Abstract
Temporal regulation of gene expression is a hallmark of cellular differentiation pathways, yet the mechanisms controlling the timing of expression for different classes of differentiation-specific genes are not well understood. We previously demonstrated that the class II arginine methyltransferase Prmt5 was required for skeletal muscle differentiation at the early stages of myogenesis (C. S. Dacwag, Y. Ohkawa, S. Pal, S. Sif, and A. N. Imbalzano, Mol. Cell. Biol. 27:384-394, 2007). Specifically, when Prmt5 levels were reduced, the ATP-dependent SWI/SNF chromatin-remodeling enzymes could not interact with or remodel the promoter of myogenin, an essential early gene. Here we investigated the requirement for Prmt5 and the class I arginine methyltransferase Carm1/Prmt4 in the temporal control of myogenesis. Both arginine methyltransferases could bind to and modify histones at late-gene regulatory sequences. However, the two enzymes showed sequential requirements for gene expression. Prmt5 was required for early-gene expression but dispensable for late-gene expression. Carm1/Prmt4 was required for late- but not for early-gene expression. The reason for the requirement for Carm1/Prmt4 at late genes was to facilitate SWI/SNF chromatin-remodeling enzyme interaction and remodeling at late-gene loci. Thus, distinct arginine methyltransferases are employed at different times of skeletal muscle differentiation for the purpose of facilitating ATP-dependent chromatin-remodeling enzyme interaction and function at myogenic genes.
Skeletal muscle differentiation involves cooperation between myogenic basic helix-loop-helix transcription factors (MyoD, Myf5, myogenin, Mrf4), ubiquitous E proteins, myocyte-enhancer factor 2 proteins, histone-modifying enzymes, and ATP-dependent chromatin-remodeling enzymes. The involvement and requirement for individual chromatin-modifying and -remodeling enzymes during skeletal muscle differentiation has been intensely investigated in recent years. However, the interdependence of different enzymes affecting chromatin structure during myogenesis has not received as much attention. In addition, regulation of myogenic gene expression is further complicated by the temporal regulation that exists and separates myogenic genes into different classes based on when they are activated relative to the onset of differentiation. Whether chromatin-altering enzymes specifically and differentially contribute to aspects of temporal regulation is largely unexplored.
We and others have previously demonstrated that SWI/SNF chromatin-remodeling enzymes containing the Brg1 ATPase are directly required for the induction of myogenesis because they remodel chromatin structure at the regulatory regions of both early and late myogenic genes (13, 14, 30, 40). Numerous histone-modifying enzymes have also been implicated in the regulation of myogenic genes, including acetyltransferases, deacetylases, lysine methyltransferases, and arginine methyltransferases (reviewed in references 15, 38, and 41). Of particular interest to us are the arginine methyltransferases. Type I arginine methyltransferases generate asymmetric dimethyl arginines on substrate proteins, while type II arginine methyltransferases catalyze the formation of symmetric dimethyl arginines (reviewed in references 1, 2, and 43). Both Prmt5, a type II arginine methyltransferase, and Carm1/Prmt4, a type I methyltransferase, have been shown to act as coregulators for numerous gene activation and repression events (reviewed in references 2, 32, and 43), and both have been independently purified in large protein complexes containing Brg1 (33, 34, 45). The connections between Prmt5 and Brg1 led us to investigate possible cooperativity between these different types of chromatin-altering enzymes in cell differentiation systems shown to be Brg1 dependent.
Our previous work demonstrated that the class II arginine methyltransferase Prmt5 was required for myogenesis (10). Prmt5 associated with the myogenin promoter and locally dimethylated H3R8. Knockdown of Prmt5 protein levels resulted in a reduction of dimethylation of H3R8 at the myogenin promoter and, importantly, a nearly total loss of Brg1 binding, which prevented chromatin remodeling of the promoter. All subsequent transcription factor binding events and the initiation of myogenin expression were inhibited. Thus, the arginine methyltransferase was required for the function of the ATP-dependent chromatin-remodeling enzyme.
To further probe the relationships between different classes of chromatin-altering enzymes and to explore potential differences between the regulation of myogenin, encoded by a myogenic early gene, and the regulation of genes expressed later in the differentiation process, we investigated the requirement for Prmt5 in the expression of myogenic late genes and also examined the involvement of Carm1/Prmt4, which had previously been linked to myogenesis via regulation of myogenin expression (7). Our data demonstrate that both Prmt5 and Carm1/Prmt4 are associated with regulatory elements of representative late myogenic genes in vivo and in culture. We also found a concomitant enrichment in dimethylation of H3R8 and H3R17, substrates for Prmt5 and Carm1/Prmt4, respectively, at these loci. Despite the presence of Prmt5 at late-gene promoters, it is dispensable for transcriptional activation of late myogenic genes. In contrast, Carm1/Prmt4 was absolutely required for activation of late myogenic targets. In the absence of Carm1/Prmt4, Brg1 association with these promoter elements was lost, as were changes in nuclease accessibility that are concurrent with gene activation. These data support the assertion that a preferential requirement exists for Carm1/Prmt4 during the later stages of myogenic differentiation while Prmt5 governs the activation of early myogenic targets. The results indicate a differential requirement for two distinct protein arginine methyltransferases during the different stages of myogenic differentiation to facilitate loading and function of an ATP-dependent chromatin-remodeling enzyme. This relationship between the different classes of enzymes may represent a paradigm for cooperation between Prmts and ATP-dependent remodeling enzymes.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cells, Prmt5 antisense construct-expressing cell lines (33), and immortalized mouse embryo fibroblasts (MEFs) from wild-type (WT) or knockout (KO) Carm1 embryos (42, 47) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% calf serum. Prmt5 antisense cell lines were maintained in 2.5 μg/ml puromycin. Cells were grown to ∼50% confluence and then transdifferentiated into the skeletal muscle lineage by ectopic expression of MyoD or myogenin in combination with Mef2D1b, using the pBABE retroviral vector system (27, 28), as previously described (13, 16, 30, 37). The viral inoculum was applied to the subconfluent cells for 24 h, during which time the cells became confluent. Cells were then differentiated using DMEM supplemented with 2% horse serum and 10 μg/ml insulin. Mock-infected cells were also treated with DMEM and 2% horse serum, and all samples were maintained in the differentiation medium for up to 24 h. Samples were harvested at indicated times for RNA, protein, and chromatin immunoprecipitation (ChIP) analysis.
RNA isolation and reverse transcriptase PCR.
RNA was isolated using Trizol (Invitrogen) according to the manufacturer's instructions. Reverse transcriptase reactions performed to generate cDNA used 1 μg of RNA as previously described (10, 14, 30). Amplification of transcripts was quantified by quantitative PCR (Q-PCR) using the Opticon Engine (MJ Research) and primers previously described (10, 14, 30).
ChIP.
ChIP was performed and quantified as described previously using primers that were also previously described (10, 14, 30, 31). Primers spanning the desmin and muscle creatine kinase (MCK) promoters and enhancers were used, with no significant differences in binding to enhancer or promoter sequences observed. The immunoprecipitation step utilized rabbit polyclonal antisera raised against Brg1 (12), dimethylated H3R8, and Prmt5 (33, 34) and commercial antibodies raised against Prmt5 (611539 [Becton Dickinson]), Carm1/Prmt4 (A300-421A [Bethyl Labs]; 07-080 [Upstate]), and dimethylated H3R17 (07-214 [Upstate]). As an additional negative control, every sample was analyzed for the presence of the immunoglobulin H (IgH) enhancer sequences; no specific enrichment of IgH sequences was ever observed. Sequential ChIP (re-ChIP) analysis was performed as previously described (26) with antibodies against Prmt5, MyoD (12), and myogenin (sc576 [Santa Cruz]).
Western analysis.
Samples were harvested at various time points by scraping into 1 ml of phosphate-buffered saline (PBS) followed by brief centrifugation to obtain a cell pellet. Carm1 Western analysis was performed by using cell lysates from immortalized MEFs using the ReliaBlot protocol (Bethyl Labs), while all other cell pellets were resuspended in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 20% glycerol, 1 mM dithiothreitol), with freshly added protease inhibitors (1 μg/ml of pepstatin A, 4 μg/ml of leupeptin, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and lysed by brief sonication.
Following centrifugation at 10,000 × g for 10 min, cell debris was discarded, and lysate concentrations were quantified by spectrophotometry. Fifty micrograms of protein was loaded and electrophoresed on a sodium dodecyl sulfate (SDS) gel and then transferred overnight onto nitrocellulose membranes. Membranes were blocked for 1 h in 5% milk in 1× TBST (Tris-buffered saline plus Tween) and incubated in primary antibody diluted in 5% milk in 1× TBST overnight. Antibodies used included Brg1 antisera (12), Prmt5 antisera (34), Carm1 (Bethyl Labs), and PI3K (Upstate). Membranes were then washed three times for 5 min each time in 1× TBST, incubated in secondary antibody diluted in 5% milk in 1× TBST, washed three times for 5 min each time, and visualized by enhanced chemiluminescence (Amersham).
Tissue isolation and nucleus preparation.
Isolation of nuclei from liver and myofibers was performed as described previously (10, 31). Briefly, skeletal muscle tissue from the hind limbs of 4- to 6-week-old BL6 mice was dissected and minced on ice into 1-mm3 pieces. Minced samples were pooled and digested with 110 U of collagenase supplemented with 1 mM CaCl2 in PBS at 37°C for 1 h with agitation. Separation of satellite cells and myotubes was achieved by use of a 70-μm-pore-size filter (Becton Dickinson). Separated populations of cells were resuspended in lysis buffer (10 mM HEPES-KOH [pH 7.3], 10 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 0.2 mM PMSF, 3 μg/ml cytochalasin B, 10 μg/ml leupeptin) and nuclei were released by homogenization followed by Dounce homogenization. Integrity of nuclei was ascertained by light microscopy. This nucleus mixture was overlaid onto a sucrose gradient and centrifuged as previously described (10, 30, 31). Following centrifugation, nuclei were cross-linked for ChIP and snap-frozen using liquid nitrogen. All samples were then stored at −80°C until use.
REAA.
A restriction enzyme accessibility assay (REAA) was performed as described previously (14, 30). Briefly, nuclei were released from cells by Dounce homogenization, and DNA was quantified by spectrophotometry. One hundred micrograms of DNA was subjected to limited digestion with PvuII for 1 h at 37°C. Following digestion, DNA was ligated to a linker previously described (14, 30). Ligation-mediated PCR was used to quantify the amount of nuclease-accessible DNA using primers corresponding to linker DNA and specific gene loci as previously described (14, 30). Results were normalized to input genomic DNA using Q-PCR.
Plasmid construction.
pCDNA3.1-MyoD was constructed by isolating the EcoRI fragment encoding the MyoD cDNA from pEMSV-MyoD (11) and cloning it into EcoRI-digested pCDNA3.1 (Invitrogen).
GST pull-down experiments.
Competent BL21 Escherichia coli was transformed with pGEX-2TK, pGEX-Prmt5 (34), and pGEX-Carm1 (6). Cells were grown to an optical density of 0.8 and induced with 1.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Invitrogen catalog no. 15529019) for 6 h at 37°C. Cells were pelleted and resuspended in STE buffer (20 mM Tris [pH 7.6], 5 mM MgC12, 100 mM NaCl, 1 mM EDTA) containing 4 mg/ml lysozyme and incubated on ice for 15 min. Then, 5 mM dithiothreitol, 0.5 mM PMSF, and 1% (wt/vol) aprotinin were added sequentially, followed by brief vortexing. 1.5% Sarkosyl was added to the cells, followed by brief vortexing. Lysis of bacterial cells was accomplished by sonication and the protein lysate was centrifuged at 14,500 × g for 15 min at 4°C. The sonicated lysate was incubated with a slurry of glutathione beads resuspended in PBS for 30 min at 4°C while rocking. Beads were washed five times for 5 min each time with STE buffer and resuspended in a 50% slurry. Beads were resuspended in sample buffer and subjected to SDS-polyacrylamide gel electrophoresis, followed by Coomassie staining to quantify immobilized glutathione S-transferase (GST) or GST fusion proteins.
pCDNA3.1-MyoD, pCS2-myogenin (a kind gift from S. Tapscott), and pCDNA1.1-MEF2D1b (a kind gift from E. Olson) were incubated with the TNT quick coupled transcription/translation system (Promega catalog no. 1171, 2081) to generate [35S]methionine-labeled proteins. In order to preclear, radiolabeled proteins were resuspended in 500 μl of NETN buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.01% NP-40) and incubated with 35 μl of glutathione beads for 45 min at 4°C with rocking. Beads were discarded, and glutathione-immobilized GST, GST-Prmt5, and GST-Carm1 were then incubated with 35S-radiolabeled full-length MyoD, myogenin, and Mef2D1b for a minimum of 2 h at 4°C with rocking. Beads were washed five times for 5 min each time with NETN buffer, resuspended in sample buffer, boiled, and subjected to SDS-polyacrylamide gel electrophoresis. Gels were dried and were visualized using a PhosphorImager (Molecular Dynamics).
RESULTS
Binding of Prmt5 and Carm1 to regulatory regions of late myogenic genes.
Our previous studies showed that the protein arginine methyltransferase Prmt5 is required to facilitate the activation of the myogenin gene, an essential gene that is induced during the early stages of skeletal muscle differentiation (10). In that study, we also showed that genes expressed later during differentiation were not activated when levels of Prmt5 were reduced. However, it was unclear if the lack of late-gene expression was a direct consequence of Prmt5 reduction or was an indirect consequence of the failure to induce myogenin, which is required for late-gene activity and terminal differentiation (20, 29).
If specific arginine methyltransferases function at myogenic late genes, the enzymes and histones modified by these enzymes should be localized at regulatory sequences controlling late-gene expression. To address physiological relevance, we first performed ChIPs using primary skeletal muscle tissue. We isolated hind limb muscle from 4- to 6-week-old BL6 mice and prepared mature myofibers for ChIP analysis, as we described previously (10, 31). Liver was isolated as a negative control. ChIP results were quantified by real-time PCR. The data demonstrated that Prmt5 was bound to the regulatory regions of the MCK locus in mature myofibers, while no significant association was observed in the control liver tissue (Fig. 1A). This induction of Prmt5 binding was accompanied by enrichment in the amount of dimethylated H3R8 at the MCK locus in myofibers, thereby correlating the presence of both Prmt5 and a histone modification known to result from Prmt5 activity (10, 17, 32, 33).
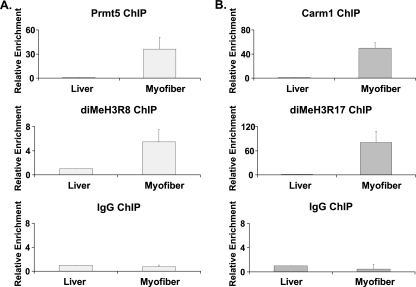
FIG. 1.
Prmt5 and Carm1 both bind to the MCK regulatory sequences in vivo. Skeletal muscle was dissected from the hind limbs of 4- to 6-week-old BL6 mice. Nuclei were isolated from muscle tissues and livers. ChIPs were performed using antibodies recognizing Prmt5, dimethylated H3R8 (diMeH3R8), Carm1, and diMeH3R17. Specific binding of Prmt5 and Carm1/Prmt4 was seen at gene regulatory regions of MCK in myofibers but not in liver. Corresponding increases in diMeH3R8 and diMeH3R17 were also observed specifically in myofibers. Values for binding in liver samples were set at 1. ChIPs with purified IgG generated background signals equivalent to the signals obtained with specific antibodies in liver samples. Data are averages plus standard deviations for four independent experiments.
Previously, the Carm1/Prmt4 enzyme was shown to localize to the MCK locus in differentiating C2C12 myoblasts, though the functional significance of this event was not addressed (7). Similarly, a considerable increase in the amount of Carm1/Prmt4 binding was seen at the MCK locus in myofibers, which correlated with an induction of dimethylated H3R17, a known epigenetic mark resulting from Carm1/Prmt4 activity (39). These results are the first demonstration that distinct protein arginine methyltransferases and histone modifications caused by these enzymes are present at regulatory regions of a late myogenic gene in terminally differentiated primary muscle tissue.
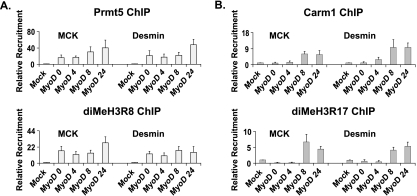
MyoD was originally identified as a cDNA that could convert immortalized mouse embryo fibroblast cell lines to the myogenic lineage (11). We turned to this well-established, manipulable cell culture system for myogenic differentiation to determine whether the association of arginine methyltransferases and specifically modified histones at late myogenic gene promoters was recapitulated in culture. Confluent, MyoD-expressing NIH 3T3 mouse embryo fibroblast cells were exposed to a low-serum differentiation cocktail for various times, and samples were evaluated by ChIP. Both Prmt5 and dimethylated H3R8 were already bound to regulatory regions of both the MCK gene and the desmin gene, which is expressed as a late gene in this and similar tissue culture systems (3, 30) at time zero, which is when the differentiation medium was added to the cultures (Fig. 2A). The presence of Prmt5 and dimethylated H3R8 was entirely dependent upon MyoD expression, since these proteins were absent from the loci in mock-differentiated cells (Fig. 2A). In contrast, Carm1/Prmt4 and dimethylated H3R17 did not show significant binding at late-gene loci until 8 h postdifferentiation (Fig. 2B), which in this culture system corresponds to the time when desmin and MCK gene expression is induced (30). As observed with Prmt5 and dimethylated H3R8, binding of Carm1 and dimethylated H3R17 did not occur in mock-differentiated cells and required MyoD expression. Once bound, both arginine methyltransferases and modified histones remained present throughout the differentiation time course (Fig. 2A and B). Thus, Prmt5 and dimethylated H3R8 binding preceded early- and late-gene expression, while Carm1/Prmt4 and dimethylated H3R17 binding correlated with the onset of late-gene expression.
FIG. 2.
Prmt5 and Carm1 bind to and methylate histones at the regulatory regions of late myogenic target genes in cell culture. MyoD-differentiated NIH 3T3 cells were harvested at different times for ChIP analysis, using antibodies recognizing Prmt5, dimethylated H3R8 (diMeH3R8), Carm1, and diMeH3R17 to analyze the temporal binding of these Prmts and the deposition of their corresponding histone modifications at late skeletal muscle target genes. The kinetics of binding of Prmt5 and diMeH3R8 (A) and Carm1 and diMeH3R17 (B) at the regulatory sequences controlling MCK and desmin in mock and MyoD-differentiated cells are shown. All values are relative to the values obtained for binding in mock-differentiated NIH 3T3 cells, which were set at 1. Data are averages plus standard deviations for four independent experiments.
Prmt5 associates with promoter elements of late myogenic targets but is not required for gene activation.
To address the requirement for Prmt5 in the induction of myogenic late genes, we utilized NIH 3T3-derived cell lines (c12 and c15) that stably express an antisense construct to Prmt5 (33). Figure 3A demonstrates that Prmt5 levels are significantly reduced under both mock and MyoD-induced differentiation conditions, in agreement with previous results (10). We previously demonstrated that ectopic expression of myogenin and the muscle-specific isoform of Mef2d (Mef2D1b) (25) in combination was sufficient to activate myogenic late genes and to drive myogenesis to completion in culture without inducing endogenous MyoD expression (30). Thus, we can utilize ectopic expression of myogenin and Mef2D1b to address whether Prmt5 and Carm1/Prmt4 are directly required for late-gene expression, because this method (i) bypasses the requirement for MyoD to synthesize myogenin and (ii) provides the muscle-specific Mef2D1b isoform to cooperate with myogenin in the activation of late-gene loci. We would expect that if either arginine methyltransferase is indirectly required for late-gene expression because of a requirement to synthesize early genes, lack of the arginine methyltransferase would not impact late-gene expression when myogenin and Mef2D1b are ectopically expressed. In contrast, if there is a direct requirement for either arginine methyltransferase during late-gene induction, simply providing myogenin and Mef2D1b should not be sufficient to induce late-gene expression. Before experimentally addressing these issues, we confirmed that the cell lines were still deficient for Prmt5 when differentiation was induced by myogenin and Mef2D1b (Fig. 3A).
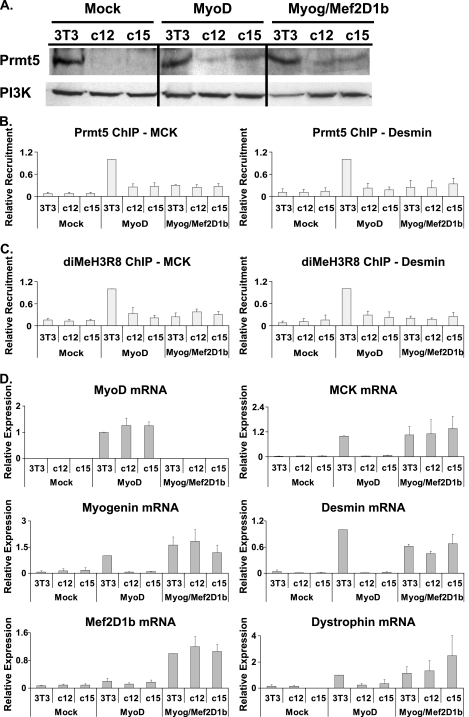
FIG. 3.
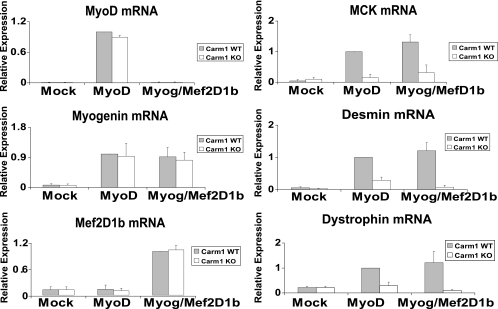
Prmt5 binds to late myogenic target genes but is not required for gene activation. (A) Western blot demonstrating the extent of Prmt5 expression in WT (3T3) and Prmt5 antisense (c12 and c15) cell lines that were mock differentiated or differentiated with MyoD or with myogenin plus Mef2D1b (Myog/Mef2D1b) for 24 h. Phosphoinositide 3-kinase (PI3K) levels are shown as a control. (B and C) ChIP experiments were performed using antibodies recognizing Prmt5 and dimethylated H3R8 (diMeH3R8) and were analyzed by Q-PCR. Values are relative to those obtained for binding in MyoD-differentiated NIH 3T3 cells, which were set at 1. (D) mRNA expression analysis for the indicated genes was performed by reverse transcriptase PCR and quantified by Q-PCR. Quantification of transcripts was normalized to the total amount of EF1-α mRNA. Values are relative to the expression values obtained in MyoD-differentiated NIH 3T3 cells, which were set at 1, except for the evaluation for Mef2D1b expression, where the value obtained for myogenin-Mef2D1b-differentiated cells was set at 1. Data in panels B to D are averages plus standard deviations for four independent experiments.
We first examined whether Prmt5 and dimethylated H3R8 were present on late-gene regulatory sequences in WT and Prmt5 antisense cell lines differentiated under the different conditions described above. Induced binding of Prmt5 at the MCK and desmin promoters was observed in MyoD-differentiated NIH 3T3 cells (Fig. 3B). As expected, Prmt5 antisense construct-expressing cell lines showed reduced levels of Prmt5 binding (Fig. 3B). Similarly, dimethylation of histone H3R8 was observed at the MCK and desmin promoters in the WT MyoD-differentiated cells and was significantly reduced in the MyoD-differentiated, Prmt5 antisense construct-expressing lines (Fig. 3C). Prmt5 and dimethylated H3R8 association with late myogenic gene sequences was specific to MyoD-differentiated cells, as no significant binding was seen in mock-differentiated samples or, interestingly, in myogenin-Mef2D1b-differentiated cells (Fig. 3B and C). These results support the assertion that Prmt5 is able to bind late-gene promoter regions only in the presence of MyoD, which binds to these loci prior to gene activation (30), and are consistent with previous studies showing that Prmt5 and MyoD could be coimmunoprecipitated from MyoD-differentiated cell extracts (10).
Analysis of gene expression in these cells was quantified by real-time PCR. Results for controls are presented in the first column of Fig. 3D. MyoD was equivalently expressed in each of the cell lines where it was introduced; as previously reported, no MyoD was detected in mock- or myogenin-Mef2D1b-differentiated samples (15, 30). Myogenin was equivalently expressed in lines that were myogenin-Mef2D1b differentiated. We note that the levels of introduced myogenin differed from the level normally induced by MyoD no more than twofold; thus, introduced myogenin was not grossly overexpressed. As previously reported (10), in MyoD-differentiated cells, myogenin expression was compromised in both of the Prmt5 antisense lines. Equivalent levels of Mef2D1b were present in the myogenin-Mef2D1b-differentiated cells (Fig. 3D).
We then examined late-gene expression. In WT MyoD-differentiated cells, induction of MCK, desmin, and dystrophin (encoded by another late gene), was observed, while MyoD-differentiated Prmt5 antisense construct-expressing cells failed to activate these late myogenic targets (Fig. 3D, right). In cells differentiated with myogenin-Mef2D1b, all of the cell lines were able to induce each of the late genes equivalently (Fig. 3D). These results, coupled with the ChIP data in Fig. 3B and C, indicate that Prmt5 and dimethylation of H3R8 are not directly required for the activation of myogenic late genes. Instead, Prmt5 is required for the activation of myogenin (10) (Fig. 3D) and thus is indirectly required for late-gene expression.
Carm1/Prmt4 associates with gene regulatory sequences of late myogenic targets and is required for transcriptional activation at these loci.
Since Prmt5 is not directly required for the activation of late myogenic targets, this prompted us to examine if Carm1/Prmt4, the other arginine methyltransferase physically present on myogenic late-gene regulatory sequences, could be coactivating late myogenic genes. To address the requirement for Carm1/Prmt4 in late-gene expression, we utilized immortalized MEFs derived from WT or Carm1/Prmt4-deficient mice (47). As expected, the cells were deficient for Carm1/Prmt4 under mock-, MyoD-, and myogenin-Mef2D1b-induced differentiation conditions (Fig. 4A).
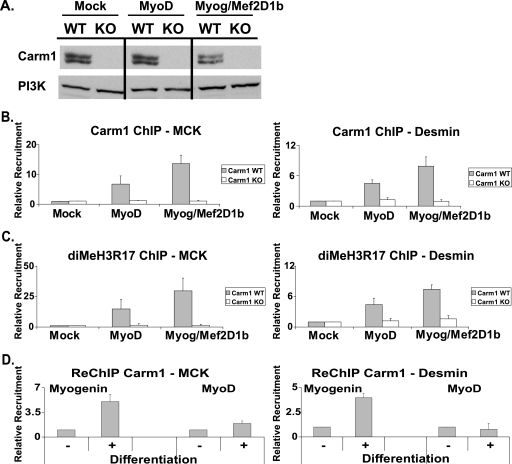
FIG. 4.
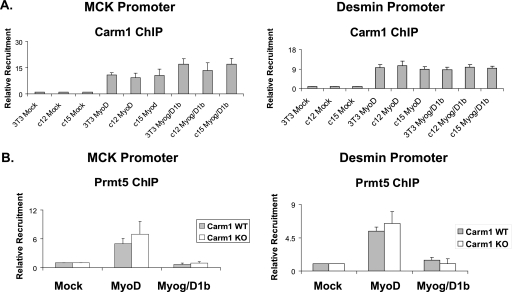
Carm1/Prmt4 and dimethylated H3R17 (diMeH3R17) bind to late myogenic gene regulatory regions. (A) Western blot showing Carm1/Prmt4 protein levels in immortalized WT or Carm1/Prmt4 KO MEFs that were mock differentiated or differentiated with MyoD or with myogenin plus Mef2D1b (Myog/Mef2D1b) for 24 h. Phosphoinositide 3-kinase (PI3K) levels are shown as a control. (B and C) ChIP experiments were performed using antibodies recognizing Carm1/Prmt4 and diMeH3R17 and were analyzed by Q-PCR. Values are relative to those obtained for binding in mock-differentiated WT MEFs, which were set at 1. (D) Re-ChIP experiments were performed and quantified by Q-PCR. Material immunoprecipitated with Carm1/Prmt4 antibodies was subsequently immunoprecipitated with either a MyoD or a myogenin antibody. Data are averages plus standard deviations for five (B and C) or four (D) independent experiments.
ChIP experiments were performed in mock-, MyoD-, or myogenin-Mef2D1b-differentiated WT and Carm1/Prmt4 KO immortalized MEFs. Carm1/Prmt4 binding to the MCK and desmin promoters was observed in MyoD-differentiated as well as in myogenin-Mef2D-differentiated WT MEFs (Fig. 4B). A concomitant enrichment in the amount of dimethylated H3R17 was observed at both promoters (Fig. 4C). As expected, no detectable Carm1/Prmt4 binding or enrichment in dimethylated H3R17 was observed in the mock-differentiated cells (Fig. 4B and C). In the KO MEFs, neither Carm1/Prmt4 nor dimethylated H3R17 was present, demonstrating that dimethylation of H3R17 at these loci is due to Carm1/Prmt4 (Fig. 4B and C). These findings demonstrate that Carm1/Prmt4 is able to directly bind late muscle promoter regions regardless of how gene activation is achieved; expression of either MyoD or myogenin-Mef2D1b is sufficient to promote binding and histone modification.
The binding of Carm1/Prmt4 to late-gene regulatory sequences at 8 h postdifferentiation (Fig. 2) corresponds to the timing of binding of myogenin and Mef2 at these promoters, as well as the interaction of the Brg1 ATPase of SWI/SNF chromatin-remodeling enzymes and concomitant increases in nuclease accessibility (30). The simultaneous occurrence of these events suggests that Carm1/Prmt4 serves as a coactivator of myogenin and Mef2D1b. We therefore performed re-ChIP analysis to determine whether myogenin and Carm1/Prmt4 were present together at late-gene regulatory sequences. We immunoprecipitated Carm1/Prmt4 from cross-linked chromatin isolated from MyoD-expressing cells before or after differentiation was induced and subsequently immunoprecipitated the Carm1/Prmt4-associated chromatin with antibodies against either myogenin or MyoD. The results demonstrate that Carm1/Prmt4 and myogenin were colocalized on both MCK and desmin regulatory sequences in differentiated but not undifferentiated cells (Fig. 4D). No colocalization of Carm1/Prmt4 with MyoD was observed (Fig. 4D), in agreement with previous observations that MyoD binding to late-gene regulatory sequences diminishes at the onset of late-gene expression (30). The data support the idea that Carm1/Prmt4 is simultaneously present with myogenin at myogenic late-gene loci.
Gene expression analysis was then performed by real-time PCR to assess the functional significance of Carm1/Prmt4 binding. MyoD was induced and equivalently expressed in MyoD-differentiated WT and KO MEFs but was not detectable in mock- or myogenin-Mef2D1b-differentiated samples (Fig. 5). Myogenin and Mef2D1b were equivalently expressed in myogenin-Mef2D1b-differentiated samples, and the levels of introduced myogenin were roughly equivalent to the levels normally induced by MyoD (Fig. 5). Interestingly, in MyoD-differentiated samples, myogenin expression was robust and was not compromised in KO MEFs (Fig. 5), indicating that Carm1 was not required for the activation of the myogenin gene.
FIG. 5.
Carm1/Prmt4 is required for myogenic late-gene expression. mRNA expression analysis of the indicated genes by Q-PCR in immortalized WT or Carm1/Prmt4 KO MEFs that were mock differentiated or differentiated with MyoD or with myogenin plus Mef2D1b (Myog/Mef2D1b) for 24 h. Transcript levels were normalized to the total amount of EF1-α mRNA. Values are relative to the expression values obtained in MyoD-differentiated WT MEFs, which were set at 1, except for the evaluation for Mef2D1b expression, where the value obtained for myogenin-Mef2D1b-differentiated WT cells was set at 1. Data are averages plus standard deviations for five independent experiments.
When late-gene activation was examined, MCK, desmin, and dystrophin gene expression was induced in MyoD and myogenin-Mef2D1b-differentiated WT MEFs, but not in mock-differentiated WT MEFs (Fig. 5). In contrast, induction of late myogenic genes was severely compromised in KO MEFs, regardless of whether the cells were differentiated with MyoD or with myogenin-Mef2D1b (Fig. 5). The lack of late-gene expression in myogenin-Mef2D1b-differentiated cells means that there is a direct requirement for Carm1/Prmt4 during late-gene induction, because even when the myogenin and Mef2D1b regulators were provided to the cell, the lack of Carm1/Prmt4 prevented late-gene expression. In combination with the analysis of myogenin expression in MyoD-differentiated cells (Fig. 4D), the data indicate that Carm1/Prmt4 is required for late-gene expression but not for expression of the early myogenin gene.
Carm1/Prmt4 binding to myogenic late-gene regulatory sequences is independent of Prmt5.
The data indicate that Carm1/Prmt4 is required for the induction of late myogenic gene expression whereas Prmt5 is not. This suggests that Carm1/Prmt4 binding should be independent of Prmt5 function. To address this question, we examined Carm1/Prmt4 binding in the Prmt5-deficient cell lines using ChIP. The data in Fig. 6A demonstrate that Carm1/Prmt4 bound equivalently well in the presence and absence of Prmt5, whether differentiation was induced by MyoD or by myogenin-Mef2D1b. Although Prmt5 binding to late myogenic regulatory sequences precedes Carm1/Prmt4 binding, we also performed the converse experiment to determine whether Prmt5 binding required Carm1/Prmt4. Such an experiment would exclude the possibility that Prmt5 binding was dependent upon a Carm1/Prmt4 function that did not involve its ability to bind to myogenic late-gene sequences. Examination of Prmt5 binding in MyoD-differentiated WT and Carm1/Prmt4-null MEF lines showed that Prmt5 binding did not require Carm1/Prmt4 (Fig. 6B). Consistent with the data presented in Fig. 3B, Prmt5 did not bind to myogenic late-gene regulatory sequences when differentiation was induced by myogenin-Mef2D1b, due to the absence of MyoD (Fig. 6B). Thus, the binding of each of the two distinct Prmts to myogenic late-gene loci is independent of the other.
FIG. 6.
Binding of Carm1/Prmt4 at the regulatory regions of late myogenic targets is not dependent on Prmt5, while binding of Prmt5 to these sequences does not require Carm1/Prmt4. (A) NIH 3T3 cells or c12 and c15 cells, which express antisense constructs against Prmt5, were mock differentiated or differentiated with MyoD or with myogenin plus Mef2D1b (Myog/D1b) for 24 h and were used for ChIP analysis using an antibody recognizing Carm1. (B) Immortalized MEFs derived from Carm1 WT or KO mice were mock differentiated or differentiated with MyoD or with myogenin plus Mef2D1b for 24 h and used for ChIP analysis using an antibody recognizing Prmt5. Values are relative to those obtained for binding in mock-differentiated cells, which were set at 1. Data are averages plus standard deviations for three independent experiments.
In vitro interactions between myogenic regulatory factors and Prmt5 and Carm1/Prmt4.
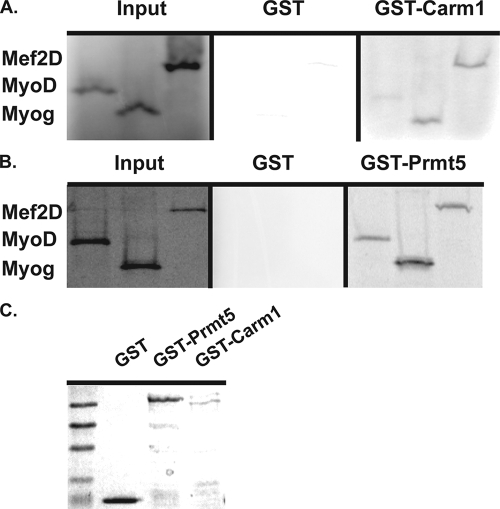
We utilized GST fusions with Prmt5 and with Carm1/Prmt4 to investigate whether each Prmt was capable of directly interacting with the myogenic regulators implicated in controlling early and late myogenic gene expression. GST-Prmt5, GST-Carm1/Prmt4, and GST were expressed in and purified from bacteria (Fig. 7C) and incubated with 35S-labeled, in vitro-translated (IVT) MyoD, myogenin, or Mef2D1b. GST alone did not interact with any of the tested myogenic regulators (Fig. 7A and B). GST-Carm1/Prmt4 interacted with both IVT myogenin and IVT Mef2D1b but only weakly interacted with IVT MyoD (Fig. 7A). These results are consistent with the re-ChIP data presented in Fig. 4D indicating that Carm1/Prmt4 and myogenin co-occupy myogenic late-gene regulatory sequences and with the functional data presented in Fig. 5 showing that Carm1/Prmt4 was required for myogenin and Mef2D1b to activate myogenic late-gene expression. The weak interaction with IVT MyoD is consistent with the observation that Carm1/Prmt4 was not required for activation of the myogenin gene at early times (Fig. 5), which is a MyoD-dependent event.
FIG. 7.
In vitro interactions exist between Prmt5 and Carm1/Prmt4 and muscle regulatory factors. GST, GST-Prmt5, and GST-Carm1/Prmt4 were expressed and purified from BL21 cells by binding to glutathione beads. (A) Bead-bound GST and GST-Prmt5 were incubated with IVT, full-length, 35S-radiolabeled MyoD, myogenin, or Mef2D1b to determine if interactions between these factors exist. Following extensive washing, pelleted beads and any bound proteins were electrophoresed on an SDS-polyacrylamide gel, and the gels were dried and exposed to film. (B) The same experiment was repeated with GST and GST-Carm1/Prmt4. (C) Purified GST and GST fusion proteins were resolved on SDS-polyacrylamide gels and visualized by Coomassie staining.
In contrast, GST-Prmt5 interacted with IVT MyoD, myogenin, and Mef2D1b (Fig. 7B). The interaction between Prmt5 and MyoD is consistent with physical and functional data that demonstrated a requirement for Prmt5 in MyoD-mediated activation of the myogenin gene during myogenesis (10). It is also consistent with the observation that Prmt5 is present on myogenic late-gene regulatory sequences at early times of differentiation prior to the expression of the late genes (Fig. 2A), a time when MyoD and HDAC2 are also present (30). Although interactions of Prmt5 with myogenin and Mef2D1b were observed in vitro, differentiation mediated by myogenin-Mef2D expression did not involve recruitment of Prmt5 to the late-gene loci (Fig. 3B). This suggests that one or more of the proteins may be modified in vivo, or that the interactions that were revealed in vitro are occluded in the context of the factor binding at myogenic regulatory sequences in differentiating cells.
Carm1/Prmt4 binding at myogenic late-gene regulatory sequences permits binding of the Brg1 ATPase of SWI/SNF chromatin-remodeling enzymes and subsequent chromatin remodeling.
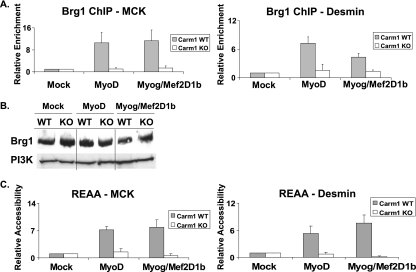
Our earlier work established that transcriptional activation of late myogenic genes requires the SWI/SNF chromatin-remodeling enzymes and, in particular, the activity of the Brg1 ATPase subunit (30). The ChIP assay results presented above revealed that Carm1/Prmt4 binds to the regulatory elements of late myogenic genes and is responsible for dimethylating H3R17, while gene expression analysis indicated that Carm1/Prmt4 was required for late-gene expression. To probe the mechanism of the requirement for Carm1/Prmt4, we determined whether the loss of Carm1/Prmt4 affected Brg1 binding and function at late muscle genes. We observed that in both MyoD- and myogenin-Mef2D1b-differentiated Carm1/Prmt4 KO cells, the binding of Brg1 at the regulatory sequences of late myogenic targets was severely diminished (Fig. 8A). This reduction in Brg1 binding was not due to a reduction in the levels of Brg1 protein, as shown by Western blotting (Fig. 8B). Thus, Carm1/Prmt4 is responsible for facilitating Brg1 binding. The association of an ATP-dependent chromatin-remodeling enzyme with a regulatory sequence implies a localized chromatin structural change. To document any structural changes in chromatin at these loci in Carm1/Prmt4 WT and KO MEFs, an REAA was performed. Upon initiation of differentiation with MyoD or myogenin-Mef2D1b, but not in mock-differentiated cells, there was an induction of restriction enzyme accessibility at PvuII sites present in the MCK and desmin promoter regulatory regions in the WT MEFs (Fig. 8C). We previously demonstrated that these changes in chromatin accessibility were Brg1 dependent (30). However, in Carm1/Prmt4 KO MEFs, no increase in accessibility was observed (Fig. 8B), and the promoter chromatin structure at these loci remained in a more inaccessible state. These results demonstrate that the Carm1/Prmt4 methyltransferase is required for myogenic late-gene expression because it facilitates binding of the Brg1 ATP-dependent chromatin-remodeling enzyme and subsequent chromatin remodeling at these regulatory sequences. Therefore, the histone-modifying enzyme is required for the activity of the ATP-dependent chromatin-remodeling enzyme. In conjunction with our earlier study, the results indicate a common molecular explanation for why different arginine methyltransferases are required for transcriptional activation at different stages of the skeletal muscle differentiation process.
FIG. 8.
Carm1/Prmt4 is required to facilitate the binding and function of the chromatin-remodeling enzyme Brg1. Immortalized MEFs derived from Carm1 WT and KO mice were mock differentiated or differentiated with MyoD or with myogenin plus Mef2D1b (Myog/Mef2D1b) for 24 h and were used for ChIP analysis using an antibody recognizing Brg1 (A), Western blot analysis to examine Brg1 and PI3K protein levels in each sample (B), or REAA to assess the extent of chromatin accessibility at PvuII restriction sites in the indicated gene regulatory regions (C). Values (A and C) are relative to the values obtained for binding or accessibility in mock-differentiated WT MEFs, which were set at 1. Data are averages plus standard deviations for five independent experiments.
DISCUSSION
Shared function by different Prmts at different stages of myogenesis.
Since both Prmt5 and Carm1/Prmt4 had been implicated in myogenesis (7, 10), we sought to determine whether these different arginine methyltransferases might cooperate in the activation of specific myogenic loci. Although we demonstrated that both enzymes were physically located at regulatory sequences controlling the expression of myogenic late genes and that both enzymes modified histones at these loci, we also demonstrated that only Carm1/Prmt4 was necessary for late-gene activation. Thus, while the contribution made by Prmt5 at myogenic late-gene loci remains to be defined, it is clearly not a direct requirement for late-gene expression.
We also noted that the absence of Carm1/Prmt4 had no impact on the ability of MyoD to induce the expression of myogenin (Fig. 5), which contrasts the previous results obtained by studying differentiation under conditions where Prmt5 levels were reduced (10). It remains possible that Carm1 might contribute to activation at earlier time points when myogenin expression is first initiated. However, if true, this hypothetical contribution of Carm1 is overcome at later stages during differentiation. Thus, even though late-gene expression was dependent upon Carm1/Prmt4, expression of the early myogenin gene was not. Instead of cooperativity at specific loci, what we observed was a sequential requirement for Prmt5 and Carm1/Prmt4 that correlated with the temporal class of gene being activated. To be specific, Prmt5 was required to facilitate activation of the early myogenin gene, whereas Carm1 was required for late myogenic gene induction.
Our results contradict a conclusion of an earlier study where antisense constructs were used to reduce the levels of Carm1/Prmt4 (7). Those investigators observed that reduction of Carm1/Prmt4 inhibited differentiation but attributed the effect to a decrease in the induction of myogenin. The reasons for the discrepancy between this report and our data are undetermined; our results clearly show that myogenin is robustly induced in the absence of Carm1/Prmt4 (Fig. 5).
Having defined a series of protein-DNA interactions that occur at late-gene regulatory sequences (30), we sought to determine which of these events might be compromised by the absence of Carm1/Prmt4 as a means to explain the lack of late-gene expression observed in the absence of Carm1/Prmt4. The results showed that interaction of Brg1, the ATPase of SWI/SNF chromatin-remodeling enzymes previously demonstrated to be required for myogenesis, was compromised at late-gene regulatory sequences (Fig. 6A). The functional consequence of this loss was then demonstrated by the lack of chromatin remodeling at these loci (Fig. 6C). We therefore conclude that Carm1/Prmt4 is required to promote Brg1 binding and chromatin remodeling at late myogenic genes.
On a more general scale, the results indicate that an arginine methyltransferase is required for an ATP-dependent chromatin-remodeling enzyme to function. Our prior demonstration that Prmt5 facilitates Brg1 and SWI/SNF enzyme function at the myogenin promoter during gene activation (10) is extended by the observation that Carm1 is required at late myogenic gene regulatory regions for the same reason, suggesting that this may be a general mechanism. Combined, these studies indicate that Prmt5 and Carm1/Prmt4, two distinct arginine methyltransferases, promote the same step of gene activation, which is the recruitment of the SWI/SNF ATP-dependent chromatin-remodeling enzyme. Prmt5 and Carm1/Prmt4 are not known to methylate the same sites on histones; Prmt5 dimethylates H3R8 and H4R3 (32, 33), while Carm1/Prmt4 dimethylates H3R17 and H3R26 (39), however, both enzymes are capable of modifying nonhistone substrates as well (9, 18, 21-23, 44). Importantly, Prmt5 and Carm1/Prmt4 can methylate the same nonhistone substrates, including CA150, SmB, PABP1, U1C, and SF3b4 (8), although it is still not clear whether the same arginine residues are methylated in these common substrates (8). The exact mechanism by which Prmt5 and Carm1/Prmt4 promote Brg1 interaction at different gene regulatory sequences during myogenesis remains to be determined, but one attractive hypothesis is that the dimethylation of different histone residues makes the chromatin a better substrate for the ATP-dependent remodeling enzyme.
Another question that remains unanswered is why different arginine methyltransferases would be required at different times of myogenesis to facilitate the same step in the activation process. One possible explanation is that the combination of activators and cofactors present at early and late myogenic gene regulatory sequences are different (discussed further below) and that the specific arginine methylations mediated by each enzyme are specific for promoting gene activation by the different sets of regulatory proteins in ways that remain to be defined. An additional possibility is that the arginine methyltransferases modify different transcriptional regulatory proteins in addition to modifying histone tails. Finally, it must be noted that despite the presence of Carm1/Prmt4 and dimethylated H3R17 at late-gene regulatory sequences both in tissue culture and in vivo, Carm1/Prmt4-deficient mice, in which the mutation is perinatal lethal for undetermined reasons, show no gross defect in skeletal muscle appearance (47). This implies either that there is a functional defect in one or more muscles that contribute to breathing or feeding or that there are undefined redundant mechanisms in vivo to compensate for the deficiency of Carm1/Prmt4. Given the widespread redundancies between MyoD, Myf5, and Mrf4 during development (reviewed in references 5 and 35), additional redundant mechanisms to ensure skeletal muscle formation and function are possible.
Mechanisms relating to Prmt5 and Carm1/Prmt4 function.
We previously determined that Prmt5 functions directly in the induction of the myogenin gene (10). It is well established that MyoD is critical in the activation of the myogenin promoter (reviewed in references 5, 19, 38, and 41); demonstration by re-ChIP analysis that MyoD and Prmt5 are colocalized to myogenin promoters in primary cells that are actively transcribing the myogenin gene supports the idea that Prmt5 acts as a coactivator for MyoD (10). Additional experiments indicating that MyoD and Prmt5 can be coimmunoprecipitated from differentiated cells further support this conclusion (10). Here we report that Prmt5 can directly interact with MyoD in an in vitro interaction assay, providing further mechanistic explanation for the cooperativity exhibited by these factors. In addition, we show that Prmt5 is localized to myogenic late-gene regulatory sequences at the onset of differentiation, prior to the initiation of late-gene expression. Previous work indicates that MyoD is also present at late-gene loci before activation (30); it is accompanied by a class I HDAC, which has been shown to maintain MyoD in a transcriptionally inactive state in myoblast cultures (24, 36). We propose that MyoD targets Prmt5 to gene regulatory sequences, regardless of whether it is functioning in an activating or repressing capacity.
In contrast, Carm1/Prmt4 binding to late-gene loci correlates with the time of late-gene activation. In both developing embryonic skeletal muscle tissue and MyoD-differentiated fibroblasts, the time of late-gene activation is marked by coincident binding of myogenin, Mef2, and the SWI/SNF ATPase Brg1 and by changes in chromatin accessibility (30). The addition of Carm1/Prmt4 to this complex of regulators that functions at the time of transcription initiation suggests that Carm1/Prmt4 acts as a coactivator of myogenin and/or Mef2 proteins. Transient-transfection studies imply cooperativity between Carm1/Prmt4 and Mef2, and Carm1/Prmt4 interacts with the Mef2C isoform in in vitro interaction assays (7). The demonstration by re-ChIP analysis that Carm1/Prmt4 is colocalized with myogenin, but not MyoD, at late-gene regulatory sequences (Fig. 4D) also supports this hypothesis. The additional demonstration that Carm1/Prmt4 can directly interact with the Mef2D1b isoform and with the myogenin protein in vitro (Fig. 7A) but only weakly interacts (Fig. 7A) or does not interact (7) with MyoD in vitro suggests that targeting of Carm1/Prmt4 occurs via myogenin and/or Mef2 proteins.
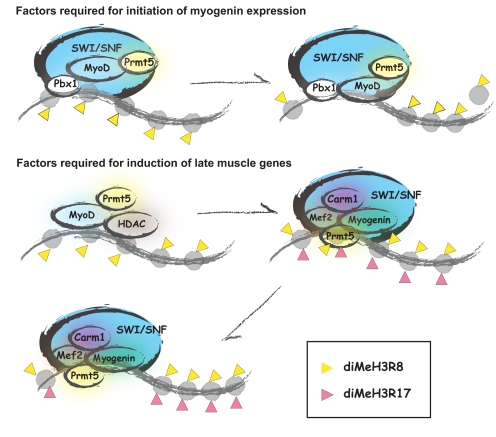
Both Prmt5 and Carm1/Prmt4 were shown to coimmunoprecipitate and copurify with Brg1 (34, 45), and coordinated activity between Brg1 and Prmt5 (10, 33, 34) and between Brg1 and Carm1/Prmt4 (45) has previously been demonstrated. Collectively, the data presented here and in previous reports (4, 10, 14) support a model (Fig. 9) where MyoD initially binds indirectly to the myogenin promoter via interaction with Pbx1 and also binds to sites at late myogenic gene regulatory sequences, thereby facilitating recruitment of Prmt5 to both classes of genes. The recruitment of Prmt5 at the myogenin gene mediates dimethylation of H3R8 and is necessary for the subsequent recruitment of Brg1-based SWI/SNF chromatin-remodeling enzymes, leading to chromatin remodeling and expression of the myogenin gene. Upon accumulation of myogenin protein, myogenin and Mef2 proteins bind to cognate binding sites at late-gene regulatory sequences, displacing MyoD and the HDAC, as shown previously (30), and recruiting Carm1/Prmt4. Carm1/Prmt4 recruitment mediates dimethylation of H3R17 and facilitates the recruitment of Brg1-based SWI/SNF chromatin-remodeling enzymes, leading to chromatin remodeling and expression of the late myogenic genes, terminal differentiation, and the formation of mature skeletal muscle tissue.
FIG. 9.
Schematic model illustrating Prmt function at the myogenin and representative late-gene regulatory sequences. Prmt5 mediates dimethylation (diMe) of H3R8 and facilitates Brg1-based SWI/SNF chromatin-remodeling enzyme interaction and function at the myogenin promoter at early times of myogenic differentiation, while Carm1/Prmt4 mediates dimethylation of H3R17 and facilitates Brg1-based SWI/SNF chromatin-remodeling enzyme interaction and function at myogenic late genes.
Acknowledgments
We gratefully acknowledge M. Stallcup for advice on the H3R17 antibody and S. Tapscott and E. Olson for reagents. We thank Y. Ohkawa and members of the Imbalzano lab for suggestions and comments on the manuscript.
This work was supported by American Cancer Society grant RSG-0418201-GMC to S.S. and by NIH grants GM56244 to A.N.I., DK62248 to M.T.B., and CA116093 to S.S.
Footnotes
Published ahead of print on 2 February 2009.
REFERENCES
- 1.Bedford, M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 1204243-4246. [DOI] [PubMed] [Google Scholar]
- 2.Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 3.Bergstrom, D. A., B. H. Penn, A. Strand, R. L. Perry, M. A. Rudnicki, and S. J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9587-600. [DOI] [PubMed] [Google Scholar]
- 4.Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14465-477. [DOI] [PubMed] [Google Scholar]
- 5.Berkes, C. A., and S. J. Tapscott. 2005. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16585-595. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 2842174-2177. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S. L., K. A. Loffler, D. Chen, M. R. Stallcup, and G. E. Muscat. 2002. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 2774324-4333. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, D., J. Cote, S. Shaaban, and M. T. Bedford. 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 2571-83. [DOI] [PubMed] [Google Scholar]
- 9.Chevillard-Briet, M., D. Trouche, and L. Vandel. 2002. Control of CBP co-activating activity by arginine methylation. EMBO J. 215457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dacwag, C. S., Y. Ohkawa, S. Pal, S. Sif, and A. N. Imbalzano. 2007. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. Biol. 27384-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, R. L., H. Weintraub, and A. B. Lassar. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51987-1000. [DOI] [PubMed] [Google Scholar]
- 12.de La Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 202839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27187-190. [DOI] [PubMed] [Google Scholar]
- 14.de la Serna, I. L., Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, and A. N. Imbalzano. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 253997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Serna, I. L., Y. Ohkawa, and A. N. Imbalzano. 2006. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 7461-473. [DOI] [PubMed] [Google Scholar]
- 16.de la Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin-remodeling enzymes. J. Biol. Chem. 27641486-41491. [DOI] [PubMed] [Google Scholar]
- 17.Fabbrizio, E., S. El Messaoudi, J. Polanowska, C. Paul, J. R. Cook, J. H. Lee, V. Negre, M. Rousset, S. Pestka, A. Le Cam, and C. Sardet. 2002. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, Q., P. Yi, J. Wong, and B. W. O'Malley. 2006. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 267846-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forcales, S. V., and P. L. Puri. 2005. Signaling to the chromatin during skeletal myogenesis: novel targets for pharmacological modulation of gene expression. Semin. Cell Dev. Biol. 16596-611. [DOI] [PubMed] [Google Scholar]
- 20.Hasty, P., A. Bradley, J. H. Morris, D. G. Edmondson, J. M. Venuti, E. N. Olson, and W. H. Klein. 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364501-506. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J., J. Lee, N. Yadav, Q. Wu, C. Carter, S. Richard, E. Richie, and M. T. Bedford. 2004. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J. Biol. Chem. 27925339-25344. [DOI] [PubMed] [Google Scholar]
- 22.Kwak, Y. T., J. Guo, S. Prajapati, K. J. Park, R. M. Surabhi, B. Miller, P. Gehrig, and R. B. Gaynor. 2003. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 111055-1066. [DOI] [PubMed] [Google Scholar]
- 23.Le Guezennec, X., M. Vermeulen, A. B. Brinkman, W. A. Hoeijmakers, A. Cohen, E. Lasonder, and H. G. Stunnenberg. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 201739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, J. F., J. M. Miano, C. M. Hustad, N. G. Copeland, N. A. Jenkins, and E. N. Olson. 1994. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 141647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115751-763. [DOI] [PubMed] [Google Scholar]
- 27.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 183587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgenstern, J. P., and H. Land. 1990. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 181068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabeshima, Y., K. Hanaoka, M. Hayasaka, E. Esumi, S. Li, I. Nonaka, and Y. Nabeshima. 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364532-535. [DOI] [PubMed] [Google Scholar]
- 30.Ohkawa, Y., C. G. Marfella, and A. N. Imbalzano. 2006. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 25490-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkawa, Y., S. Yoshimura, C. Higashi, C. G. Marfella, C. S. Dacwag, T. Tachibana, and A. N. Imbalzano. 2007. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J. Biol. Chem. 2826564-6570. [DOI] [PubMed] [Google Scholar]
- 32.Pal, S., and S. Sif. 2007. Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell Physiol. 213306-315. [DOI] [PubMed] [Google Scholar]
- 33.Pal, S., S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst, and S. Sif. 2004. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 249630-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 237475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pownall, M. E., M. K. Gustafsson, and C. P. Emerson, Jr. 2002. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 18747-783. [DOI] [PubMed] [Google Scholar]
- 36.Puri, P. L., S. Iezzi, P. Stiegler, T. T. Chen, R. L. Schiltz, G. E. Muscat, A. Giordano, L. Kedes, J. Y. Wang, and V. Sartorelli. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8885-897. [DOI] [PubMed] [Google Scholar]
- 37.Roy, K., I. L. de la Serna, and A. N. Imbalzano. 2002. The myogenic basic helix-loop-helix family of transcription factors shows similar requirements for SWI/SNF chromatin-remodeling enzymes during muscle differentiation in culture. J. Biol. Chem. 27733818-33824. [DOI] [PubMed] [Google Scholar]
- 38.Sartorelli, V., and G. Caretti. 2005. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr. Opin. Genet. Dev. 15528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurter, B. T., S. S. Koh, D. Chen, G. J. Bunick, J. M. Harp, B. L. Hanson, A. Henschen-Edman, D. R. Mackay, M. R. Stallcup, and D. W. Aswad. 2001. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 405747-5756. [DOI] [PubMed] [Google Scholar]
- 40.Simone, C., S. V. Forcales, D. A. Hill, A. N. Imbalzano, L. Latella, and P. L. Puri. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36738-743. [DOI] [PubMed] [Google Scholar]
- 41.Tapscott, S. J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 1322685-2695. [DOI] [PubMed] [Google Scholar]
- 42.Weiss, R. S., T. Enoch, and P. Leder. 2000. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 141886-1898. [PMC free article] [PubMed] [Google Scholar]
- 43.Wysocka, J., C. D. Allis, and S. Coonrod. 2006. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11344-355. [DOI] [PubMed] [Google Scholar]
- 44.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 2942507-2511. [DOI] [PubMed] [Google Scholar]
- 45.Xu, W., H. Cho, S. Kadam, E. M. Banayo, S. Anderson, J. R. Yates III, B. M. Emerson, and R. M. Evans. 2004. A methylation-mediator complex in hormone signaling. Genes Dev. 18144-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.Yadav, N., J. Lee, J. Kim, J. Shen, M. C. Hu, C. M. Aldaz, and M. T. Bedford. 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 1006464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]