Abstract
Macrophage migration-inhibitory factor (MIF) is an upstream regulator of innate immunity and a potential molecular link between inflammation and cancer. The unusual structural homology between MIF and certain tautomerases, which includes both a conserved substrate-binding pocket and a catalytic N-terminal proline (Pro1), has fueled speculation that an enzymatic reaction underlies MIF's biologic function. To address the functional role of the MIF tautomerase activity in vivo, we created a knock-in mouse in which the endogenous mif gene was replaced by one encoding a tautomerase-null, Pro1→Gly1 MIF protein (P1G-MIF). While P1G-MIF is completely inactive catalytically, it maintains significant, albeit reduced, binding to its cell surface receptor (CD74) and to the intracellular binding protein JAB1/CSN5. P1G-MIF knock-in mice (mifP1G/P1G) and cells derived from these mice show a phenotype in assays of growth control and tumor induction that is intermediate between those of the wild type (mif+/+) and complete MIF deficiency (mif−/−). These data provide genetic evidence that MIF's intrinsic tautomerase activity is dispensable for this cytokine's growth-regulatory properties and support a role for the N-terminal region in protein-protein interactions.
Macrophage migration-inhibitory factor (MIF) is a widely expressed cytokine and upstream regulator of the immune response (23). Immunoneutralization and genetic knockout studies have established a central position for MIF in the host response to infection and tissue invasion (5, 9, 15). MIF's importance in human disease also has been revealed by the association of high-expression MIF alleles with clinical severity of different autoimmune disorders (18).
An important role for MIF in tumorigenesis and in the contribution of inflammation to cancer development also has been proposed (7, 20). Different tumor types express high levels of MIF, and clinical studies have shown that MIF production correlates with tumor aggressiveness and metastatic potential (1, 22, 27). Studies using genetically engineered MIF-deficient cells and mice show that MIF contributes to the development of the malignant phenotype by several mechanisms, including enhancement of cell cycle progression by sustained mitogen-activated protein kinase (MAPK) activation (28, 30), decreased proteasomal protein degradation (33) leading to altered expression of key cell cycle-regulatory proteins (15, 21, 35), and tumor promotion by neoangiogenesis (10, 48). Importantly, MIF also inhibits the proapoptotic and cell cycle-regulatory function of the p53 tumor suppressor, thereby allowing for the accumulation of oncogenic mutations (20, 32). MIF's role in tumor progression additionally is supported by human genetic studies, and a recent report has described an association between high-expression MIF alleles and incidence of prostate cancer, which is a tumor type in which recurrent inflammation is considered to have an etiologic role (27).
Information regarding MIF structure and function has emerged only in the last few years. The mammalian protein is encoded by a unique gene, and the solution of MIF's three-dimensional crystal structure revealed that the protein defines a new structural superfamily (41, 42). Notably, MIF shows structural but not sequence homology with two prokaryotic enzymes, 4-oxalocrotonate tautomerase and 5-carboxy-methyl-2-hydroxymuconate isomerase, which act in the pathway of aromatic amino acid catabolism. Subsequent studies have shown that MIF tautomerizes model substrates such as d-dopachrome and p-hydroxyphenylpyruvate, although the measured enzymatic rate constants do not support a physiologic role for these reactions (36, 37). The d stereoisomer of dopachrome also does not occur in nature, but the l steroisomer is a substrate for the melanotic encapsulation response, which constitutes an invertebrate host defense mechanism against microbial invasion. Accordingly, some investigators have proposed that the catalytic activity of MIF is vestigial and reflects the protein's ancestral origin in innate immunity (6). Others have argued that MIF's highly conserved substrate binding pocket and catalytic N-terminal proline (Pro1), which are shared with the prokaryotic isomerases, provide strong evidence for an intrinsic, enzymatic function for mammalian MIF (44).
In vitro studies that have sought to address the precise relationship between MIF's catalytic activity and its biological function have not been conclusive. Site-directed mutagenesis approaches that have replaced MIF's catalytic, N-terminal proline with inactive residues have confirmed that Pro1 and a surrounding hydrophobic pocket mediate tautomerase activity (2). Most studies have shown a preservation of proinflammatory function for catalytically inactive, recombinantly produced MIF proteins in vitro (2, 19); however, this finding has not been consistent (34, 43), and conclusions may be limited by the semiquantitative nature of the assays employed. It also should be noted that a commercially available recombinant human MIF bears a mutation in the carboxy-terminal oligomerization domain (Asn106→Ser106) that may influence bioactivity (4, 12). Recent studies also suggest that the MIF N-terminal region makes critical contacts with CD74, a cell surface receptor that mediates the cytokine's signal transduction activity (24, 38).
To address the role of the MIF tautomerase activity in vivo, we created a knock-in mouse in which the endogenous mif gene is replaced by a tautomerase-null, Pro1→Gly1 MIF protein (P1G-MIF). We reasoned that, if the tautomerase activity was necessary for MIF biologic function, a genetically defined P1G-MIF mouse (mifP1G/P1G), or cells derived from such a mouse, would show a phenotype identical to that of the MIF knockout mouse (mif−/−). Alternatively, if the tautomerase activity was dispensable, then P1G-MIF mice should show the same phenotype as the wild type (mif+/+). An intermediate level of activity would support a structural role for Pro1, but not the tautomerase activity per se, in protein-protein interactions necessary for MIF effector function.
MATERIALS AND METHODS
Generation of mifP1G/P1G knock-in mice.
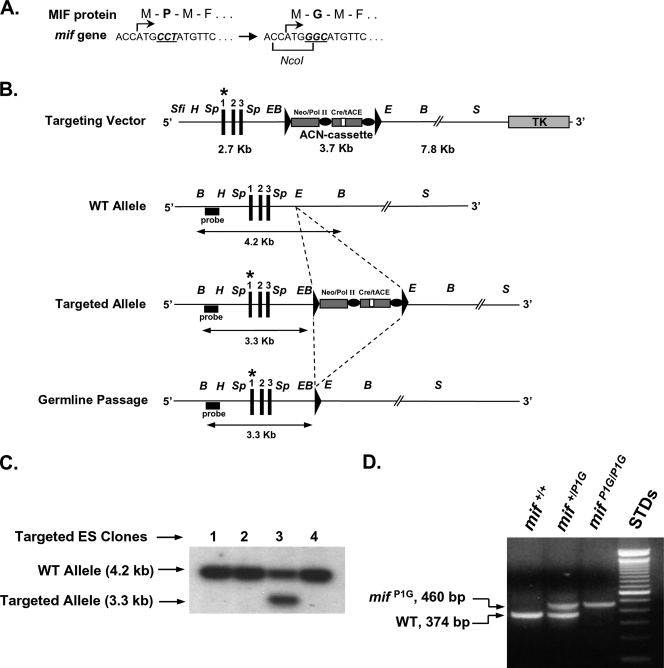
Genetically defined mouse strains (mifP1G/P1G and mif−/− strains) were created at the Institute for Genetics (Cologne, Germany) in accordance with institutional and NIH resource sharing guidelines. The targeting vector pEasyFlox was modified to contain loxP-flanked selection cassette ACN (provided by K. Thomas, University of Salt Lake City), containing the Cre recombinase gene under the control of the testis-specific promoter for angiotensin-converting enzyme (tACE-Cre) as well as the neomycin gene under the control of the polymerase II promoter (Pol II-Neo), which allows self-excision of the targeting cassette during germ line transmission (8). A 2.7-kb HindIII-EcoRI fragment containing the entire mif gene from a 129/Sv P1 library (Incyte Genomics) was cloned as the left arm of homology. A 1.8-kb SpeI subfragment containing mif exon 1 was used for PCR-directed mutagenesis to replace the codon CCT (Prol1) by GGC (Gly1). The right arm of homology was cloned as a 7.8-kb EcoRI-SalI fragment. To allow Southern screening of homologous recombinants, a BamHI site was introduced between the EcoRI site and the first loxP.
C57BL/6 embryonic stem cells (Bruce-4) were transfected with the SfiI-linearized targeting vector and subjected to double selection with G418-ganciclovir. Targeted homologous embryonic stem (ES) cell clones were identified by Southern blotting using an external upstream probe; additional nonhomologous integrants were excluded using a neo probe and HindIII digestion. Two of the correctly targeted ES cell clones were microinjected into BALB/c blastocysts at the microinjection facility at Brigham and Women's Hospital (Boston, MA). Chimeras gave germ line transmission, and offspring were bred to homozygosity to produce a population of mifP1G/P1G mice on the pure C57BL/6 background. Successful deletion of the ACN cassette during germ line transmission in heterozygous offspring was demonstrated by PCR.
The presence of the mifP1G mutation in homozygous mice was verified by PCR cloning and DNA sequencing. In addition, the novel NcoI restriction site in mutant mif exon 1 permits the facile identification of the mutation via PCR followed by NcoI restriction digestion of the PCR product. While the wild-type allele remains uncut as a 444-bp product, the mutant allele is cut by NcoI and yields two fragments of 220 and 224 bp. Primer sequences are available from the authors upon request.
All mice were kept under specific-pathogen-free conditions on a 12-h light/dark cycle and received normal mouse chow. All experiments were approved by institutional animal use and care committees. Murine embryonic fibroblasts (MEFs) were prepared from embryonic day 14.5 embryos from wild-type (mif+/+), MIF knockout (mif−/−) (15), P1G-MIF knock-in (mifP1G/P1G), and CD74−/− (24) mice, all in the C57BL/6 genetic background. Unless otherwise specified, passage 4 to 6 MEFs were used. The MIF content of MEF conditioned media was analyzed by enzyme-linked immunosorbent assay (31) using as the detector antibody a goat polyclonal anti-MIF that is insensitive to the P→G amino acid substitution (Santa Cruz Biotechnology).
Northern blotting.
Total RNA was isolated from mouse liver with the RNeasy kit (Qiagen) and blotted onto a positively charged nylon membrane (Hybond-N) according to standard procedures. After hybridization with a [32P]dCTP-labeled MIF or β-actin cDNA probe, the membrane was exposed to a BioMax autoradiography film (Kodak/Sigma) at −70°C for 2 to 8 h.
Western blotting.
Cells were lysed in radioimmunoprecipitation assay buffer and processed according to standard procedures. The primary antibodies used were polyclonal anti-MIF (Ka565; J. Bernhagen, RWTH Aachen), anti-phosphorylated extracellular signal-regulated kinase 1 and 2 (ERK1/2) and anti-total ERK1/2 (Cell Signaling), anti-β-tubulin, anti-β-actin, anti-E1A, anti-H-ras (Santa Cruz), and anti-JAB1 FL-334 (Santa Cruz). The bound antibodies were visualized with horseradish peroxidase-conjugated immunoglobulin G (IgG) antibody and enhanced chemiluminescence (Amersham). Coimmunoprecipitation studies followed the protocol specified in Lue et al. (25). The amount of specific protein present was quantified by digital densitometry (39). For quantitation of ERK1/2 phosphorylation, Western blots were scanned and the ratio of phosphorylated kinase to the total kinase protein was expressed as the change for each lane (25). The P values were calculated for each of the comparisons shown from three experiments (Student's unpaired t test).
Immunohistochemistry.
Paraformaldehyde-fixed and paraffin-embedded tissues were stained for MIF using the monoclonal anti-MIF antibody ZMD.321 (Invitrogen) and the LSAB horseradish peroxidase kit (Dako) according to the manufacturer's instructions. Pooled nonimmune serum was used as the negative control. The sections were stained with 3-amino-9-ethylcarbazole as the chromogenic substrate and counterstained with Meyer's hematoxylin.
Dopachrome tautomerization assay.
Liver lysates were analyzed for MIF tautomerase activity with l-dopachrome methyl ester as the substrate (2). The enzymatic reaction was initiated at room temperature by adding the substrate (0.3 ml) to a cuvette containing recombinant MIF or murine P1G-MIF (600 ng/ml) or liver lysate (43 μg/ml) in a total volume of 0.7 ml. The activity was determined by the decrease in absorbance at 475 nm.
MIF binding studies.
The binding between P1G-MIF and the MIF cell surface receptor (CD74) was analyzed in a soluble-receptor binding assay. Individual wells of a 96-well plate were coated with the recombinant soluble CD74 ectodomain (sCD7473-232) as described by Leng et al. (24). Biotinylated MIF was incubated in triplicate wells together with increasing concentrations of nonbiotinylated recombinant MIF, heat-denatured MIF, P1G-MIF, or a neutralizing anti-CD74 antibody (clone LN2) at room temperature for 2 h, followed by washing with TTBS (20 mM Tris-HCl, 0.5 M NaCl, 0.05% Tween 20; pH 7.4). The amount of bound, biotinylated MIF was detected with streptavidin-conjugated alkaline phosphatase (R&D Systems) and p-nitrophenylphosphate (Sigma). Absorbance at 405 nm was measured in a microplate reader, and values were plotted relative to those for wells containing biotinylated MIF alone. Each plot represents at least three independently performed assays, and each data point depicts a standard error of the mean of ≤10%.
The real-time binding interaction of P1G-MIF with sCD74 was measured by surface plasmon resonance using a model 2000 optical biosensor (BIAcore AB, Sweden). The MIF receptor ectodomain (CD7473-232) was immobilized according to the manufacturer's instructions by a standardized amine coupling method using the Biacore amine coupling kit. The derived sensor chips were washed and equilibrated in HEPES (pH 8.0) at 20 μl/min, and the ligand (P1G-MIF) was introduced at five serial dilutions in BIAcore buffer in 60- to 100-μl injection volumes at a flow rate of 20 μl/min. Binding was measured at 25°C for 5 min, followed by 15 min of dissociation. One minute of sensor chip regeneration was performed with 1 M NaCl-50 mM NaOH. The whole process was repeated three times for each dilution sample. Sensorgram response data were analyzed in the BIA evaluation kinetics package, and the equilibrium binding constants were calculated.
The interaction between MIF or P1G-MIF and JAB1 in cell lysates prepared from mif+/+ or mifP1G/P1G MEFs was assessed by immunoprecipitation using a polyclonal anti-MIF antibody (Ka345) and visualization of JAB1 by Western blotting. Ka345 reacts equally well with recombinant MIF or P1G-MIF, as confirmed by titration in control Western blotting studies (data not shown). The CXCR2 competition binding studies employed HEK293-CXCR2 transfectants and 125I-CXCL8, as recently described (47). MIF-induced ERK1/2 phosphorylation in synchronized fibroblasts was measured by Western blotting using specific phospho-ERK1/2 and total ERK1/2 antibodies as described previously (24).
One-stage skin carcinogenesis model.
Thirty-two to 36 16-week-old mice per group (mif+/+, mifP1G/P1G, or mif−/−) were shaved on their backs and treated topically with 200 μg of benzo[α]pyrene (Sigma) in 100 μl of acetone once weekly during weeks 1 to 20. Mice were killed when tumor incidence reached 100% in all groups (week 22), and tumors were processed for pathological examination. Only tumors with a size >1 mm3 were evaluated.
Cell-based assays.
MEFs were transformed using the replication-deficient retroviral vector REBNA. Proliferation assays employed either primary or transformed MEFs plated at high (9,000 cells/cm2) or low (3,000 cells/cm2) density in 6-cm dishes in Dulbecco's modified Eagle medium (DMEM)-10% fetal calf serum (FCS). At daily intervals, cells were harvested, counted, and replated at the starting density. For confluence assays, MEFs were seeded at 60% density in a 10-cm dish in DMEM-10% FCS. Cells grew to complete confluence during the subsequent 6 to 9 days and were counted on day 12. For assessment of focus formation, 1 × 103 E1A- and H-rasV12-expressing MEFs were seeded together in DMEM-5% FCS with 3 × 105 primary MEFs of the same genotype. The H-rasV12 mutant was originally a gift from Dafna Bar-Sagi (State University of New York at Stony Brook). The medium was changed every 3 days, and the macroscopically visible colonies were counted after 14 days of culture. For storage and photography, the colonies were fixed in 4% paraformaldehyde, stained in Giemsa, and allowed to air dry.
Statistics.
Data are expressed as means ± standard deviations (SD) or ± standard error of the mean. Unless indicated otherwise, statistical analysis was performed using a two-tailed Student t test. Tumor incidence was tested by Kaplan-Meier analysis with a log rank test. Tumor numbers were tested using general linear model statistics. P values that were <0.05 were considered significant.
RESULTS
Creation of a P1G-MIF knock-in mouse (mifP1G/P1G).
We recently described a MIF knockout mouse (mif−/−), created by deleting the entire gene using the Cre-loxP technique, and we reported on the defective response of this mouse to ras-mediated malignant transformation (15). Subsequent studies with different tumor models provided additional molecular insight into MIF's role in cell cycle regulation and in antagonizing the p53 tumor suppressor gene (13, 45). To better clarify the role of MIF's N-terminal region and intrinsic tautomerase activity in these biologic actions, we engineered a gene knock-in mouse in which the endogenous, wild-type mif allele was replaced by a tautomerase-null mifP1G allele (Fig. 1). This genetic strategy was guided by the principle that the MIF tautomerase activity is strictly dependent on an N-terminal proline (formed after cleavage of Met during initial protein processing) because the unprotonated imine acts as a general base to initiate proton abstraction from the substrate (40). Replacement of Pro1 with residues that lack a nucleophilic center eliminates MIF's tautomerization activity (2). We chose to replace Pro1 with glycine (Gly) because this mutation disrupts enzymatic activity and is expected to minimally perturb the protein's three-dimensional structure (43).
FIG. 1.
Creation of a P1G-MIF-expressing mouse by gene targeting. (A) The MIF N-terminal proline (Pro1) encoded by exon 1 of mif is replaced with glycine (P→G) by exchanging the codon CCT for GGC. This strategy creates a new NcoI restriction site that permits the detection of the mutated allele by RFLP. M, Met; P, Pro; G, Gly, F, Phe. (B) The targeting vector contains the mutated mif gene (mifP1G) at the left arm of homology, a loxP-flanked selection cassette (ACN) with a Pol II-neomycin resistance gene, and a Cre recombinase gene under the control of the testis-specific promoter for angiotensin-converting enzyme (tACE-Cre). WT, wild-type; Sfi, SfiI; H, HindIII; Sp, SpeI; E, EcoRI; B, BamHI; S, SalI. Asterisks indicate the mutant allele. (C) Homologous integration in C57BL/6 ES cells verified by Southern blotting of BamHI-digested genomic DNA with an external probe. (D) PCR-based detection of ACN deletion after germ line transmission. ACN deletion decreases the interprimer distance from >4 kb to <0.5 kb, thus allowing product amplification under the conditions used. The single remaining loxP site is used to differentiate the mutated mifP1G allele from the WT mif+ allele. (E) Verification of mutagenesis by PCR plus RFLP using liver genomic DNA obtained from mif+/P1G and mifP1G/P1G mice. The mifP1G allele contains a novel NcoI site. Digestion with NcoI yields a single band of 444 bp for the WT (mif+) allele and a doublet of 222/224 bp for the mifP1G allele. (F) Verification of mutagenesis (CCT→GGC) by direct sequencing of liver genomic DNA from a mifP1G/P1G mouse. (G) mRNA and protein expression from mif+/+, mif+/P1G, and mifP1G/P1G mice. (Top) Northern analysis for MIF and β-actin mRNA from 8-week-old mice. (Bottom) Western analysis of liver extracts with a polyclonal anti-MIF antibody and anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase), demonstrating the equivalent production of MIF and P1G-MIF. Recombinant MIF or P1G-MIF was loaded as the control. (H) Tautomerase activity of MIF and P1G-MIF present in liver lysates prepared from 8-week-old mice.
The mif codon for Pro1 (CCT) was replaced by GGC. GGC is a frequently used codon in mice, and its substitution for CCT produces a novel restriction site for NcoI, which facilitates the screening of recombinants (Fig. 1A). A targeting vector that contained the mutant codon and a loxP-flanked excisable neomycin selection cassette (ACN) was constructed and placed downstream of the mif gene. The ACN selectable cassette contains the Cre recombinase gene under the control of the testis-specific promoter for angiotensin-converting enzyme (tACE). This arrangement leads to Cre expression and deletion of the selectable marker during spermatogenesis and germ line transmission of the targeted ES cells (8) (Fig. 1B). This construct was transfected into C57BL/6 ES cells, and homologous recombinants were identified by restriction digestion with EcoRI and Southern blotting with an external probe (Fig. 1C). After injection of one clone into BALB/c blastocysts, the ES cells gave germ line transmission and all of the offspring had the expected germ line deletion of the selectable cassette with one remaining loxP site (allele mif+/P1G) (Fig. 1D). The presence of the GGC codon in the murine genome was verified by PCR amplification followed by restriction fragment length polymorphism (RFLP) with NcoI (Fig. 1E), as well as by direct sequencing from genomic DNA (Fig. 1F). The P1G-MIF gene (mifP1G) was well expressed, as evidenced by tissue analysis of mRNA expression and protein production (Fig. 1G). Tissue analysis of d-dopachrome tautomerization revealed the complete absence of enzymatic activity in mifP1G/P1G knock-in mice, which is in accord with the loss of this activity in pure, recombinant P1G-MIF (Fig. 1H). P1G-MIF mice were found to be viable and fertile and to produce normal-size litters. No congenital defects were noted on necropsy, and histopathologic examination did not reveal any abnormalities. To date, no differences in lifespan or spontaneous disease or tumor development have been noted (data not shown).
P1G-MIF retains appreciable interaction with MIF binding proteins and signal transduction activity.
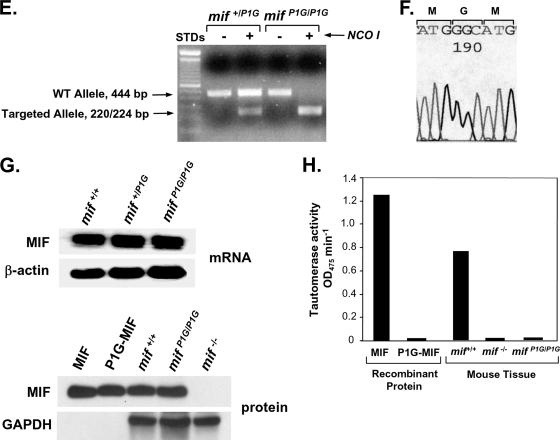
While P1G-MIF is enzymatically inactive (Fig. 1H), there is evidence that chemical modifications of Pro1 may inhibit MIF interaction with its cell surface receptor (38). Accordingly, we first sought to determine if the Pro1→Gly1 substitution influences the interaction between MIF and its two well-characterized binding partners, the cell surface receptor CD74 (24) and the intracellular transcriptional regulator and COP9 signalosome component JAB1/CSN5 (JAB1) (21). Both the CD74 and JAB1 effector proteins have been implicated in MIF-dependent growth control by pathways involving MAPK activation, the regulation of p53, and the control of cell cycle regulators (17, 21, 24-26, 33, 39). For instance, MIF induction of ERK1/2 activation and regulation of p53-dependent apoptosis are strictly dependent on the cell surface receptor CD74 (39). MIF interaction with JAB1 appears necessary for the sustained phase of ERK1/2 activation, and JAB1 in turn regulates proteasome-mediated degradation of cell cycle components (25).
We examined the comparative binding activities of MIF and P1G-MIF in a competition-based, receptor binding assay employing the CD74 ectodomain (CD7473-232 or sCD74). As shown in Fig. 2A, P1G-MIF showed significant activity for binding to CD74, although not fully reaching the level observed for wild-type MIF. We then determined the equilibrium dissociation constant for the binding of P1G-MIF to sCD74 by surface plasmon resonance, a technique that measures real-time binding interactions by changes in the refractive index of a biospecific surface (Fig. 2B). Optical biosensor surfaces were prepared, and BIAcore analysis of the binding interaction between P1G-MIF and sCD74 showed an equilibrium dissociation constant (Kd) of 39 nM. This contrasts with a Kd for the binding of wild-type MIF with sCD74 of 9 nM (24). These data indicate that P1G-MIF binds to the MIF cell surface receptor CD74, albeit with some loss of affinity due to the Pro→Gly amino acid substitution.
FIG. 2.
Comparative protein binding and signal transduction activity of MIF and P1G-MIF. (A) Comparative binding of recombinant, wild-type MIF and P1G-MIF to soluble CD74 in an in vitro capture assay employing immobilized soluble CD7473-232 (sCD74) and biotinylated MIF as the competitor. Heat-denatured MIF and a blocking anti-CD74 antibody were used as controls. Data shown are the means of quadruplicate measurements, with each point having an SD of <10%. (B) High-affinity binding of P1G-MIF to sCD74, as measured by real-time, surface plasmon resonance (BIAcore analysis). Representative biosensorgrams of the interaction between sCD74 and increasing concentrations of the P1G-MIF sensor chip were obtained as described in Materials and Methods. (C) ERK1/2 phosphorylation induced by MIF or P1G-MIF. Levels of phospho-ERK1/2 (p-ERK1/2) and total ERK1/2 in synchronized, wild-type MEFs stimulated for 2 h with recombinant MIF or P1G-MIF at the indicated concentrations are shown. (D) ERK1/2 phosphorylation in synchronized, immortalized mif−/− MEFs stimulated for 15 min with supernatants from mif+/+ (wild-type), mifP1G/P1G, or mif−/− MEF cultures. The Western blots shown are representative of four separate experiments. The numerical p-ERK/total ERK ratios are from a densitometric analysis of Western blots from three experiments showing the increase in p-ERK1/2 relative to total ERK1/2 (P < 0.05 for mif+/+ MEFs versus mifP1G/P1G or mif−/− MEFs). The bottom row shows enzyme-linked immunosorbent assay values for the MIF or P1G-MIF content in mif+/+, mifP1G/P1G, and mif−/− supernatants (P was not significant for MIF content of mif+/+ versus mifP1G/P1G supernatants). (E) Competitive binding of MIF or P1G-MIF to CXCR2-expressing HEK293 cells assessed by competition with radiolabeled CXCL8. Values shown are the means ± SD of triplicate measurements. (F) Interaction of JAB1 with P1G-MIF or MIF, as assessed by coimmunoprecipitation of mif+/+ or mifP1G/P1G MEFs with anti-MIF (top) or anti-JAB1 (bottom). IP, immunoprecipitating antibody; WB, Western blotting antibody; IgG-HC, IgG heavy chain band.
The MAPK pathway is a major regulator of malignant transformation (11), and MIF is known to activate the ERK1/2 MAPK pathways in a CD74-dependent manner (24). Like MIF, P1G-MIF activated ERK1/2 phosphorylation in fibroblasts, although the level of phosphorylation also appeared less than that for wild-type MIF (Fig. 2C). Cultured fibroblasts secrete MIF, which contributes to baseline ERK1/2 phosphorylation (30), and a reduced level of ERK1/2 phosphorylation was evident upon testing the supernatants from mifP1G/P1G versus mif+/+ MEFs on mif−/− target MEFs, despite the similar levels of production of MIF or P1G-MIF by these cell lines (Fig. 2D).
Recent studies also have identified a role for MIF in leukocyte migration by activation of the CXCR2 and CXCR4 chemokine receptors (3, 47). This effect may involve presentation by CD74 as well as an interaction between the chemokine receptor and a pseudo-(E)LR motif within MIF that is similar to that found in the N-terminal region of interleukin-8 (CXCL8) (3, 47). Using a previously characterized cell-based assay for CXCR2 binding (47), we compared the binding of MIF with that of P1G-MIF using radiolabeled CXCL8. The binding of P1G-MIF was less complete than that of MIF and reached a plateau at 55% competition, with a Kd for P1G-MIF that was similar to that for wild-type MIF (MIF, 1.4 nM; P1G-MIF, 1.0 nM) (Fig. 2E). These data suggest a structural contribution of Pro1 to these protein-protein interactions.
Accruing evidence supports an important intracellular role for MIF in regulating growth control by binding and inhibiting the COP9 signalosome component JAB1 (21, 25, 32). We observed a significant intracellular interaction between JAB1 and P1G-MIF in cell lysates obtained from mifP1G/P1G fibroblasts, although there appeared to be less activity with P1G-MIF than wild-type MIF produced by mif+/+ fibroblasts (Fig. 2F). These data, taken together, indicate that, while P1G-MIF is completely inactive enzymatically, it nevertheless retains a significant level of interaction with the two MIF effector proteins, CD74 and JAB1.
Reduced development of benzo[α]pyrene-induced skin tumors in mifP1G/P1G mice.
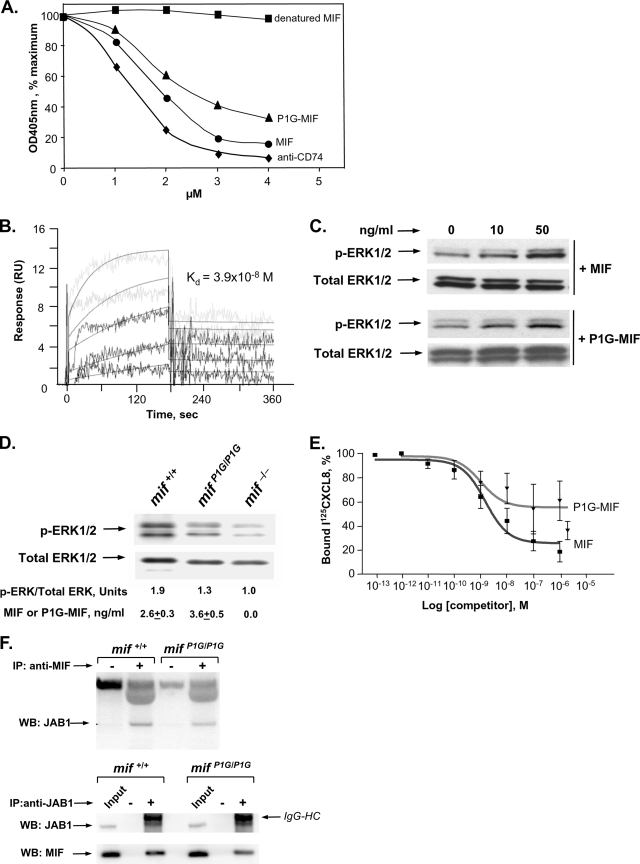
To assess the functional phenotype of the tautomerase-null P1G-MIF protein in vivo, we focused our studies on established models of MIF-dependent growth regulation that have been linked to the CD74- and JAB1-mediated effector pathways (13, 15, 17, 26, 32, 48). Mice with a complete deletion of the mif gene show altered rates of skin tumorigenesis when subjected to one- or two-stage skin carcinogenesis protocols (13, 15). We first verified that levels of MIF protein production in the epidermis in mifP1G/P1G and mif+/+ mice do not differ (Fig. 3A) and then treated age- and sex-matched mif−/−, mifP1G/P1G, and mif+/+ mice with a topical application of benzo[α]pyrene for 20 weeks. Compared to that in mif+/+ mice, tumor incidence was accelerated by approximately 1 week in mifP1G/P1G mice and by 2 weeks in mif−/− mice (Fig. 3B). Both the mifP1G/P1G and the mif−/− mice also developed significantly more tumors over time than the mif+/+ mice (Fig. 3C). Average tumor sizes for mice of the different genotypes did not differ (data not shown). The increase in tumor numbers in the mif−/− mice was found on histologic analysis to be due to an increase in the number of benign skin tumors, and the ratio of benign to malignant tumors was greatly skewed toward the benign phenotype (Table 1). This increase in the ratio of benign to malignant tumors also was present in mifP1G/P1G mice, but to a lesser extent than in the mif−/− mice (mif−/− versus mif+/+, P < 0.0001; mifP1G/P1G versus mif+/+, P = 0.0001). These data indicate that the in vivo phenotype of the mifP1G/P1G mouse with respect to skin tumorigenesis is not one of complete MIF deficiency. Given that P1G-MIF is completely lacking in tautomerase activity (Fig. 1H) yet retains appreciable activity for binding with its effector proteins (Fig. 2A to F), we postulate that the MIF tautomerization activity is dispensable for MIF regulation of growth control during benzo[α]pyrene-induced skin carcinogenesis.
FIG. 3.
P1G-MIF-expressing mice show an intermediate phenotype in skin tumorigenesis. (A) Immunohistochemical evidence of equivalent production of P1G-MIF and native MIF in skin of mifP1G/P1G and mif+/+ mice (magnification, ×400). (B) Tumor incidence in one-stage benzo[α]pyrene-induced skin tumorigenesis (n = 32 to 36 mice per group). Kaplan-Meier analysis and a log rank test yield the following: P < 0.001 for mif−/− versus mif+/+ mice; P < 0.029 for mifP1G/P1G versus mif+/+ mice; P not significant for mif−/− versus mifP1G/P1G mice. (C) Tumors per mouse in one-stage benzo[α]pyrene-induced skin tumorigenesis. Statistics using the general linear model yields the following: *, P < 0.001 for mif−/− versus mif+/+ mice, P < 0.011 for mifP1G/P1G versus mif+/+ mice, and P < 0.001 for mif−/− versus mifP1G/P1G mice.
TABLE 1.
Frequencies of benign and malignant tumors in mif−/−, mifP1G/P1G, and mif+/+ mice in the one-stage, benzo[α]pyrene-induced skin tumorigenesis modela
| Mouse genotype | % of miceb with:
|
Benign/malignant tumor ratio | |
|---|---|---|---|
| Benign tumors | Malignant tumors | ||
| mif−/− | 94 (30/32) | 6 (2/32) | 16.0c |
| mifP1G/P1G | 75 (24/32) | 25 (8/32) | 3.0d |
| mif+/+ | 60 (18/30) | 40 (12/30) | 1.5 |
Thirty tumors from each mouse group were examined and scored according to WHO classification criteria(16).
Parentheses show numbers of mice with tumors/numbers of mice tested.
P < 0.0001 by χ2 test.
P = 0.0001 by χ2 test.
Pro1 is not required for MIF-dependent Ras-mediated transformation.
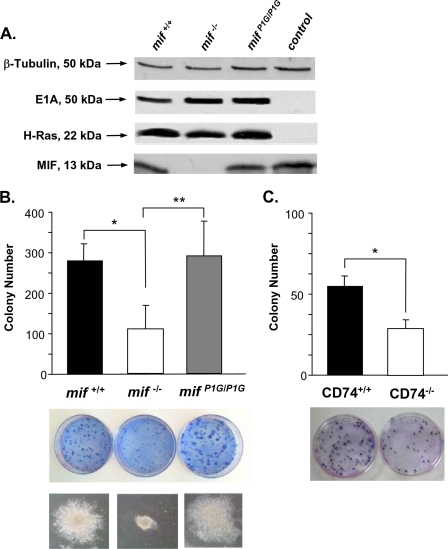
We next assessed the oncogenic potential of MEFs derived from mifP1G/P1G, mif−/−, and mif+/+ mice. The transduction of MEFs with genes encoding the adenoviral oncoprotein E1A and constitutively active Ras (H-rasV12 mutant or H-Ras) fully transforms these cells into malignant fibroblasts, leading to cell-autonomous proliferation, loss of contact inhibition, and visible colony formation in focus formation assays (11). In the absence of a concomitant deficiency or mutation in p53, mif deletion reduces the ability of MEFs to undergo ras-mediated transformation in vitro (15, 32, 45). Retroviral transfer of E1A and H-Ras into mif+/+, mif−/−, and mifP1G/P1G MEFs resulted in equivalent expression of these proteins in the different cell lines (Fig. 4A.). The mif−/− MEFs produced fewer and smaller colonies than mif+/+ cells, while the mifP1G/P1G MEFs exhibited the full transforming ability of mif+/+ MEFs and both the numbers and the sizes of the malignant colonies were similar (Fig. 4B). In separate studies, we also observed that mifP1G/P1G feeder cells rescued the transformation defect of mif−/− cells to the same extent as mif+/+ cells (data not shown). These data indicate that the tautomerase-null, P1G-MIF-expressing MEFs retain the malignant potential of wild-type MEFs under the experimental conditions of transformation with the E1A/H-rasV12 oncogenes. We also verified a direct role for the MIF cell surface receptor, CD74, in fibroblast growth control, which has not previously been documented. Primary MEFs were prepared from CD74−/− mice and transformed by successive retroviral infection with the E1A and H-rasV12 oncogenes. As shown in Fig. 4C, CD74−/− MEFs phenocopy genetic MIF deficiency; these cells show reduced focus formation compared to their wild-type counterparts (30 ± 5 CD74−/− colonies versus 54 ± 8 CD74+/+ colonies; P < 0.001) (Fig. 4C).
FIG. 4.
Ras-mediated transformation of MEFs. (A) Western blotting analysis demonstrating equivalent production of the E1A and H-Ras oncoproteins after retrovirus-mediated transfection of mif+/+, mif−/−, and mifP1G/P1G MEFs. Uninfected mif+/+ MEFs were used as the control. (B) Colony formation in focus formation assays of E1A/H-rasV12-transfected mif+/+, mif−/−, and mifP1G/P1G MEFs. (Top) Colony enumeration for the different transfected MEF lines. Data shown are from 10 independent experiments. *, P < 0.00001 for mif+/+ versus mif−/− MEFs; **, P < 0.001 for mifP1G/P1G versus mif−/− MEFs. (Middle) Macroscopic appearance of three representative plates. (Bottom) Microscopic appearance of a typical colony from each plate (magnification, ×20). (C) Colony formation in focus formation assays of E1A/H-rasV12-transfected wild-type (CD74+/+) and CD74−/− MEFs. Colonies were enumerated from 10 independent experiments (*, P < 0.001). (Bottom) Macroscopic appearance of two representative plates (magnification, ×20).
We next assessed the growth characteristics of both the E1A/H-rasV12-transformed and primary MEFs since previous analysis had shown that MIF deficiency leads to an increase in contact inhibition and subsequently to growth retardation (15). The malignant mifP1G/P1G MEFs showed a modest proliferative defect compared to mif+/+ cells (doubling times: mif+/+, 18.7 h; mifP1G/P1G, 19.8 h; mif−/−, 21.7 h; mif+/+ versus mif−/−, P < 0.001; mif+/+ versus mifP1G/P1G, P not significant) (Fig. 5A). Examination of the growth characteristics of untransformed mif+/+, mifP1G/P1G, and mif−/− MEFs under different density conditions also showed that the P1G-MIF-expressing cells reached a confluence density that was intermediate between those of wild-type and MIF-deficient cells (Fig. 5B). These data, taken together, indicate that the tautomerase-null P1G-MIF protein retains significant bioactivity with respect to MIF's action in regulating oncogenesis. We interpret these findings as being consistent with the conclusion that structural features in the MIF N-terminal region, which are influenced by the P→G substitution, and not tautomerase activity mediate these MIF-dependent biologic functions.
FIG. 5.
Representative growth characteristics of E1A/H-rasV12-transformed and primary MEFs. (A) Proliferative rate of E1A/H-rasV12-transformed mif−/−, mifP1G/P1G, and mif+/+ MEFs. For mif+/+ versus mif−/− MEFs, P was <0.001; for mif+/+ versus mifP1G/P1G MEFs, P was not significant. (B) Confluent cell density of mif−/−, mifP1G/P1G, and mif+/+ MEFs (mean ± SD of seven independent assays). *, P < 0.02 for mif−/− versus mif+/+ MEFs; **, P not significant for mifP1G/P1G versus mif+/+ MEFs; ***, P < 0.05 for mifP1G/P1G versus mif−/− MEFs.
DISCUSSION
MIF's enigmatic tautomerase activity has been the subject of significant interest since its discovery in 1996 and subsequent structural characterization (2, 36, 37, 40). While some investigators have argued that the catalytic Pro1 supports an enzymatic function for MIF, others have suggested that it is a vestigial property that may be attributed to MIF's genetic origin in the innate, melanotic encapsulation response (6, 44). Physiologic substrates for MIF have not been convincingly described, and conclusions from site-directed studies have been limited by the semiquantitative nature of the particular assays studied and by recent evidence that the catalytic, N-terminal proline has a structural role in receptor binding (2, 19, 34, 38, 43). Indeed, covalent modification of MIF's Pro1 has been shown to reduce MIF cell surface binding activity in a flow cytometry-based assay (38).
To better illuminate the role of tautomerization in MIF's biologic functions, we pursued a genetic approach and created a tautomerase-null MIF knock-in mouse. We elected to examine MIF's known action with respect to growth control and tumorigenesis because these activities involve two well-characterized high-affinity MIF binding proteins: the cell surface receptor CD74, which initiates ERK1/2 activation (23, 39), and JAB1, which binds MIF intracellularly and regulates the DNA damage checkpoint response via control of proteasomal activity (21, 32). With the exception of MIF interaction with CXCR2 in vitro, which mediates migration activity (3, 47), we did not evaluate immunologic functions because of evidence that MIF's role in the immunity varies markedly with genetic background (14, 31).
We replaced the MIF N-terminal proline with glycine, which lacks the nucleophilic imine necessary for catalysis (2, 40), and we show that the recombinant P1G-MIF protein retains high-affinity binding to CD74 and JAB1, although some reduction in binding and signal transduction activity as a result of the Pro1→Gly1 substitution is evident. Our targeting strategy involved Cre-mediated excision of the selection cassette, leading to removal of nearly all of the extraneous DNA sequence and to a gene and flanking region structure that is almost identical to that of wild-type mif. P1G-MIF was expressed in mifP1G/P1G mice at levels similar to that at which MIF is expressed in wild-type mice, verifying the success of the knock-in gene replacement strategy. Our comparative studies were performed with mice on the genetically pure C57BL/6 background, and the mif−/− mouse employed was one that was produced by a similar Cre-loxP strategy in which the entire mif gene—promoter, three exons, and intronic regions—was deleted.
In the epithelial skin tumorigenesis model, the mifP1G/P1G mice showed a level of chemically induced skin tumors that was significantly lower than that of completely MIF-deficient mice but also significantly greater than that of wild-type mice. Transformation assays of fibroblasts also showed that, upon retroviral introduction of the E1A and H-ras oncoproteins, the mifP1G/P1G fibroblasts possess full transforming capacity, like mif+/+ fibroblasts. The in vitro growth properties of these cells before and after E1A/H-rasV12 transformation showed characteristics that appeared intermediate in phenotype: a higher growth rate and reduced sensitivity to contact inhibition for the mifP1G/P1G cells compared to mif−/− cells but lower growth rate and sensitivity than mif+/+ cells. The most likely interpretation for these findings is that the tautomerase-null P1G-MIF protein retains significant protein-protein interaction activity, which is necessary for the expression of these growth-regulatory effects in vitro and in vivo.
With respect to MIF's emerging role in growth regulation, it should be emphasized that, like those of growth regulators such as transforming growth factor β and E2F1, the MIF-dependent phenotype varies with the tissue type and tumor model under study (13). While tumor induction in mesenchymal tissue (e.g., fibroblasts and lymphocytes) demonstrates an overall protumorigenic action of MIF, in epithelial tissues MIF acts either as a tumor suppressor or tumor promoter (15, 45, 46, 48). The mechanisms behind this differential phenotype may involve the relative expression levels of MIF and MIF receptors or the tissue-specific usage of particular signaling mediators. Keratinocytes are strong producers of MIF, and chemical carcinogenesis in MIF-deficient murine skin leads to significantly increased tumor formation compared to that in wild-type mice.
While the present data provide strong in vivo support for the conclusion that MIF's catalytic activity may be dispensable for its biologic function, we cannot exclude the possibility that a physiologic function for MIF-mediated tautomerization may yet be discovered. An accruing body of data nevertheless has indicated that MIF's growth-regulatory effects have a central role in its biologic function, including host immunity (29). Moreover, recent genetic studies have shown an association between high- versus low-expression MIF alleles and human prostate cancer progression, arguing strongly that MIF's growth-regulatory effects are important in tumorigenesis (27).
The present genetic data support a model of MIF biologic function whereby structural features in the protein's N-terminal region, but not its intrinsic tautomerase activity, are important for receptor binding and downstream activation responses. These studies also focus further attention on the possibility of pharmacologically targeting the MIF N-terminal region for therapeutic benefit.
Acknowledgments
We are grateful to Klaus Rajewsky, Werner Muller, and the Institute of Genetics at University of Cologne for the support of this project and to Arlene Sharpe (Harvard Medical School) for expert assistance with ES cell injections. Michael Thiele assisted with the signaling and JAB1 binding studies.
These studies were supported by NIH grants AI042310, AR049610, and AR050498 to R.B. and grants Fi 712/2-1 and Be 1977/4-1 of the German Research Council (DFG) to G.F.R. and J.B., respectively; by the RWTH Aachen University START program (J.B.); and by the Koeln Fortune Program of the Medical Faculty of Cologne University (G.F.R.).
Footnotes
Published ahead of print on 2 January 2009.
REFERENCES
- 1.Agarwal, R., D. H. Whang, A. B. Alvero, I. Visintin, Y. L. Lai, E. A. Segal, P. Schwartz, D. Ward, T. Rutherford, and G. Mor. 2007. Macrophage migration inhibitory factor expression in ovarian cancer. Am. J. Obstet. Gynecol. 196348.e1-348.e5. [DOI] [PubMed] [Google Scholar]
- 2.Bendrat, K., Y. Al-Abed, D. J. Callaway, T. Peng, T. Calandra, C. N. Metz, and R. Bucala. 1997. Biochemical and mutational investigations of the enzymatic activity of macrophage migration inhibitory factor. Biochemistry 3615356-15362. [DOI] [PubMed] [Google Scholar]
- 3.Bernhagen, J., R. Krohn, H. Lue, J. L. Gregory, A. Zernecke, R. R. Koenen, M. Dewor, I. Georgiev, A. Schober, L. Leng, T. Kooistra, G. Fingerle-Rowson, P. Ghezzi, R. Kleemann, S. R. McColl, R. Bucala, M. J. Hickey, and C. Weber. 2007. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 13587-596. [DOI] [PubMed] [Google Scholar]
- 4.Bernhagen, J., R. A. Mitchell, T. Calandra, W. Voelter, A. Cerami, and R. Bucala. 1994. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF). Biochemistry 3314144-14155. [DOI] [PubMed] [Google Scholar]
- 5.Bozza, M., A. R. Satoskar, G. Lin, B. Lu, A. A. Humbles, C. Gerard, and J. R. David. 1999. Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J. Exp. Med. 189341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucala, R. (ed.). 2007. MIF: most interesting factor, p. 19-34. World Scientific, London, United Kingdom.
- 7.Bucala, R., and S. C. Donnelly. 2007. Macrophage migration inhibitory factor: a probable link between inflammation and cancer. Immunity 26281-285. [DOI] [PubMed] [Google Scholar]
- 8.Bunting, M., K. E. Bernstein, J. M. Greer, M. R. Capecchi, and K. R. Thomas. 1999. Targeting genes for self-excision in the germ line. Genes Dev. 131524-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calandra, T., J. Bernhagen, C. N. Metz, L. A. Spiegel, M. Bacher, T. Donnelly, A. Cerami, and R. Bucala. 1995. MIF as a glucocorticoid-induced modulator of cytokine production. Nature 37768-71. [DOI] [PubMed] [Google Scholar]
- 10.Chesney, J., C. Metz, M. Bacher, T. Peng, A. Meinhardt, and R. Bucala. 1999. An essential role for macrophage migration inhibitory factor (MIF) in angiogenesis and the growth of a murine lymphoma. Mol. Med. 5181-191. [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon, A. S., S. Hagan, O. Rath, and W. Kolch. 2007. MAP kinase signalling pathways in cancer. Oncogene 263279-3290. [DOI] [PubMed] [Google Scholar]
- 12.El-Turk, F., M. Cascella, H. Ouertatani-Sakouhi, R. L. Narayanan, L. Leng, R. Bucala, M. Zweckstetter, U. Rothlisberger, and H. A. Lashuel. 2008. The conformational flexibility of the carboxy terminal residues 105-114 is a key modulator of the catalytic activity and stability of macrophage migration inhibitory factor. Biochemistry 4710740-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fingerle-Rowson, G., and O. Petrenko. 2007. MIF coordinates the cell cycle with DNA damage checkpoints. Lessons from knockout mouse models. Cell Division 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fingerle-Rowson, G., A. Satoskar, and R. Bucala. 2003. Macrophage migration inhibitory factor (MIF)-deficient mice, p. 361-378. In G. Fantuzzi (ed.), Cytokine knockouts. Humana Press, Totowa, NJ.
- 15.Fingerle-Rowson, G., O. Petrenko, C. N. Netz, T. G. Forsthuber, R. Mitchell, R. Huss, U. Moll, W. Müller, and R. Bucala. 2003. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc. Natl. Acad. Sci. USA 1009354-9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldschmidt, M. H., R. W. Dunstan, A. A. Stannard, C. von Tscharner, E. J. Walder, and J. A. Yager (ed.). 1998. Histological classification of epithelial and melanocytic tumors of the skin of domestic animals, 2nd series. Armed Forces Institute of Pathology, Washington, DC.
- 17.Gore, Y., D. Starlets, N. Maharshak, S. Becker-Herman, U. Kaneyuki, L. Leng, R. Bucala, and I. Shachar. 2008. Macrophage migration inhibitory factor (MIF) induces B cell survival by activation of a CD74/CD44 receptor complex. J. Biol. Chem. 2832784-2792. [DOI] [PubMed] [Google Scholar]
- 18.Gregersen, P., and R. Bucala. 2003. MIF, MIF alleles, and the genetics of inflammatory disorders: incorporating disease outcome into the definition of phenotype. Arthritis Rheum. 481171-1176. [DOI] [PubMed] [Google Scholar]
- 19.Hermanowski-Vosatka, A., S. S. Mundt, J. M. Ayala, S. Goyal, W. A. Hanlon, R. M. Czerwinski, S. D. Wright, and C. P. Whitman. 1999. Enzymatically inactive macrophage migration inhibitory factor inhibits monocyte chemotaxis and random migration. Biochemistry 3812841-12849. [DOI] [PubMed] [Google Scholar]
- 20.Hudson, J. D., M. A. Shoaibi, R. Maestro, A. Carnero, G. J. Hannon, and D. H. Beach. 1999. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J. Exp. Med. 1901375-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleemann, R., A. Hausser, G. Geiger, R. Mischke, A. Burger-Kentischer, O. Flieger, F. J. Johannes, T. Roger, T. Calandra, A. Kapurniotu, M. Grell, D. Finkelmeier, H. Brunner, and J. Bernhagen. 2000. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature 408211-216. [DOI] [PubMed] [Google Scholar]
- 22.Legendre, H., C. Decaestecker, N. Nagy, A. Hendlisz, M. P. Schuring, I. Salmon, H. J. Gabius, J. C. Pector, and R. Kiss. 2003. Prognostic values of galectin-3 and the macrophage migration inhibitory factor (MIF) in human colorectal cancers. Mod. Pathol. 16491-504. [DOI] [PubMed] [Google Scholar]
- 23.Leng, L., and R. Bucala. 2006. Insight into the biology of macrophage migration inhibitory factor (MIF) revealed by the cloning of its cell surface receptor. Cell Res. 16162-168. [DOI] [PubMed] [Google Scholar]
- 24.Leng, L., C. Metz, Y. Fang, J. Xu, S. Donnelly, J. Baugh, T. Delonery, Y. Chen, R. A. Mitchell, and R. Bucala. 2003. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 1971467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lue, H. Q., A. Kapurniotu, G. Fingerle-Rowson, T. Roger, L. Leng, M. Thiele, T. Calandra, R. Bucala, and J. Bernhagen. 2006. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell. Signal. 18688-703. [DOI] [PubMed] [Google Scholar]
- 26.Meyer-Siegler, K. L., K. A. Iczkowski, L. Leng, R. Bucala, and P. L. Vera. 2006. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 1778730-8739. [DOI] [PubMed] [Google Scholar]
- 27.Meyer-Siegler, K. L., P. L. Vera, K. A. Iczkowski, C. Bifulco, A. Lee, P. K. Gregersen, L. Leng, and R. Bucala. 2007. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 8646-652. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell, R. A. 2004. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell. Signal. 1613-19. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, R. A., H. Liao, J. Chesney, G. Fingerle-Rowson, J. Baugh, J. David, and R. Bucala. 2002. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc. Natl. Acad. Sci. USA 99345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, R. A., C. N. Metz, T. Peng, and R. Bucala. 1999. Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J. Biol. Chem. 27418100-18106. [DOI] [PubMed] [Google Scholar]
- 31.Mizue, Y., S. Ghani, L. Leng, C. McDonald, P. Kong, J. Baugh, S. J. Lane, J. Craft, J. Nishihira, S. C. Donnelly, Z. Zhu, and R. Bucala. 2005. Role for macrophage migration inhibitory factor (MIF) in asthma. Proc. Natl. Acad. Sci. USA 10214410-14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemajerova, A., P. Mena, G. Fingerle-Rowson, U. M. Moll, and O. Petrenko. 2007. Impaired DNA damage checkpoint response in MIF-deficient mice. EMBO J. 26987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemajerova, A., U. M. Moll, O. Petrenko, and G. Fingerle-Rowson. 2007. Macrophage migration inhibitory factor coordinates DNA damage response with the proteasomal control of the cell cycle. Cell Cycle 61030-1034. [DOI] [PubMed] [Google Scholar]
- 34.Onodera, S., K. Kaneda, Y. Mizue, Y. Koyama, M. Fujinaga, and J. Nishihira. 2000. Macrophage migration inhibitory factor up-regulates expression of matrix metalloproteinases in synovial fibroblasts of rheumatoid arthritis. J. Biol. Chem. 275444-450. [DOI] [PubMed] [Google Scholar]
- 35.Petrenko, O., and U. M. Moll. 2005. Macrophage migration inhibitory factor MIF interferes with the Rb-E2F pathway. Mol. Cell 17225-236. [DOI] [PubMed] [Google Scholar]
- 36.Rosengren, E., P. Aman, S. Thelin, C. Hansson, S. Ahlfors, P. Bjork, L. Jacobsson, and H. Rorsman. 1997. The macrophage migration inhibitory factor MIF is a phenylpyruvate tautomerase. FEBS Lett. 41785-88. [DOI] [PubMed] [Google Scholar]
- 37.Rosengren, E., R. Bucala, P. Aman, L. Jacobsson, G. Odh, C. N. Metz, and H. Rorsman. 1996. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol. Med. 2143-149. [PMC free article] [PubMed] [Google Scholar]
- 38.Senter, P. D., Y. Al-Abed, C. N. Metz, F. Benigni, R. A. Mitchell, J. Chesney, J. Han, C. G. Gartner, S. D. Nelson, G. J. Todaro, and R. Bucala. 2002. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc. Natl. Acad. Sci. USA 99144-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, X., L. Leng, T. Wang, W. Wang, X. Du, C. McDonald, Z. Chen, J. W. Murphy, E. Lolis, P. Noble, W. Knudson, and R. Bucala. 2006. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity 25595-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamps, S. L., M. C. Fitzgerald, and C. P. Whitman. 1998. Characterization of the role of the amino-terminal proline in the enzymatic activity catalyzed by macrophage migration inhibitory factor. Biochemistry 3710195-10202. [DOI] [PubMed] [Google Scholar]
- 41.Sun, H. W., J. Bernhagen, R. Bucala, and E. Lolis. 1996. Crystal structure at 2.6-Å resolution of human macrophage migration inhibitory factor. Proc. Natl. Acad. Sci. USA 935191-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki, M., H. Sugimoto, A. Nakagawa, I. Tenaka, J. Nishihira, and M. Sakai. 1996. Crystal structure of the macrophage migration inhibitory factor from rat liver. Nat. Struct. Biol. 3259-266. [DOI] [PubMed] [Google Scholar]
- 43.Swope, M., H. W. Sun, P. R. Blake, and E. Lolis. 1998. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 173534-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swope, M. D., and E. Lolis. 1999. Macrophage migration inhibitory factor: cytokine, hormone, or enzyme. Rev. Physiol. Biochem. Pharmacol. 1391-32. [DOI] [PubMed] [Google Scholar]
- 45.Talos, F., P. Mena, G. Fingerle-Rowson, U. Moll, and O. Petrenko. 2005. MIF loss impairs Myc-induced lymphomagenesis. Cell Death Differ. 121319-1328. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, J. A., G. A. Kuchel, P. Hegde, O. S. Voznesensky, K. Claffey, J. Tsimikas, L. Leng, R. Bucala, and C. Pilbeam. 2007. Null mutation for macrophage migration inhibitory factor (MIF) is associated with less aggressive bladder cancer in mice. BMC Cancer 7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber, C., S. Kraemer, M. Drechsler, H. Q. Lue, R. R. Koenen, A. Kapurniotu, A. Zernecke, and J. Bernhagen. 2008. Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc. Natl. Acad. Sci. USA 10516278-16283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, J. M., P. L. Coletta, R. J. Cuthbert, N. Scott, K. MacLennan, G. Hawcroft, L. Leng, J. B. Lubetsky, K. K. Jin, E. Lolis, F. Medina, J. A. Brieva, R. Poulsom, A. F. Markham, R. Bucala, and M. A. Hull. 2005. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology 1291485-1503. [DOI] [PubMed] [Google Scholar]