1.50, and 1.45 g/ml. The gradients were centrifuged for 24 h at 30,000 rpm in a Sorvall AH650 rotor to separate the free proteins from the DNA.
To ensure equal DNA loading, the DNA concentration in the extract was measured and this value was used to adjust the volume of the fractions loaded on the slot blot. For this purpose, the aliquots of extract which were set aside for DNA quantification were treated overnight with RNase (0.5 μg/ml), and the DNA concentration was determined fluorimetrically using PicoGreen (Molecular Probes/Invitrogen detection technologies). After centrifugation, the gradients were fractionated into 0.5-ml fractions and adjusted amounts were loaded onto a slot blot.
To detect the presence of covalently bound hemagglutinin-tagged Rec12Spo11 in the DNA fractions, the membrane was probed with a monoclonal antibody (sc-7392; Santa Cruz). The membrane was processed using standard Western blot procedures and visualized using chemiluminescence. Using this procedure, control cultures of untagged strains showed only slight cross-hybridization with the top two fractions (fractions 9 and 10) from the CsCl gradient, which contained the free proteins. These fractions did not contain any DNA, were difficult to load on a slot blot as they tended to clog the membrane, and were therefore not loaded for most experiments. Slot blots of premeiotic cells showed no Rec12Spo11 signal in any of the DNA-containing fractions (data not shown).
RESULTS
S. pombe rad50S mutant is temperature sensitive for meiotic spore viability and DSB repair and deficient for Rec12Spo11 removal.
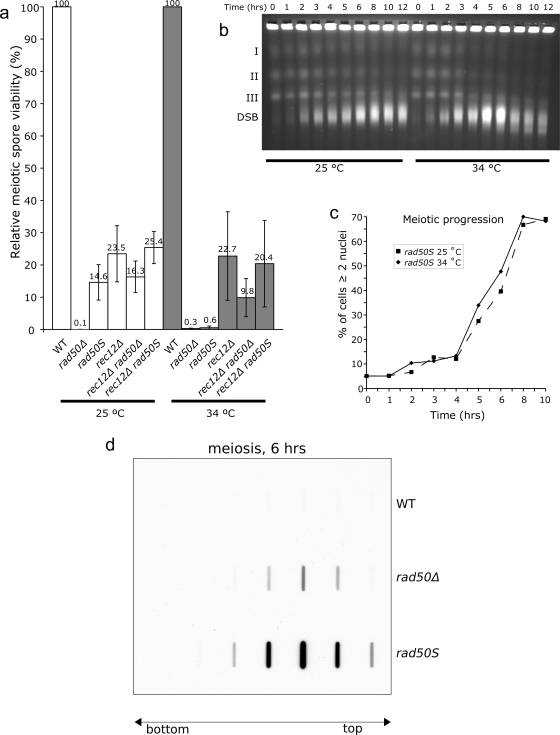
We previously created a rad50S mutant (rad50-K81I) which has been instrumental in the study of meiotic DSB formation in S. pombe (42). As previously reported (10), we found that the rad50S mutant is temperature sensitive for meiotic spore viability (Fig. 1a). At 34°C, the spore viability was 0.6% (similar to that of the rad50Δ mutant), and at 25°C, it was 14.6%. As shown previously for the rad50Δ strain (41), the low spore viability of the rad50S strain at 34°C was rescued by deletion of rec12spo11, suggesting that the reduced viability is due to a DSB repair defect in this mutant. To test if the temperature-sensitive spore viability phenotype of the rad50S mutant indeed reflects a defect in DSB repair, we looked at meiotic DSB formation and repair in a meiotic time course with the rad50S strain at permissive and restrictive temperatures, using pulsed-field gel electrophoresis (Fig. 1b). We found that at 34°C, the intact chromosomes were transformed into broken DNA fragments that remained unrepaired. At 25°C, most DNA was transformed into broken fragments, but a significant fraction of chromosomal DNA was intact near the end of the time course. After approximately 8 h, the intensity of the intact chromosomal DNA bands started to diminish. At this time, the majority of the cells had sporulated (Fig. 1c) (spore wall formation makes spores resistant to lysis, preventing the DNA from entering the gel). We concluded that the rad50S mutant is deficient for meiotic DSB repair at 34°C but partially proficient at 25°C.
FIG. 1.
The rad50S mutant is temperature sensitive for meiotic spore viability and defective for meiotic DSB repair and Rec12Spo11 removal. (a) Meiotic spore viability relative to that of the WT in different strains at 25°C and 34°C. Error bars show standard deviations, and values are averages for three independent experiments. (b) Pulsed-field gel electrophoresis of a synchronized meiotic pat1+ rad50S culture at 25°C and 34°C. The bands labeled I, II, and III correspond to the intact chromosomes, and the smears labeled DSB correspond to broken DNA fragments. (c) Meiotic progression of the time course presented in panel b, expressed as numbers of cells that have completed meiosis I at different time points. (d) Slot blot showing the presence of covalently bound Rec12Spo11 on the DNA 6 h after meiotic induction at 34°C. At time point 0, no Rec12Spo11 signals were visible (data not shown). The arrow indicates where the top and bottom fractions of the CsCl gradient were loaded. The bulk of the DNA was found in fractions 5, 6, and 7, which showed the strongest Rec12Spo11 signals for rad50 mutants.
We next asked if the low spore viability and inability to repair meiotic DSBs in rad50Δ and rad50s mutants was due to a defect in removing covalently bound Rec12Spo11 from the DSB ends. Based on previously published procedures (17, 34), we developed an assay (DNA-linked protein detection assay) to detect the presence of covalently bound Rec12Spo11 on the DNA (see Materials and Methods). As shown in Fig. 1d, both rad50Δ and rad50S mutants showed a strong presence of covalently bound Rec12Spo11 6 h after the initiation of meiosis, whereas in WT cells Rec12Spo11 was removed from the DNA. We consistently found higher covalently bound Rec12Spo11 levels in the rad50S strain than in the rad50Δ strain (please see Discussion for possible explanations). We concluded from these experiments that the rad50S mutant is temperature sensitive and, like the rad50Δ strain, is defective in Rec12Spo11 removal, leading to the inability to repair meiotic DSBs and a strong reduction in spore viability at restrictive temperature.
The rad50S strain is a separation-of-function mutant which is proficient for meiotic recombination functions independent of Rec12Spo11 removal.
To find out if the rad50S mutant is deficient only for Rec12Spo11 removal or also for other recombination-related functions which are defective in the rad50Δ mutant, we compared different meiotic phenotypes between the rad50Δ and rad50S strains.
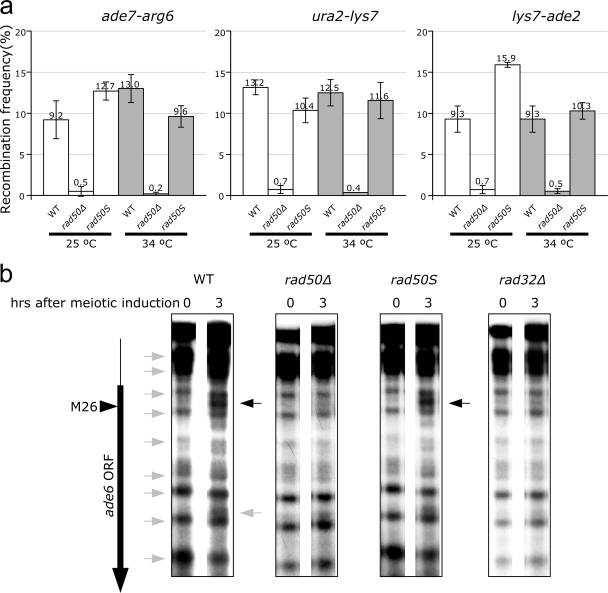
The repair of meiotic DSBs results in genetically detectable recombination when the homologous chromosome is used as a repair template, while recombination with the sister chromatid is usually silent. We determined meiotic recombination levels (in surviving spores) at three genetic intervals in rad50Δ and rad50S mutants at both 25°C and 34°C. As shown in Fig. 2a, meiotic intergenic recombination levels in the rad50Δ mutant were strongly reduced at all intervals tested at both 25°C and 34°C (a 28-fold reduction on average). Surprisingly, recombination levels in the rad50S mutant were similar to those of WT cells at both temperatures (1.0-fold on average).
FIG. 2.
The rad50S mutant is proficient for meiotic recombination and meiosis-specific nucleosome remodeling, but both functions are defective in the rad50Δ mutant. (a) Meiotic intergenic recombination levels in surviving rad50Δ spores were strongly reduced at different genetic intervals at both 25°C and 34°C. However, the rad50S strain was proficient for recombination at both temperatures. (b) Meiosis-specific nucleosome remodeling at ade6-M26 (see black arrow for the WT at 3 h) was defective at 34°C in the rad50Δ strain (and the rad32mre11Δ mutant), whereas the rad50S strain was proficient.
The S. pombe ade6-M26 mutation creates a hot spot for meiotic recombination. This hot spot activity has been associated with the creation of a meiosis-specific MNase hypersensitive site through chromatin remodeling (23). To see if this meiosis-specific MNase sensitivity is affected in MRN mutants, we performed an MNase assay on a synchronized meiotic time course. As shown in Fig. 2b, meiosis-specific chromatin remodeling at ade6-M26 (see black arrow for the WT at 3 h) was defective in both rad50Δ and rad32mre11Δ mutants, whereas remodeling was proficient in the rad50S strain.
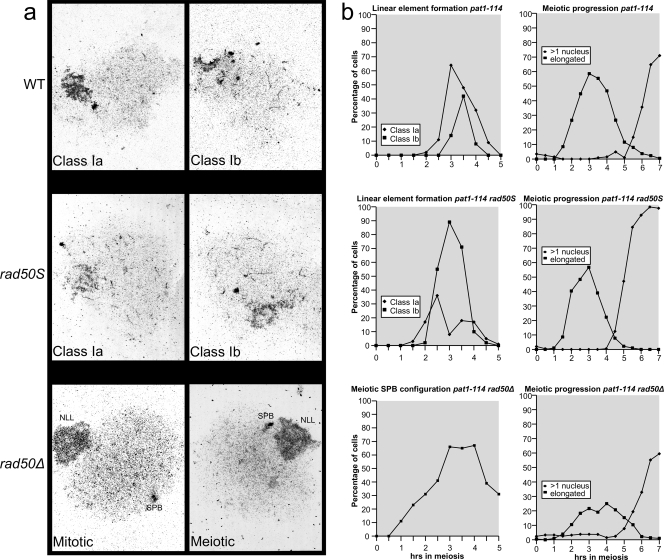
S. pombe LEs are similar to the axial elements of the SC in other eukaryotes and are believed to play a role in meiotic chromosome organization and recombination (4). To assess the role of the MRN complex in LE formation, we looked at LEs in meiotic spread preparations of rad50Δ and rad50S mutants (Fig. 3). We found that LEs in pat1-114 meiosis (used for its high degree of synchrony and relative stability of rad50Δ diploids [see Discussion]) (Fig. 1a) are shorter and lack the networks, bundles, and long elements normally found in pat1+ meiosis (4). We therefore classified pat1-114 LEs into classes 1a (short LEs) and 1b (longer LEs). LEs in the rad50S mutant were slightly longer and more abundant than those in the WT (signified by the increase of class 1b LEs in this mutant compared to the WT) (Fig. 3b), but LEs were totally absent in the rad50Δ mutant. We quantified the position of the spindle pole body (SPB) relative to the nucleolus (which is associated with the ribosomal DNA near both ends of chromosome 3) in rad50Δ cells to confirm that these cells went into meiosis. In mitotic cells, the SPB is found next to the centromeres, but upon induction of meiosis, the centromeres and telomeres switch positions and the telomeres associate with the SPB (9). As shown in the bottom left panel of Fig. 3b, the majority of the rad50Δ cells showed a meiotic SPB configuration, confirming that these cells went into meiosis.
FIG. 3.
The rad50S mutant is proficient for linear element formation, whereas LEs are absent in the rad50Δ mutant. (a) Electron micrographs of lysed and spread meiotic nuclei. LEs in pat1-114 meiosis (used for its high degree of synchrony) were shorter than those in the WT (pat1+) (4), whereas networks and bundles were not detected. LEs in pat1-114 meiosis were classified into classes 1a (short LEs) and 1b (long LEs). LEs in rad50S cells were slightly longer and more abundant than those in the WT. LEs were absent in rad50Δ cells. The bottom two panels (rad50Δ) illustrate typical SPB orientations in mitotic (opposite the nucleolus [NLL]) and meiotic (next to the nucleolus) cells. This allows the distinction between mitotic and meiotic cells and confirms that rad50Δ cells underwent meiosis in the absence of LEs. (b) (Left top and middle) Quantification of LE classes 1a and 1b at different time points. Class 1b was more abundant in rad50S cells than in the WT. (Bottom left) Quantification of rad50Δ cells (without LEs) containing an SPB configuration indicative of meiosis. At later time points, cells started to form ascus and spore walls, making the cells resistant to lysis, which led to an artifactual underrepresentation of meiotic cells. (Right) Quantification of DAPI (4′,6-diamidino-2-phenylindole)-visualized elongated (horsetail) nuclei indicative of meiotic prophase. The percentage of cells with more than one nucleus indicates progression through the first and second meiotic divisions. All quantifications are based on at least 100 cells per time point.
We concluded from this set of experiments that the rad50S strain is a separation-of-function mutant which is deficient only for Rec12Spo11 removal but proficient for other Rad50-dependent functions related to meiotic DSB repair.
Rad32Mre11 nuclease activity is required for Rec12Spo11 removal.
Although various observations suggest that the nuclease activity of Mre11 is responsible for Spo11 removal in S. cerevisiae, these experiments could not distinguish between a role for Mre11 in Spo11 removal and the subsequent exonucleolytic resection, and an involvement of the Mre11 nuclease activity in Spo11 removal has not been confirmed experimentally (19). The possibility that a nuclease other than Mre11 could be responsible for Spo11 removal is illustrated by the observation in S. pombe that the nuclease activity that degrades the C-rich strand at the telomeres (to create a 3′ G-rich strand overhang) is dependent on but not provided by the MRN complex and that a second single-stranded-DNA-specific endonuclease, Dna2 (3), is recruited by MRN and provides the nuclease activity (37, 38).
To distinguish between the possibilities that Rec12Spo11 is removed by Rad32Mre11 or by another nuclease recruited and/or controlled by MRN, we created a rad32mre11-D65N nuclease-dead mutant. This mutant is the equivalent of the well-characterized S. cerevisiae mre11-D56N mutant, which has been shown to be deficient for nuclease activity and proficient for MRN complex formation (18). Also, in S. pombe, the rad32mre11-D65N mutant is proficient for MRN complex formation (Nick Rhind, personal communication).
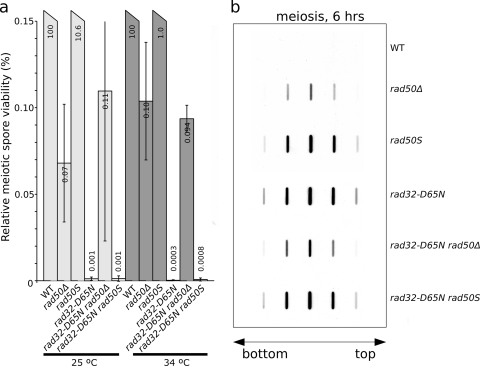
We first studied the meiotic spore viability of the rad32mre11-D65N mutant in combination with rad50Δ and rad50S mutations (Fig. 4a). The viability of rad32mre11-D65N and rad32mre11-D65N rad50S spores was strongly reduced compared to that of rad50Δ spores, whereas rad32mre11-D65N rad50Δ spore viability was rescued to approximately rad50Δ strain levels. We believe that these observations might reflect MRN complex stability in rad50S and rad32mre11-D65N mutants versus instability in the rad50Δ mutant. An intact but nuclease-deficient complex might block access of DSB ends to (as yet unknown) alternative removal activities, further decreasing spore viability. The double mutants with rad50S (and rad50Δ) did not show a lower spore viability than the single rad32mre11-D65N mutant, suggesting that they are all defective in the same Rec12Spo11 removal pathway.
FIG. 4.
The rad32mre11-D65N mutant is defective for Rec12Spo11 removal. (a) Analysis of spore viability epistasis between different rad32mre11-D65N mutants. Note that the graph shows only the lower range (<0.15%) of spore viability. Error bars show standard deviations, and values are averages for three independent experiments. (b) Analysis of Rec12Spo11 removal in different mutants at 34°C. Levels of covalently bound Rec12Spo11 were increased in rad32mre11-D65N mutant strains. The arrow indicates where the top and bottom fractions of the CsCl gradient were loaded. The bulk of the DNA was found in fractions 5, 6, and 7, which showed the strongest Rec12Spo11 signals in rad50 mutants.
The extremely low spore viability of the rad32-D65N mutant precluded a reliable measurement of meiotic recombination. Among 280 survivors, not a single recombinant was detected. However, prolonged snail enzyme digestion (which is used to kill vegetative cells) further reduced the number of survivors (unpublished observation), and it is therefore likely that these survivors did not result from meiotic spores but represent a small fraction of vegetative cells that resisted snail enzyme treatment.
Using the DNA-linked protein detection assay to detect covalently bound Rec12Spo11 (Fig. 4b), we found that the rad32mre11-D65N mutant was indeed defective in Rec12Spo11 removal, to a degree similar to that of the rad50S mutant. As in the spore viability assay, the defect was less pronounced in the rad50Δ and rad32mre11-D65N rad50Δ mutants, whereas the defect in the rad32mre11-D65N rad50S strain was comparable to that of the rad32mre11-D65N and rad50S mutants. Taken together, these data show that the Rad32Mre11 endonuclease activity is required for Rec12Spo11 removal in meiosis.
Ctp1CtIP is required for Rec12Spo11 removal.
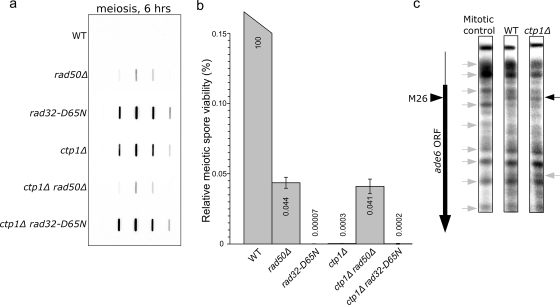
Sae2 shows only weak homology to Ctp1CtIP (21). S. cerevisiae sae2 mutants exhibit a rad50S mutant-like phenotype and are deficient for Spo11 removal (28, 32). Like the rad50S strain, the sae2Δ strain is only mildly methyl methanesulfonate sensitive (21). In contrast, the sensitivities of the S. pombe ctp1Δ mutant to ionizing radiation (21) and methyl methanesulfonate (13) are identical to those of MRN null mutants and much higher than those of the rad50S strain (13). Using pulsed-field gel electrophoresis, it was previously shown that a ctp1Δ mutant is deficient for meiotic DSB repair (1). To test for functional conservation between Sae2 and Ctp1CtIP, we analyzed the ctp1Δ strain for the ability to remove covalently bound Rec12Spo11 from the DNA. As shown in Fig. 5a, the ctp1Δ strain was as defective in Rec12Spo11 removal as the rad32mre11-D65N strain, suggesting a functionally conserved role for Ctp1/Sae2 homologues in Spo11 removal. Whereas covalently bound Rec12Spo11 levels were similar in the ctp1Δ mutant and a ctp1Δ rad32-D65N double mutant, these levels were comparatively lower in the rad50Δ mutant and a ctp1Δ rad50Δ double mutant.
FIG. 5.
(a) A ctp1 deletion mutant is deficient in removing Rec12Spo11 from the DNA in meiotic cells. The defect is comparable to that of rad50S and rad32mre11-D65N strains. (b) Meiotic spore viability is strongly reduced in the ctp1Δ strain, similar to that of the rad32-D65N strain. (c) The ctp1Δ strain is proficient for ade6-M26 chromatin remodeling (black arrow).
As shown in Fig. 5b, meiotic spore viability in the ctp1Δ strain was reduced to below rad50Δ strain levels, similar to that of the rad32-D65N strain. The spore viability was rescued to rad50Δ levels in a ctp1Δ rad50Δ double mutant but not in a ctp1Δ rad32-D65N double mutant, possibly reflecting the fact that the MRN complex remains intact in ctp1Δ strains. As explained above for the rad32-D65N strain, the extremely low viability of the ctp1Δ spores precluded a reliable measurement of meiotic recombination.
We also studied meiosis-specific chromatin remodeling and found that in the ctp1Δ mutant the ade6-M26 MNase hypersensitive site was present during meiosis (Fig. 5c) and thus that Ctp1CtIP is not required for this chromatin remodeling event.
DISCUSSION
Rad32Mre11 nuclease activity and Ctp1CtIP are required for Rec12Spo11 removal.
Several studies have suggested that Mre11 nuclease activity might be responsible for Spo11 removal. Moreau et al. (27) found that an S. cerevisiae mre11 nuclease-dead mutant was deficient in meiotic DSB end resection and proposed that Mre11 is responsible for removing Spo11. However, this study could not distinguish between a role of Mre11 in (endo)nucleolytic Spo11 removal and a role in (exo)nucleolytic resection downstream of Spo11 removal. Similarly, the presence of the Spo11 removal product (Spo11 with a covalently attached nucleotide) has not been studied with an mre11 nuclease-dead mutant (28). In this study, we thus provide the first direct demonstration that the Rad32Mre11 nuclease activity is indeed required for Rec12Spo11 removal in meiosis.
We have shown that rad50Δ, rad50S, rad32mre11-D65N, and ctp1Δ mutants are defective in removing Rec12Spo11 from the DNA. We consistently found higher levels of covalently bound Rec12Spo11 in rad50S, rad32mre11-D65N, and ctp1Δ mutants than in the rad50Δ mutant (e.g., see Fig. 1d, 4b, and 5a). This might be (partially) due to a reduced viability of rad50Δ cells (approximately 25% of rad50Δ cells were dead) (12). However, this reduced viability is unlikely to account fully for the threefold reduction in meiotic DSB formation in the rad50Δ strain (41). Also, levels of covalently bound Rec12Spo11 in the ctp1Δ strain were higher than those in the rad50Δ strain and only very slightly reduced compared to those in the rad50S and rad32mre11-D65N strains, while the growth defect of the ctp1Δ strain was comparable to that of MRN null mutants (1, 21). These observations suggest that Rad50 is required for WT levels of meiotic DSBs. In S. cerevisiae, RAD50 is absolutely required for meiotic DSB formation (6, 16).
The almost identical Rec12Spo11 removal defects of the rad50S and rad32mre11-D65N mutants suggest that Rad50 somehow controls the Rad32Mre11 nuclease activity. Based on structural studies, it has previously been proposed that ATP-driven directional switching of Rad50 controls the Mre11 nuclease activity (14). Interestingly, the rad50S mutation is found in a putative protein interaction site, and based on structural studies, it has previously been proposed that this site might interact with Sae2 (15). A recent study (33) showed that CtIP interacts directly with the MRN complex. Since the Rec12Spo11 removal defect in the ctp1Δ mutant is also similar to that of the rad32mre11-D65N mutant, this opens up the possibility that CtIP/Sae2 controls the Mre11 nuclease activity through its interaction with Rad50.
The most straightforward interpretation of our data is that the Rad32Mre11 nuclease is directly responsible for Rec12Spo11 removal. However, a recent study (20) showed that purified S. cerevisiae Sae2 possesses a nuclease activity which cleaves hairpin DNA structures in vitro, cooperatively with the MRN complex (called MRX in S. cerevisiae). Purified MRX promotes cleavage by enlarging a single-strand gap in the DNA opposite the Sae2 cleavage site. This raises the possibility that the coordinated action of Mre11 and Sae2 nuclease activities might be required and that Sae2 is ultimately responsible for Spo11 removal.
MRN null mutants are defective for meiosis-specific chromatin remodeling, LE formation, and recombination.
We found that meiotic recombination in the rad50Δ mutant was reduced approximately 28-fold, in line with the previously reported reduction in meiotic recombination in a rad32mre11Δ mutant (36). This reduction might be partially due to reduced DSB formation in this mutant (see the previous section). We showed that in rad50Δ and rad32Δ mutants, ade6-M26 chromatin remodeling is almost completely abolished. In contrast, in S. cerevisiae, only Mre11 is required for meiosis-specific chromatin remodeling at meiotic recombination hot spots, whereas Rad50 is dispensable for this process (29). The role of meiosis-specific chromatin remodeling and the role of MRN therein are not well understood, but they are probably involved in meiotic DSB formation and/or subsequent recombinational repair. S. cerevisiae Mre11 has also been implicated in chromatin remodeling during mitotic DSB repair (39).
We found that LE formation was totally abolished in the rad50Δ mutant. A potential caveat is that these experiments were performed with a pat1-114 mutant, which shows shortened LEs, while networks, bundles, and longer LEs (as found in a pat1+ strain) (4) are absent. Because of the extreme instability of h+/h− rad50Δ/rad50Δ diploids (12), we were not able to perform these experiments for pat1+ meiosis. In an S. cerevisiae rad50Δ mutant, shortened axial cores are formed, but they never form a tripartite SC structure (2). Most recombination-defective mutants studied so far do form (often aberrant) LEs (24, 25, 26). LE formation is also abolished is the rec10Δ mutant (26), and it has been shown that Rec10 is an LE component (22). Our observations raise the possibility that Rad50 fulfils a structural role or might regulate an early step in LE formation.
Rec12Spo11 removal is not required for meiosis-specific chromatin remodeling, LE formation, and recombination.
Whereas the rad50S strain is defective in Rec12Spo11 removal, we found that it is proficient for meiotic recombination, meiosis-specific chromatin remodeling, and LE formation, which are all defective in the rad50Δ mutant.
Meiotic recombination levels and levels of MNase sensitivity at ade6-M26 are very similar in the rad50S mutant and the WT. However, we found that LEs in rad50S cells were elongated compared to those in the WT. We speculate that this might be related to the prolonged presence of meiotic DSBs in the rad50S strain, maybe allowing more time for the LEs to mature. In an S. cerevisiae rad50S mutant, as in the rad50Δ mutant, no fully formed SC is found. However, axial cores in the rad50S strain are longer than those in the rad50Δ strain, and sometimes short stretches of tripartite structure are formed (2).
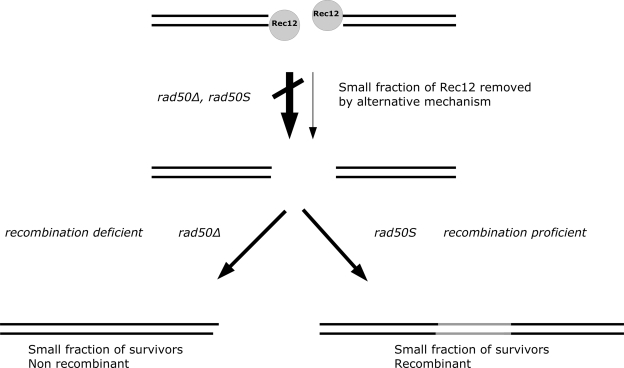
Figure 6 shows a diagram which explains our interpretation of the relationships between Rec12Spo11 removal, meiotic spore viability, and meiotic recombination in rad50Δ and rad50S mutants. Both the rad50Δ and rad50S mutants (at a restrictive temperature) are deficient for the removal of covalently bound Rec12Spo11 after meiotic DSB formation, leading to low spore viability. However, the presence of viable spores of these mutants suggests that a small fraction of cells are able to remove Rec12Spo11 (through an as yet unknown alternative mechanism), allowing repair of the DSB and viable spore formation. For the rad50Δ strain, these survivors show a strong reduction in recombination rates, suggesting that they survive through a nonrecombinogenic survival mechanism (possibly nonhomologous end joining or recombinational repair using the sister chromatid as a template). The rad50S cells are proficient for meiotic recombination (once Rec12Spo11 has been removed), and the surviving spores therefore show normal meiotic recombination levels.
FIG. 6.
Interpretation of the observed meiotic phenotypes of rad50Δ and rad50S mutants. Both rad50Δ and rad50S mutants (at a restrictive temperature) are deficient for the removal of covalently bound Rec12Spo11 after meiotic DSB formation, leading to low spore viability. However, a small fraction of cells are able to remove Rec12Spo11 (through an unknown mechanism), allowing repair of the DSBs and viable spore formation. For the rad50Δ mutant, these survivors show a strong reduction in recombination rates, suggesting that they survive through a nonrecombinogenic survival mechanism. The rad50S cells are proficient for meiotic recombination (once Rec12Spo11 has been removed), and the surviving spores therefore show normal meiotic recombination levels.
Conclusions and outlook.
Although it has been predicted that S. cerevisiae Mre11 nuclease activity is responsible for Spo11 removal, this has not been confirmed experimentally. This study demonstrates for the first time that the Rad32Mre11 nuclease activity is required for Rec12Spo11 removal. The Rec12Spo11 removal defect in the ctp1Δ strain suggests a functional conservation between the distantly related Sae2 and Ctp1CtIP proteins. We also confirmed that the S. pombe rad50S mutant is defective in Rec12Spo11 removal. Our finding that the temperature-sensitive rad50S mutant is proficient for meiotic recombination, meiosis-specific chromatin remodeling, and LE formation, functions which are all defective in the rad50Δ strain, suggests that the involvement of MRN in these functions is independent of Rec12Spo11 removal.
The conservation of the involvement of the MRN complex and Ctp1CtIP in Rec12Spo11 removal, SC/LE formation, and meiosis-specific chromatin remodeling in the distantly related yeasts S. cerevisiae and S. pombe suggests that these MRN functions might be conserved throughout the eukaryotic kingdom. The analysis of MRN and CtIP functions in meiosis of higher eukaryotes has been hampered by the inviability of relevant mutants. Whereas a mouse Rad50R83I mutant (with an allele equivalent to the rad50-K81I allele used in this study) is inviable, a Rad50K22M mutant (equivalent to the less-well-characterized S. cerevisiae R20M rad50S mutant) (2) is viable but shows only mild meiotic phenotypes (5). These observations might reflect either that the similar but not identical amino acid change (K22M in the mouse versus R20M in S. cerevisiae) might not confer a Spo11 removal defect or that the MRN complex is not involved in Spo11 removal in mice. Another study (8) suggests that the mouse MRN complex is involved in meiotic prophase progression, chromosome synapsis, and recombination.
The findings presented in this study have important implications for our understanding of the role of the MRN complex and CtIP in meiotic recombination, as their defects lead to chromosome nondisjunction, infertility, and chromosomal abnormalities.
Acknowledgments
We thank Gerry Smith for discussions and for sharing reagents and Eva Hoffmann and Alan Lehmann for comments on the manuscript.
This work was funded by the MRC (A.M.C.) and by a CRUK grant to E.H. (CRUK C20600/A6620).
Footnotes
Published ahead of print on 12 January 2009.
REFERENCES
- 1.Akamatsu, Y., Y. Murayama, T. Yamada, T. Nakazaki, Y. Tsutsui, K. Ohta, and H. Iwasaki. 2008. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 283639-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61419-436. [DOI] [PubMed] [Google Scholar]
- 3.Bae, S. H., and Y. S. Seo. 2000. Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 27538022-38031. [DOI] [PubMed] [Google Scholar]
- 4.Bähler, J., T. Wyler, J. Loidl, and J. Kohli. 1993. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121241-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, C. F., M. L. Sikes, R. Sullivan, L. E. Huye, M. M. Le Beau, D. B. Roth, O. K. Mirzoeva, E. M. Oltz, and J. H. J. Petrini. 2002. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 162237-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 611089-1101. [DOI] [PubMed] [Google Scholar]
- 7.Cervantes, M. D., J. A. Farah, and G. R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5883-888. [DOI] [PubMed] [Google Scholar]
- 8.Cherry, S. M., C. A. Adelman, J. W. Theunissen, T. J. Hassold, P. A. Hunt, and J. H. J. Petrini. 2007. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr. Biol. 17373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikashige, Y., D. Q. Ding, Y. Imai, M. Yamamoto, T. Haraguchi, and Y. Hiraoka. 1997. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 16193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farah, J. A., G. Cromie, W. W. Steiner, and G. R. Smith. 2005. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics 1691261-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutz, H., H. Leslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In Handbook of genetics, vol. 1. Plenum Press, New York, NY. [Google Scholar]
- 12.Hartsuiker, E., E. Vaessen, A. M. Carr, and J. Kohli. 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 206660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartsuiker E., M. J. Neale, and A. M. Carr. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33117-123. [DOI] [PMC free article] [PubMed]
- 14.Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105473-485. [DOI] [PubMed] [Google Scholar]
- 15.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101789-800. [DOI] [PubMed] [Google Scholar]
- 16.Johzuka, K., and H. Ogawa. 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 1391521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88375-384. [DOI] [PubMed] [Google Scholar]
- 18.Krogh, B. O., B. Llorente, A. Lam, and L. S. Symington. 2005. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 1711561-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38233-271. [DOI] [PubMed] [Google Scholar]
- 20.Lengsfeld, B. M., A. J. Rattray, V. Bhaskara, R. Ghirlando, and T. T. Paull. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limbo, O., C. Chahwan, Y. Yamada, R. A. M. de Bruin, C. Wittenberg, and P. Russell. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz, A., J. L. Wells, D. W. Pryce, M. Novatchkova, F. Eisenhaber, R. J. McFarlane, and J. Loidl. 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 1173343-3351. [DOI] [PubMed] [Google Scholar]
- 23.Mizuno, K., Y. Emura, M. Baur, J. Kohli, K. Ohta, and T. Shibata. 1997. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 11876-886. [DOI] [PubMed] [Google Scholar]
- 24.Molnar, M., J. Bähler, M. Sipiczki, and J. Kohli. 1995. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 14161-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molnar, M., S. Parisi, Y. Kakihara, H. Nojima, A. Yamamoto, Y. Hiraoka, A. Bozsik, M. Sipiczki, and J. Kohli. 2001. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157519-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molnar, M., E. Doll, A. Yamamoto, Y. Hiraoka, and J. Kohli. 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 1161719-1731. [DOI] [PubMed] [Google Scholar]
- 27.Moreau, S., J. Ferguson, and L. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale, M. J., J. Pan, and S. Keeney. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 4361053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta, K., A. Nicolas, M. Furuse, A. Nabetani, H. Ogawa, and T. Shibata. 1998. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc. Natl. Acad. Sci. USA 95646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta, K., T. Shibata, and A. Nicolas. 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 135754-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page, S. L., and R. S. Hawley. 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20525-558. [DOI] [PubMed] [Google Scholar]
- 32.Prinz, S., A. Amon, and F. Klein. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146781-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartori, A. A., C. Lukas, J. Coates, M. Mistrik, S. Fu, J. Bartek, R. Baer, J. Lukas, and S. P. Jackson. 2007. Human CtIP promotes DNA end resection. Nature 450509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw, J. L., J. Blanco, and G. C. Mueller. 1975. Simple procedure for isolation of DNA, RNA and protein fractions from cultured animal cells. Anal. Biochem. 65125-131. [DOI] [PubMed] [Google Scholar]
- 35.Sipiczki, M. 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1REVIEWS1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavassoli, M., M. Shayeghi, A. Nasim, and F. Z. Watts. 1995. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomita, K., T. Kibe, H. Kang, Y. Seo, M. Uritani, T. Ushimaru, and M. Ueno. 2004. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol. Cell. Biol. 249557-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomita, K., A. Matsuura, T. Caspari, A. M. Carr, Y. Akamatsu, H. Iwasaki, K. Mizuno, K. Ohta, M. Uritani, T. Ushimaru, K. Yoshinaga, and M. Ueno. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 235186-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukuda, T., A. B. Fleming, J. A. Nickoloff, and M. A. Osley. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, R. S., J. S. Williams, and J. A. Tainer. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochim. Biol. Cell. 85509-520. [DOI] [PubMed] [Google Scholar]
- 41.Young, J. A., R. W. Hyppa, and G. R. Smith. 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young, J. A., R. W. Schreckhise, W. W. Steiner, and G. R. Smith. 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9253-263. [DOI] [PubMed] [Google Scholar]