Abstract
Changes in chromatin composition are often a prerequisite for gene induction. Nonallelic histone variants have recently emerged as key players in transcriptional control and chromatin modulation. While the changes in chromatin accessibility and histone posttranslational modification (PTM) distribution that accompany gene induction are well documented, the dynamics of histone variant exchange that parallel these events are still poorly defined. In this study, we have examined the changes in histone variant distribution that accompany activation of the inducible CD69 and heparanase genes in T cells. We demonstrate that the chromatin accessibility increases that accompany the induction of both of these genes are not associated with nucleosome loss but instead are paralleled by changes in histone variant distribution. Specifically, induction of these genes was paralleled by depletion of the H2A.Z histone variant and concomitant deposition of H3.3. Furthermore, H3.3 deposition was accompanied by changes in PTM patterns consistent with H3.3 enriching or depleting different PTMs upon incorporation into chromatin. Nevertheless, we present evidence that these H3.3-borne PTMs can be negated by recruited enzymatic activities. From these observations, we propose that H3.3 deposition may both facilitate chromatin accessibility increases by destabilizing nucleosomes and compete with recruited histone modifiers to alter PTM patterns upon gene induction.
The ability of a multicellular organism to coordinate the behavior of distinct cell types and respond to environmental changes critically depends on regulated gene expression. In the nuclei of eukaryotic cells, gene transcription takes place in the context of a supramolecular structure called chromatin. The smallest unit of chromatin is the nucleosome, which consists of 147 bp of DNA wrapped around a histone octamer. Each octamer typically contains two of each of the “core” histones (namely, H2A, H2B, H3, and H4); however, nonallelic isoforms of the core histones (referred to as histone variants) are also expressed by cells (44). Chromatin is not merely an inert DNA packaging complex, and in recent years it has become clear that dynamic changes in chromatin structure play a key role in regulating genome functions, including transcription. Positioned nucleosomes and higher-order chromatin compaction can preclude the access of the transcriptional machinery to target genes, meaning that chromatin must often be reorganized on gene regulatory regions before transcription will occur (26). Chromatin also serves as an interface to recruit regulatory cofactors, meaning that changes in chromatin composition can alter the complexes recruited to a specific regulatory region (42). With regard to transcriptional regulation, distinct chromatin states exist, including the silenced state, the poised state, and the active transcribed state. The means by which chromatin is reorganized into these distinctively different states is still poorly understood.
Thus far, three main mechanisms of chromatin modulation have been characterized. First, ATP-dependent chromatin remodeling complexes can utilize ATP hydrolysis to directly evict, slide or rotate nucleosomes across gene regulatory regions (14, 43). Through nucleosome reorganization, remodeling complexes can affect the access of transcriptional regulatory factors to target sequences (26). Second, the amino-terminal tails of histones are subject to a plethora of posttranslational modifications (PTMs) mediated by enzymes recruited to gene regulatory regions (24, 50). Such PTMs can subsequently recruit other factors that either activate or repress transcription (4, 24). PTMs can also directly change chromatin architecture by modifying internucleosomal contacts and the formation of higher-order chromatin structures (48). Finally, chromatin composition can be reorganized by exchanging the canonical histones with one or more of a number of histone variants (22, 44). Variant histone deposition can directly change chromatin structure by altering nucleosome stability (21) and higher-order chromatin structure (12). Histone variants can also indirectly alter chromatin structure by carrying specific PTMs onto the targeted region (30).
Recent findings have illustrated the central role that histone variant distribution is likely to play in orchestrating chromatin structure. While a number of histone variants exist, most work has focused on the variants H3.3 and H2A.Z. Importantly, unlike the core histones, both of these variants can be deposited onto target gene regions in a replication-independent manner (1, 2, 33, 36, 51). Several genome-wide chromatin immunoprecipitation (ChIP) studies with Saccharomyces cerevisiae have convincingly demonstrated that H2A.Z (termed Hzt1 in yeast) is preferentially enriched on transcriptionally inactive promoters while being depleted from highly transcribed genes (17, 27, 39, 54). Furthermore, inducible genes poised for transcription appear to contain a defined nucleosome-free region within their promoters that is flanked by two well-positioned Hzt1-containing nucleosomes (39, 53), with similar observations recently described in human CD4+ T cells (3). Collectively, these data suggest that H2A.Z plays a role in maintaining genes in a poised state. In contrast to H2A.Z, studies with epitope-tagged versions of H3.3 have elegantly demonstrated the association of H3.3 with transcriptionally active chromatin both at promoters and in the transcribed region (32, 52). Consistent with a role in promoting gene transcription, H3.3 is often enriched in PTMs associated with active chromatin (18, 30, 31). These findings suggest that such replication-independent H3.3 deposition potentiates and/or consolidates the active state.
While many of the genome-wide studies have provided a static picture of histone variant distribution, few studies have examined the temporal changes in histone variant distribution over gene regulatory regions during the transition between a poised and an active state. It is also worth noting that the lack of an H3.3-specific antibody has precluded ChIP studies of endogenous H3.3 distribution, instead relying on the transfection of epitope-tagged H3.3 into target cells. In this study, we sought to determine the changes in histone variant distribution and chromatin structure that accompany rapid gene induction in T cells. As many inducible genes are rapidly expressed upon T-cell activation, T cells represent an ideal model system to dissect out the chromatin changes that accompany rapid gene induction in mammalian cells. With this system, we have examined both the temporal and spatial changes in H3.3 and H2A.Z distribution that are associated with transcriptional activation and chromatin remodeling of the T-cell-inducible CD69 and heparanase genes. To allow direct measurements of endogenous H3.3 levels, we have developed an H3.3-specific antibody. Using both this antibody and existing ChIP antibodies, we have demonstrated that, in the resting state, the transcription start site (TSS) of both genes is flanked by H2A.Z- and H3-enriched regions with little or no detectable H3.3. Moreover, this poised chromatin conformation appears to be associated with docked RNA polymerase II (Pol II), even on the CD69 gene, which has low transcription. As expected, transcriptional activation was accompanied by a rapid increase in chromatin accessibility and RNA Pol II occupancy. Importantly, though, activation also provoked depletion of H2A.Z and H3 and a concomitant deposition of H3.3 with no net nucleosome loss. This process coincided with an increase in chromatin accessibility, suggesting a potential role in remodeling. Sequential ChIP experiments demonstrated that H3.3 deposited upon activation was enriched in the activating PTM acetylated H3K9 and depleted of the repressive PTM dimethylated H3K9. However, we also provide evidence that the recruitment of enzymatic activities onto regulatory regions can counteract PTM deposition by H3.3. Collectively, these results provide a detailed insight into the chromatin changes that accompany the transition from a poised to an active chromatin state and represent the first study of endogenous H3.3 distribution.
MATERIALS AND METHODS
Cell culture.
Human Jurkat T cells and the human mammary adenocarcinoma lines MDA-231 and MCF-7, were maintained as described previously (20, 49). Jurkat T cells in suspension at a density of 5 × 105/ml were stimulated with 20 ng/ml phorbol 12-myristate 13-acetate and 1 μM Ca 2+ ionophore A23187 (PI) for the times indicated (20). For actinomycin D treatment studies, cells were treated with 10 μg/ml actinomycin D for the times indicated. For trichostatin A (TSA) treatment studies, cells were pretreated with 500 ng/ml TSA for 5 h prior to stimulation.
Primary T-cell preparation and stimulation.
Human blood was obtained from healthy volunteers. Human CD4+ T cells were purified with MACS CD4+ beads according to the manufacturer's guidelines (Miltenyi Biotec). The cells were subsequently stained and analyzed by flow cytometry with CD4+ T-cell populations shown to be greater than 90% pure with CD4-, CD8-, B220 (B cell)-, and CD11b (macrophage)-specific antibodies (BD Biosciences). T cells were stimulated with immobilized anti-human CD3 antibodies and a soluble CD28 antibody (BD Biosciences) at a final concentration of 5 μg/ml as previously described (40). Six-well plates were coated with 10 μg/ml anti-CD3 antibody at 37°C for 3 h or overnight at 4°C. The plates were subsequently washed (four or five times) with phosphate-buffered saline before the addition of cells at a density of 1 × 106/ml.
RNA extraction and real-time PCR analysis.
Total RNA was extracted from nonstimulated (NS) and stimulated T cells with TRIzol reagent (Sigma-Aldrich) as described previously (40). RNA (1 μg) was subsequently reverse transcribed with the SuperScript III RNase H− reverse transcriptase kit (Invitrogen). TaqMan real-time PCR was performed in a total volume of 10 μl containing a 1:100 cDNA dilution, as detailed in the manufacturer's guidelines (Applied Biosystems). Each PCR was performed in duplicate. An aliquot of each sample was analyzed by quantitative PCR with the housekeeping cyclophilin A gene. The following human TaqMan primer sets were used: heparanase, Hs00180737_m1; CD69, Hs00156399_m1; cyclophilin A, Hs99999904_m1 (Applied Biosystems). Threshold cycle values from the PCR amplification plots were converted to arbitrary copy numbers as described previously (40), and mRNA changes were then expressed as n-fold changes relative to NS T cells.
Chromatin accessibility by real-time PCR (CHART-PCR) assay.
Accessibility to digestion by micrococcal nuclease (MNase) was analyzed with the CHART-PCR assay as described previously (41). Briefly, NS and stimulated T cells (5 × 106/sample) were pelleted by centrifugation, washed in phosphate-buffered saline, and lysed in NP-40 lysis buffer on ice. The cell suspension was then centrifuged to pellet the nuclei, and the pellet was washed in MNase digestion buffer (without CaCl2) and resuspended at 5 × 106 nuclei/100 μl. MNase digestion was then performed on the nuclear suspension. Genomic DNA obtained after digestion was used for SYBR green real-time PCR, which was performed with an ABI PRISM 7700 sequence detector (Applied Biosystems). A control without the accessibility agent was used to monitor endogenous endonuclease activity. To convert the threshold cycle values from the CHART-PCR amplification plots to percent accessibility, a standard curve was generated with genomic DNA. For MNase accessibility, the percentage of cutting was calculated by expressing the amount of genomic DNA remaining as a percentage of the amount of genomic DNA in cells that were not treated with MNase. The data were then converted to percent accessibility as follows: percent accessibility = 100 − percent cutting. PCR products were visualized on a 4% Nusieve 3:1 agarose gel (PE Applied Biosystems) to ensure that a single product was generated by the PCR. All samples were subjected to real-time PCR analysis with primer set F on the human interleukin-2 (IL-2) gene (41) to ensure equal loading of DNA in all PCRs. For the primer set sequences, see Tables S1 and S2 in the supplemental material.
Generation of a histone H3.3-specific antibody.
Synthetic peptides (10 mg) were synthesized by Auspep Pty. Ltd., Australia. The amino acid sequence of the peptide used for immunization was CGGRKSAPSTGGVKK, which corresponds to amino acids 26 to 37 of the H3.3 protein plus a CGG sequence at the N terminus added to aid conjugation. Peptides were conjugated to keyhole limpet hemocyanin prior to immunization by the ACRF Biomolecular Resource Facility, Australian National University, Canberra, Australia. Polyclonal antibodies were raised in New Zealand White rabbits by subcutaneous immunization in accordance with protocols approved by the Australian National University Animal Experimentation Ethics Committee. Blood samples were checked by enzyme-linked immunosorbent assay for reactivity with 96-well plates precoated at 100 ng/ml with the H3.3 peptide, a serine 31-phosphorylated form of the H3.3 peptide, an irrelevant control peptide, and recombinant histone H3.1 (New England BioLabs). Terminal blood samples were then purified by protein A immunoglobulin G purification by the Biomolecular Resource Facility, Australian National University, Canberra, Australia, before affinity purification on peptide-coupled columns with a SulfoLink kit (Pierce) according to the manufacturer's guidelines.
ChIP assays.
ChIP assays were performed in accordance with the protocol supplied by Upstate Biotechnology. Fixation steps were performed as detailed, and fixed chromatin was sonicated with an Ultrasonic processor (Cole/Parmer) under optimized conditions that gave average DNA fragments of approximately 500 bp, as determined by 2% agarose gel electrophoresis. Prior to antibody addition, samples were precleared with salmon sperm DNA-protein A-agarose and the soluble chromatin fraction was incubated overnight at 4°C with 5 to 10 μg of anti-histone H3 (Abcam), anti-histone H2A.Z (Abcam), anti-histone H3.3 (see H3.3 antibody generation), anti-acetyl-H3K9 (Upstate), anti-dimethyl-H3K4 (Upstate), anti-dimethyl-H3K9 (Upstate), anti-H4 (Upstate), or anti-RNA Pol II CTD repeat YSPTSPS (8WG16; Abcam) antibody or without antibody as a control. Immune complexes were bound to salmon sperm DNA-protein A-agarose and then washed and eluted as described. Protein-DNA cross-links were reversed by incubation at 65°C overnight, and the DNA in each sample was recovered by phenol-chloroform extraction and ethanol precipitation. DNA pellets were washed in 70% ethanol, resuspended in Tris (pH 8.0), and subsequently used for SYBR green real-time PCR amplification (Applied Biosystems). Standard curves were generated for each primer set to correct for differences in primer efficiency. For the primer set sequences, see Tables S1 and S2 in the supplemental material. ChIP enrichment ratios were calculated as described previously (38). Sequential ChIP assays were performed as described previously (23). Briefly, samples were resuspended in 10 mM dithiothreitol after primary ChIP and incubated for 1 h at 37°C. Samples were subsequently diluted 1:40 in ChIP dilution buffer, and aliquots were taken for use as total genomic input controls. The second antibody was added to these samples for immunoprecipitation, which was again performed as described above for the primary immunoprecipitation. For a description of the method used to analyze sequential ChIP data, see Table S3 in the supplemental material.
RESULTS
Heparanase and CD69 transcript induction kinetics.
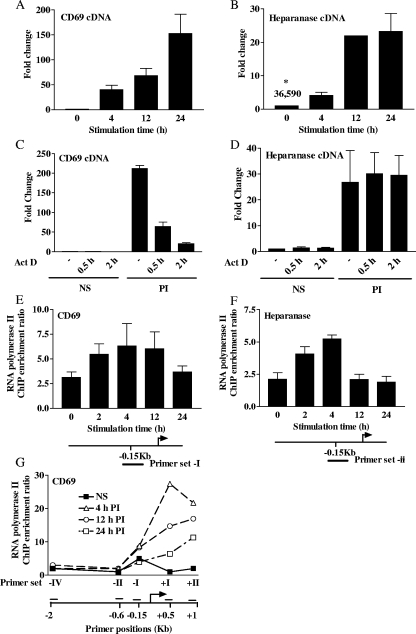
The human Jurkat T-cell line was chosen as a model system for this study. In order to examine the chromatin changes associated with transcription of the CD69 and heparanase genes, we first determined the transcript induction kinetics of these two genes in Jurkat T cells. The CD69 gene is involved in T-cell migration (47) and exhibits little basal expression but is rapidly induced upon T-cell activation (8). Consistent with these observations, we observed no detectable transcript in NS Jurkat T cells; however, pharmacological stimulation with PI induced a rapid transcript accumulation that peaked after 24 h (Fig. 1A). The heparanase gene is involved in extracellular matrix destruction and cell invasion of tissues (35). In contrast to the CD69 gene, the heparanase gene displays significant basal expression levels in T cells but can be further induced upon T-cell activation (10). Similarly, we found that the Jurkat T-cell line displayed a detectable basal heparanase gene expression that could be substantially increased by PI stimulation (Fig. 1B). These patterns of transcript accumulation were not an artifact of the transformed Jurkat T-cell line, as similar profiles have been obtained from primary human CD4+ T cells (data not shown) (11).
FIG. 1.
Heparanase and CD69 transcript induction kinetics and Pol II recruitment. (A to D) Heparanase and CD69 mRNA kinetics were measured by TaqMan real-time PCR of cDNA prepared from resting (NS or 0) Jurkat T cells and cells stimulated with PI for the times indicated. mRNA levels are expressed as n-fold changes relative to those of NS samples. For actinomycin D (Act D) experiments, NS cells or cells stimulated for 24 h with PI were treated with actinomycin D for either 0.5 or 2 h prior to the 24-h time point. Panels A and B show the transcript induction kinetics of CD69 and heparanase, while panels C and D show actinomycin data for CD69 and heparanase, respectively. Data shown are the mean ± the standard error of three replicate experiments. The asterisk refers to the number of copies of heparanase detected per microgram of total RNA. (E to G) RNA Pol II ChIP assays were performed with Jurkat T cells that were either left NS or stimulated with PI for the times indicated. The names and positions of the primer sets are indicated beneath the graphs. ChIP data are shown for the CD69 gene promoter (E), the heparanase gene promoter (F), and across the CD69 gene region (G). The results represent either the mean ± the standard error of three independent experiments (E and F) or a representative experiment from three replicates (G).
We next examined whether transcript accumulation of both genes upon stimulation was a result of sustained transcription or transcript persistence due to high mRNA stability. To this end, we stimulated Jurkat T cells for 24 h with PI but this time incubated the cells with the transcriptional inhibitor actinomycin D for either 30 min or 2 h prior to the 24-h time point. While CD69 mRNA levels were substantially diminished by inhibitor treatment (Fig. 1C), heparanase transcript levels were maintained in the presence of the inhibitor (Fig. 1D). Thus, CD69 transcript accumulation is solely due to sustained transcription while heparanase mRNA accumulation occurs as a result of both augmented transcription and transcript persistence.
RNA Pol II recruitment is a necessary step in transcriptional induction. ChIP was next used to examine Pol II recruitment to the CD69 and heparanase gene promoter regions. On both promoters, Pol II levels increased upon stimulation in a manner that paralleled gene induction, further demonstrating the transcriptional induction of these genes upon T-cell activation (Fig. 1E and F). However, it should be noted that Pol II levels did decrease at later time points, particularly on the heparanase gene promoter. This decrease in the levels of promoter-bound Pol II may be due, in part, to movement of the polymerase into the transcribed region during transcriptional elongation. Consistent with this, when Pol II levels were fine mapped at a higher resolution in the CD69 gene region, Pol II was found to be redistributed to the transcribed region upon T-cell stimulation (Fig. 1G). Unfortunately, the high G/C content of the sequences flanking the heparanase gene promoter precluded the development of primer sets to similarly examine Pol II distribution across the heparanase gene region. However, the most interesting observation was the presence of substantial levels of Pol II on the CD69 gene promoter in NS T cells (Fig. 1E). While the presence of Pol II on the heparanase gene promoter (Fig. 1F) was expected in NS T cells given the high basal expression of this gene, it was unexpected that Pol II would be present on the CD69 gene in the basal state when this gene has low levels of transcription. Pol II was similarly observed on the promoter of the inactive CD69 gene in resting primary human CD4+ T cells (data not shown). Mapping studies demonstrated that Pol II was largely restricted to the CD69 gene promoter in NS T cells but redistributed to the transcribed region upon gene induction (Fig. 1G). The presence of Pol II on the CD69 gene promoter and the associated low levels of transcription in the basal state are reminiscent of recent observations that poised Pol II is docked on the promoters of inducible genes prior to rapid transcription (16, 28, 45). Therefore, while the heparanase gene can be classified as a basally transcribing gene whose expression can be upregulated, the CD69 gene is a gene that is poised for rapid induction.
Chromatin accessibility changes across the promoters of the CD69 and heparanase genes.
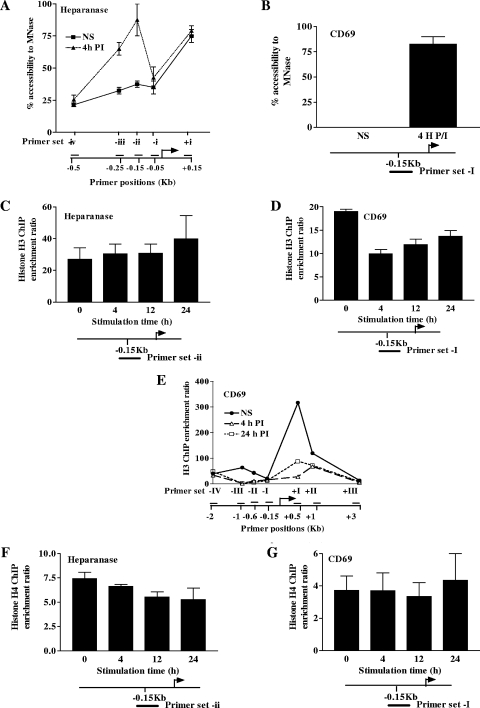
Having determined the mRNA induction kinetics of both genes, we next sought to characterize the chromatin changes that accompanied their transcriptional activation. Chromatin accessibility increases were measured by CHART-PCR, a technique in which quantitative real-time PCR is used to assess the MNase accessibility of chromatin across a gene region (41). As the promoter region of the heparanase gene has not been extensively investigated, chromatin accessibility changes upon PI stimulation were mapped to a high resolution with multiple primer sets across the entire promoter. This revealed that, in the basal state, the transcribed region of the heparanase gene is highly accessible to nuclease digestion while the upstream promoter region exhibits a low but detectable level of nuclease cutting (Fig. 2A). Interestingly, PI stimulation was capable of causing a substantial increase in chromatin nuclease accessibility selectively within the promoter region (Fig. 2A). This increase was observable at 4 h after stimulation, a time point that coincides with the upregulation of heparanase mRNA associated with T-cell activation. The CD69 gene promoter also exhibited an increase in chromatin accessibility across the promoter region upon gene activation (Fig. 2B). In contrast to the promoter region of the heparanase gene, however, the promoter region of this gene was inaccessible to digestion in the basal state.
FIG. 2.
Chromatin accessibility and core histone occupancy changes upon induction of the heparanase and CD69 genes. (A and B) CHART assays were performed with Jurkat T cells left NS or stimulated for 4 h with PI either across the heparanase gene promoter (A) or on the CD69 gene promoter (B). The names and positions of the primer sets are indicated beneath the graphs. Data are graphed as percent accessibility with respect to the sample minus the accessibility agent. Data are pooled from three independent experiments and show the mean ± the standard error. (C to G) ChIP assays were performed with Jurkat T cells either left NS or stimulated with PI for the times indicated. ChIP was conducted with either anti-H3 antibody (C to E) or anti-H4 antibody (F and G). Data are shown for the heparanase gene promoter (C and F), the CD69 gene promoter (D and G), and across the CD69 gene region (E). The names and positions of the primer sets are indicated beneath the graphs. The results represent the mean ± the standard error of 11 independent experiments for panels C to E and 3 independent experiments for panels F and G.
Nucleosome loss was recently demonstrated as a mechanism for chromatin remodeling across the T-cell-inducible IL-2 and granulocyte-macrophage colony-stimulating factor genes (7, 9). We next examined if a similar mechanism was responsible for the chromatin accessibility increases observed on the promoters of the CD69 and heparanase genes upon their induction. Changes in levels of the core histone H3 were first measured by ChIP on the promoters of the CD69 and heparanase genes upon their induction. While there was no detectable change in H3 levels on the heparanase gene promoter during its induction (Fig. 2C), a significant decrease in H3 levels occurred upon CD69 transcriptional activation (Fig. 2D). When H3 levels were assessed across the gene regions flanking the CD69 gene promoter, even greater losses of H3 were observed specifically in the transcribed region but also upstream of the promoter (Fig. 2E). Interestingly, a greater H3 signal was detected in the transcribed region than in the promoter region in resting T cells. This may be due to the low nucleosome density that is a feature of promoter regions (5, 6, 25, 45). Collectively, these data suggested that while chromatin remodeling on the heparanase gene promoter was not accompanied by nucleosome loss, nucleosome loss on the CD69 gene promoter might account for its increased accessibility upon gene induction. To further investigate whether this was the case, the levels of another core histone, H4, were also determined on the promoters of these genes during activation. Surprisingly, there was no change in H4 levels on the promoter of either the heparanase (Fig. 2F) or the CD69 (Fig. 2G) gene during its activation. One way in which to reconcile the conflicting H3 and H4 ChIP data on the CD69 gene promoter is that the exchange of H3 with H3.3, rather than nucleosome loss, is occurring across the CD69 gene promoter during transcriptional activation.
Generation of an affinity-purified histone H3.3-specific antibody.
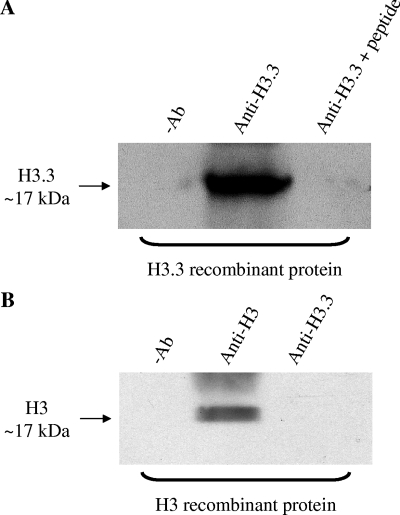
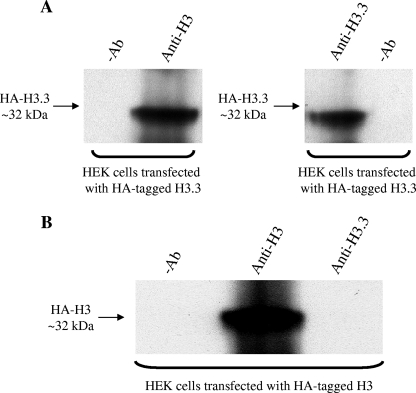
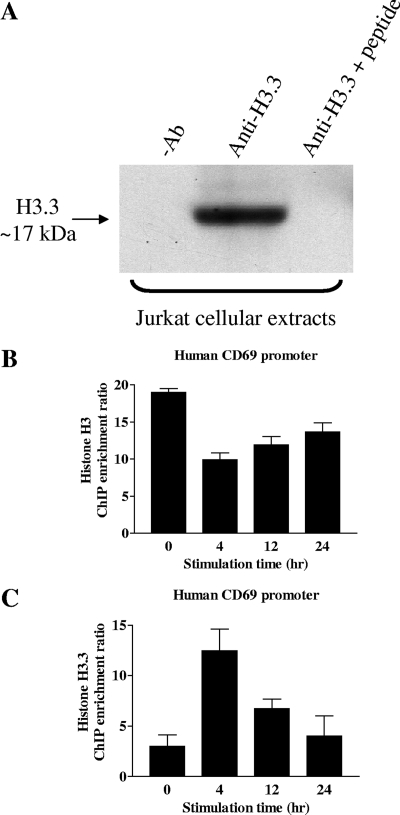
The lack of a commercially available H3.3-specific antibody has precluded ChIP studies of endogenous H3.3 distribution, instead relying on the transfection of epitope-tagged H3.3 into target cells (32, 52). Therefore, an antibody specific to histone H3.3 was raised. Both in vitro and in vivo data demonstrate that this antibody is highly specific to histone H3.3 and does not cross-react with the core histone H3. First, in vitro immunoprecipitation experiments with recombinant histone H3.3 and H3 proteins show that anti-histone H3.3 immunoprecipitated recombinant histone H3.3 (Fig. 3A [for a full scan of Fig. 3, see Fig. S3 in the supplemental material]) but failed to immunoprecipitate the H3 recombinant protein (Fig. 3B). Furthermore, preblocking of the anti-histone H3.3 antibody with H3.3-specific peptides successfully prevented the anti-histone H3.3 antibody from immunoprecipitating the recombinant histone H3.3 (Fig. 3A). Second, in vivo immunoprecipitation experiments with human embryonic kidney (HEK) cells transiently transfected with hemagglutinin (HA)-tagged histone H3.3 and H3 proteins show that anti-histone H3.3 specifically immunoprecipitates HA-tagged histone H3.3 (Fig. 4A [for a full scan of Fig. 4, see Fig. S1 in the supplemental material]) but not the HA-tagged histone H3 (Fig. 4B). In contrast, anti-histone H3 immunoprecipitated both HA-tagged H3.3 and H3, confirming that this H3 antibody cross-reacts with both H3 and H3.3. The commercial anti-histone H3 (ab1791; Abcam) antibody used in this study recognizes both histones H3 and H3.3 (Abcam, personal communication). These experiments were carried out with HEK cells because of the high transfection efficiency of these cells. Third, the anti-histone H3.3 antibody immunoprecipitated endogenous H3.3 from Jurkat cellular extracts and this interaction was inhibited by preblocking the anti-histone H3.3 antibody with H3.3-specific peptides (Fig. 5A [for a full scan of Fig. 5, see Fig. S2 in the supplemental material]). Importantly, the ChIP experiments with the anti-histone H3.3 antibody were carried out under immunoprecipitation conditions identical to those of these validation experiments. Fourth, ChIP assays show distinct H3 (Fig. 5B) and H3.3 (Fig. 5C) occupancy profiles on the promoter of the human CD69 gene. Additionally, the affinity-purified H3.3 antibody demonstrated specific binding to H3.3 as measured by enzyme-linked immunosorbent assay and fluorescence microscopy (see Fig. S4 in the supplemental material). These data also show how our H3.3-specific antibody is different from the highly specific histone H3.3 (phospho-S31) antibody (ab2889; Abcam), which recognizes histone H3.3 only when serine-31 is phosphorylated (19). In summary, the affinity-purified anti-H3.3 antibody generated in this study is specific for the histone variant H3.3 (recognizing both the phosphorylated and unphosphorylated S31 forms) and, importantly, does not cross-react with the core histone H3.
FIG. 3.
The affinity-purified anti-histone H3.3 antibody immunoprecipitates recombinant histone H3.3 but not recombinant histone H3. Recombinant H3 and H3.3 (25 μg/ml; New England BioLabs) were subjected to immunoprecipitation with 4 μg of either anti-histone H3 (ab1791; Abcam) or anti-histone H3.3 (generated in this investigation) antibody. As a negative control (−Ab), the immunoprecipitation was performed in the absence of either antibody. Alternatively, the anti-histone H3.3 antibody was preincubated with the H3.3-specific peptides that were synthesized by Auspep, Pty. Ltd., Australia. Each H3.3-specific peptide (CGGRKSAPSTGGVKK and CGGRKSAPS*TGGVKK) was made to a final concentration of 10 μM before incubation with the anti-histone H3.3 antibody for 0.5 h at room temperature with constant rotation. Subsequent immunoprecipitation with the anti-histone H3.3 antibody was performed as described above. Samples were then subjected to Western blotting with the anti-histone H3 antibody. Note that the anti-histone H3 antibody (ab1791; Abcam) raised against the C-terminal region of histone H3 is known to cross-react with both histones H3 and H3.3 because of the homology of this region in both proteins (Abcam, personal communication). The asterisk denotes the site of phosphorylation.
FIG. 4.
The anti-histone H3.3 antibody immunoprecipitates HA-tagged histone H3.3 but not HA-tagged histone H3. Immunoprecipitation with the anti-histone H3 antibody (ab1791; Abcam) or our anti-histone H3.3 antibody was carried out with sonicated lysates of HEK cells that had been transiently transfected with recombinant HA-tagged H3.3 or H3 (constructs kindly donated by Chunyuan Jin and Gary Felsenfeld). Cells (∼5 × 106) were transiently transfected with FuGENE HD at a ratio of 3:2 (microliters of FuGENE to micrograms of DNA) according to the manufacturer's instructions (Roche Diagnostics). At 72 h posttransfection, cells were collected for immunoprecipitation experiments. As a negative control (−Ab), the immunoprecipitation assay was performed in the absence of either antibody. Samples were then subjected to Western blotting with an anti-HA tag-specific polyclonal antibody (ab9110; Abcam).
FIG. 5.
The anti-histone H3.3 antibody reacts with endogenous histone H3.3 in Jurkat T cells. (A) Immunoprecipitation with our anti-histone H3.3 antibody was carried out with sonicated whole-cell extracts from NS Jurkat T cells. As a negative control (−Ab), immunoprecipitation was performed in the absence of the anti-histone H3.3 antibody. Alternatively, the anti-histone H3.3 antibody was preincubated with the H3.3-specific peptides (see Fig. 3 legend) prior to immunoprecipitation with the anti-histone H3.3 antibody. Samples were then subjected to Western blotting with the anti-histone H3 antibody (ab1791; Abcam). Note that the anti-histone H3 antibody (ab1791; Abcam) raised against the C-terminal region of histone H3 is known to cross-react with both histones H3 and H3.3 because of the homology of this region in both proteins (Abcam, personal communication). Anti-histone H3 and anti-histone H3.3 antibodies produce distinct ChIP profiles. In panels B and C, ChIP assays were performed with Jurkat T cells either left NS or stimulated with PI for the times indicated. Real-time SYBR green PCR analysis was performed with immunoprecipitated DNA recovered with the anti-histone H3 antibody (ab1791; Abcam) (B) or our anti-histone H3.3 antibody (C) and primer sets covering the CD69 proximal promoter (−0.15 kb). Data were calculated as the n-fold enrichment ratio of immunoprecipitated DNA relative to the no-antibody control and normalized against the total input DNA. The results represent the mean ± the standard error of 11 independent experiments in the case of the anti-histone H3 antibody and 3 independent experiments in the case of the anti-histone H3.3 antibody.
H3.3 and H2A.Z histone variant exchange during induction of the CD69 and heparanase genes.
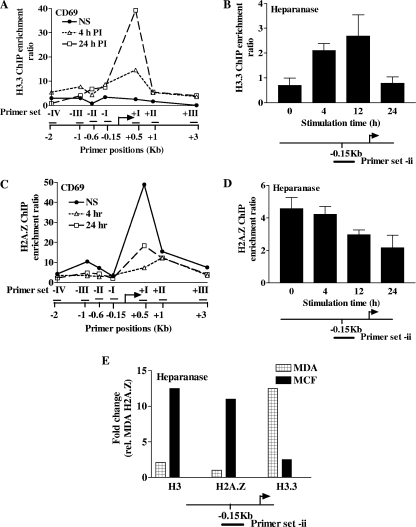
To test the possibility that H3 is replaced by H3.3 during the induction of the CD69 gene, we used our H3.3-specific antibody to measure endogenous H3.3 distribution by ChIP. When H3.3 levels were assessed with this antibody, relatively low levels of H3.3 were detected within the CD69 gene promoter and flanking regions in NS T cells. However, upon CD69 gene induction, extensive H3.3 deposition was observed within the CD69 gene promoter and flanking regions (Fig. 6A), with H3.3 predominantly enriched within regions found to be depleted of H3 (Fig. 2E). As with H3, the higher H3.3 density in the transcribed region is likely due to a higher nucleosome density. Thus, chromatin remodeling across the CD69 gene promoter is marked by a loss of H3 and a concomitant deposition of H3.3. Similar results were obtained with primary human CD4+ T cells (data not shown). Interestingly, H3.3 deposition was also observed on the heparanase gene promoter upon T-cell activation (Fig. 6B). Nevertheless, the failure to detect H3 loss on this promoter (Fig. 2C) suggests that H3.3 deposition is relatively modest on the heparanase gene promoter. Interestingly, and unlike the CD69 gene promoter, H3.3 levels dropped back to basal levels on the heparanase gene promoter at 24 h (Fig. 6B). However, our actinomycin C data (Fig. 1D) suggest that transcription of the heparanase gene may have returned to basal levels at this time point. Thus, H3.3 deposition on both promoters appears to correlate with active transcription.
FIG. 6.
H3.3 and H2A.Z changes upon transcriptional activation of CD69 and heparanase. (A to E) ChIP was conducted with Jurkat T cells, MDA cells, and MCF cells with an anti-H3.3 antibody (A, B, and E), an anti-H2A.Z antibody (C to E), or an anti-H3 antibody (E). H3.3 levels are shown across the CD69 gene region (A) and on the heparanase gene promoter (B) in Jurkat T cells left NS or stimulated with PI for the times indicated. Similar data are shown for H2A.Z and the CD69 and heparanase genes in panels C and D, respectively. Panel E depicts the H2A.Z, H3.3, and H3 occupancy in MDA and MCF cells. The names and positions of the primer sets are indicated beneath all of the graphs. Representative data from one of three independent experiments are shown.
The rapid histone H3 variant exchange observed on the CD69 gene promoter prompted us to examine whether other core histones are exchanged for variants during gene activation. In particular, we were interested in whether levels of the H2A histone variant H2A.Z changed upon gene induction based on the observation that H2A.Z is often located on the promoters of inducible genes (3, 26, 39, 53). Since we were unsuccessful in performing ChIP experiments with commercially available H2A antibodies (data not shown), it was not possible to correlate changes in H2A levels with those in H2A.Z. Nonetheless, H2A.Z was detected by ChIP at relatively high levels on the CD69 gene within NS T cells, particularly upstream of the promoter and immediately downstream of the TSS. T-cell stimulation provoked the specific loss of the H2A.Z histone variant within these two highly enriched regions (Fig. 6C). A similar loss of H2A.Z occurred on the heparanase gene promoter upon PI treatment, although again these changes were not as dramatic as those observed over the CD69 gene and occurred with more delayed kinetics (Fig. 6D). Thus, transcriptional activation and chromatin remodeling are accompanied by H2A.Z depletion.
As the shifts in H3.3 and H2A.Z were relatively modest on the heparanase gene promoter, we wished to further characterize their association with heparanase transcriptional activity. To achieve this, we took advantage of the distinct heparanase expression patterns found within two human mammary adenocarcinoma cell lines, namely, MDA-231, which is highly metastatic and exhibits high levels of heparanase transcription and protein expression (49), and MCF-7, a nonmetastatic mammary adenocarcinoma that has low levels of heparanase transcription (data not shown). When the levels of H3, H2A.Z, and H3.3 were examined on the heparanase gene promoter within these cell lines by ChIP, high H3.3 levels and low H2A.Z and H3 levels were apparent in the MDA-231 cells while the opposite was seen in the MCF-7 cells. Thus, despite the relatively minor shifts observed upon heparanase induction within T cells, substantial differences in histone variant deposition do parallel heparanase gene expression levels when comparing a nonmetastatic with a metastatic cell line.
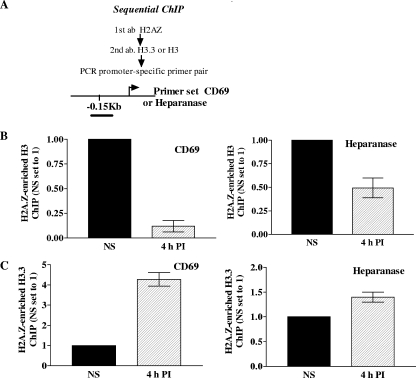
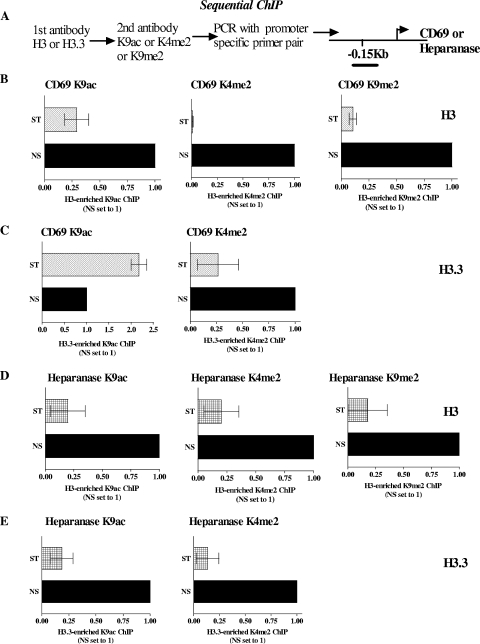
The profiles examined thus far suggest that the core histone H3 is replaced by its variant histone H3.3 during gene induction, with a likely interacting partner for both histones being H2A.Z. To test the possibility that H3.3 coexists with H2A.Z on transcriptionally active inducible genes, sequential ChIP analyses were carried out across the proximal promoter regions of the CD69 and heparanase genes in NS and stimulated Jurkat T cells. The 4-h stimulatory time point was chosen because this is when H2A.Z deposition remained relatively unchanged, whereas H3.3 levels were significantly enhanced on these two promoters compared to those in the basal state. In sequential ChIP assays, formaldehyde-cross-linked chromatin that has been immunoprecipitated with one antibody is further immunoprecipitated with a second antibody to examine whether co-occupancy of two factors on a single stretch of DNA occurs (15). As expected, sequential ChIP assays involving an anti-H2A.Z antibody followed by an anti-H3.3 antibody (Fig. 7C; see Table S3 in the supplemental material) revealed that following gene induction there was an increase in the association of H2A.Z with H3.3 within the proximal promoter regions of both the CD69 and heparanase genes. However, as shown in Fig. 7B, sequential ChIP assays involving an anti-H2A.Z antibody, followed by an anti-H3 antibody, consistently displayed a diminution in H2A.Z/H3 co-occupancy on the CD69 gene promoter following T-cell activation. The same trend was observed, but to a lesser extent, on the heparanase gene promoter (Fig. 7B; see Table S3 in the supplemental material). We expect there to be no net change in the association of H2A.Z with H3 and H3.3 between resting and activated samples because our data showed no change in histone H4, indicating no net nucleosome loss. Consequently, this result is somewhat unexpected and remains to be explained. Nevertheless, these results demonstrate that H2A.Z may predominantly coexist with the core histone H3 in the basal state on the CD69 and heparanase promoter regions while predominantly partnering with H3.3 during gene induction. The fact that H2A.Z/H3.3-containing nucleosomes are less stable than their H2A.Z/H3-containing counterparts (21) may have implications for the increased chromatin accessibility of the promoters of the CD69 and, to a lesser extent, heparanase genes seen following gene activation.
FIG. 7.
H3.3 and H2A.Z co-occupancy on the promoters of the heparanase and CD69 genes during transcriptional activation. (A) Schematic diagram indicating the experimental protocol used in the sequential ChIP assays. Note that the antibody order was reversed for all experiments and similar results were obtained (see Fig. S5 in the supplemental material). The primers used were located in the promoter regions of the CD69 and heparanase genes, as indicated. (B) CD69 and heparanase promoter DNA enrichment seen when chromatin was immunoprecipitated first with anti-H2A.Z antibody and then anti-H3 antibody. CD69 and heparanase data are shown for Jurkat T cells left NS or stimulated for 4 h with PI. Graphs show the n-fold changes in signal relative to the NS sample. Data pooled from two independent experiments are shown, with the mean ± the standard error plotted. (C) Sequential ChIP assays were performed as in panel B, except that anti-H3.3 antibody was used for secondary immunoprecipitation in place of anti-H3 antibody. Data are depicted as in panel B. Data pooled from two independent experiments are shown, with the mean ± the standard error plotted.
H3.3 deposited onto the promoters of the CD69 and heparanase genes carry PTMs associated with active transcription.
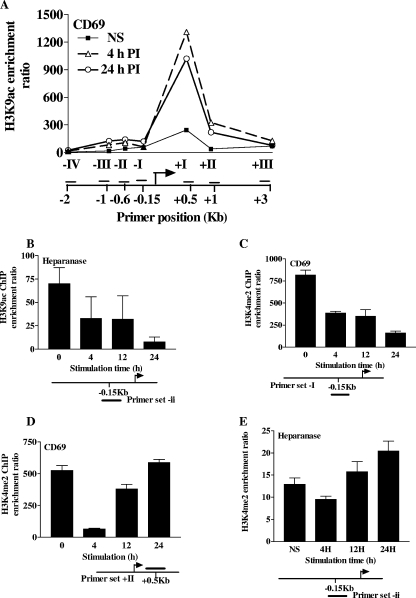
While the histone variant composition of nucleosomes can directly influence chromatin structure, histone variants can also be enriched in PTMs that modulate both chromatin structure and transcription. For example, H3.3 is often enriched in PTMs associated with gene activation, such as acetylated H3K9 (K9Ac), while being depleted of modifications associated with gene repression, such as dimethylated H3K9 (K9me2) (18, 31). In fact, free histone H3.3 is often “premodified” with certain PTMs (30) and thus may carry them directly into chromatin upon nucleosome incorporation. To examine whether a similar phenomenon occurs upon H3.3 deposition on the promoters of the CD69 and heparanase genes, the patterns of histone PTMs present on these gene promoters were determined by ChIP. CD69 gene activation was accompanied by an increase in the activating PTM, K9Ac, across the CD69 gene promoter and transcribed regions (Fig. 8A). Importantly, the appearance of K9Ac was strongly associated with regions of H3.3 deposition (Fig. 8A and 6A), suggesting that H3.3 may be directly carrying K9Ac onto the CD69 gene. Surprisingly, heparanase gene induction was actually accompanied by K9 deacetylation (Fig. 8B), suggesting the recruitment of deacetylase activity onto this gene's promoter upon transcriptional induction. The recruitment of the activating PTM K4me2 was also examined to correlate its association with H3.3. Interestingly, K4me2 was depleted from the CD69 gene promoter upon activation (Fig. 8C). Furthermore, K4me2 was also initially depleted from the transcribed region before reappearing at later time points (Fig. 8D). The heparanase gene promoter also displayed a small depletion of K4me2 at early time points; however, in contrast to the CD69 gene promoter, K4me2 was enriched at the heparanase gene promoter at later time points (Fig. 8E). Thus, H3.3 deposition on the promoters of both the CD69 and heparanase genes is initially paralleled by a loss of K4me2, with the pattern of this modification at later time points varying depending on the location or gene promoter examined. Collectively, these data suggest that H3.3 is likely to carry K9Ac, but not K4me2, onto chromatin regions.
FIG. 8.
Deposition patterns of PTMs associated with active transcription during CD69 and heparanase gene induction. (A and B) Acetylated H3K9 (H3K9Ac) ChIP was performed with Jurkat T cells left NS or stimulated with PI for the times indicated. H3K9Ac levels were assessed across the CD69 gene region (A) and on the heparanase gene promoter (B) with the indicated primer sets. Representative data from one of three independent experiments are shown. (C to E) Dimethylated H3K4 (H3K4me2) ChIP was performed with Jurkat T cells left NS or stimulated with PI for the times indicated. H3K4me2 levels were determined on the CD69 gene promoter (C), within the CD69 transcribed region (D), and on the heparanase gene promoter (E) with the indicated primer sets. Data pooled from two independent experiments are shown, with the mean ± the standard error plotted.
To further elucidate the association of PTMs with H3 and H3.3, sequential ChIP assays were performed as described in Fig. 7. As expected, sequential ChIP assays involving an anti-H3.3 antibody, followed by an anti-K9ac antibody, revealed that following gene induction there was an increase in H3.3 bearing K9Ac (Fig. 9C) within the proximal CD69 gene promoter. Surprisingly, while the little K9Ac present on the CD69 gene promoter in NS cells was predominantly associated with H3, stimulation led to a depletion of K9-acetylated H3 (Fig. 9B). Based on our primary ChIP data (Fig. 8A), we would have expected a net increase in the association of H3 with K9ac following gene induction, given that the H3 antibody recognizes both histones H3 and H3.3. Nevertheless, these findings further lend support to the idea that H3.3 carries K9Ac onto the CD69 gene promoter following T-cell activation. Consistent with the depletion of K4me2 observed on the CD69 gene promoter upon activation, K4me2 diminished on both H3 and H3.3 upon activation (Fig. 9B and C). This suggests that the little H3.3 present in NS cells possesses higher levels of K4me2 than freshly incorporated H3.3, further supporting the idea that unincorporated H3.3 has relatively low levels of K4me2. Interestingly, while a decrease was observed in H3 associated with the repressive mark K9me2 upon CD69 gene activation (Fig. 9B), no K9me2 could be detected on CD69 gene promoter-bound H3.3 in either NS or activated T cells (Fig. 9 legend). Thus, H3.3 is selectively depleted of K9me2 on the CD69 gene promoter.
FIG. 9.
Co-occupancy of H3 and H3.3 with histone PTMs. (A) Schematic diagram indicating the experimental protocol used in the sequential ChIP assays. Note that the antibody order was reversed for all experiments and similar results were obtained (see Fig. S6 in the supplemental material). The primers used were located in the promoter region of the CD69 and heparanase genes, as indicated. (B and C) Enrichment of DNA of the CD69 gene promoter seen when chromatin was immunoprecipitated first with either anti-H3 or anti-H3.3 antibody and then the indicated anti-PTM antibody. Data are shown for H3 (B) and H3.3 (C) in Jurkat T cells left NS or stimulated for 4 h with PI (ST). Graphs show the n-fold change in signal relative to the NS sample. (D and E) Sequential ChIP assays were performed as in panel B, except that the heparanase gene promoter was analyzed. Data are depicted as in panels B and C, with H3 data shown in panel D and H3.3 data depicted in panel E. Data pooled from two independent experiments are shown, with the mean ± the standard error plotted. Note that the H3.3-enriched K9me2 ChIPs for CD69 and heparanase have not been plotted, as no enrichments were detected above the background levels in these sequential ChIP experiments.
Similar trends were observed for K4me2 and K9me2 on the heparanase gene promoter. Consistent with the loss of K4me2 on the heparanase gene promoter observed at 4 h poststimulation, a depletion of K4me2-modified H3 and H3.3 was observed at this time point (Fig. 9D and E). Furthermore, similar to the observations on the CD69 gene promoter, H3.3 was completely devoid of K9me2 on the heparanase gene promoter (Fig. 9 legend) and K9me2-associated H3 diminished upon activation (Fig. 9D). Heparanase gene induction was accompanied by a loss of K9Ac-associated H3 (Fig. 9D). However, in contrast to the CD69 gene promoter, freshly incorporated H3.3 was depleted of K9Ac (Fig. 9E), suggesting active deacetylation of deposited H3.3.
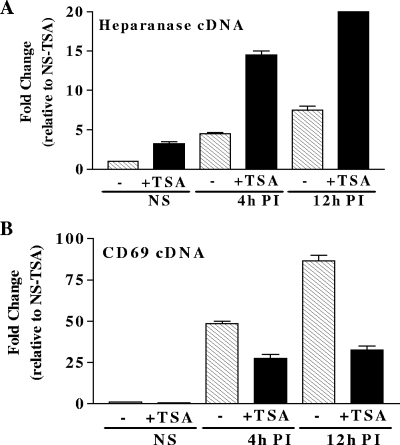
The heparanase gene data thus far suggested that active deacetylation may be regulating heparanase gene promoter activity. To confirm that deacetylation was having a functional impact upon heparanase gene transcription, the impact of the specific histone deacetylase inhibitor TSA upon heparanase gene induction was examined. Consistent with the observed promoter deacetylation, TSA treatment was able to substantially augment heparanase transcript induction (Fig. 10A). This suggests that deacetylase activity recruited to the heparanase gene promoter restrains the magnitude of heparanase mRNA induction upon T-cell activation. In agreement with the absence of any observable deacetylase activity on the CD69 gene promoter, TSA treatment had no impact upon CD69 mRNA induction kinetics; if anything, TSA treatment impaired CD69 transcript production, perhaps due to TSA-associated cytotoxicity (Fig. 10B). Thus, while H3.3 may potentiate transcription by carrying K9Ac onto gene regulatory regions, concomitant induction of deacetylase activity has the potential to negate this effect. However, it should be noted that our results could be a consequence of other effects of TSA. For example, it is well established that TSA hyperacetylates at numerous residues and also has the ability to directly influence the acetylation of chromatin components such as transcription factors.
FIG. 10.
Differential effects of TSA treatment on CD69 and heparanase transcript kinetics. (A and B) Heparanase and CD69 mRNA kinetics were measured by TaqMan real-time PCR with resting (NS) Jurkat T cells and cells stimulated with PI for the times indicated. mRNA levels are expressed as n-fold changes relative to NS samples. For TSA treatment, cells were preincubated with TSA for 5 h and then stimulated for the times indicated. Transcript induction kinetics are shown for heparanase (A) and CD69 (B). Data shown are the mean ± the standard error of three replicate experiments.
DISCUSSION
In this study, we examined the changes in histone variant distribution that accompany gene induction and attempted to determine the relationships between the exchange of H3 histones, changes in chromatin accessibility, and alterations in PTM patterns with transcriptional activation. Two genes with distinct expression patterns were examined, the CD69 gene, a poised gene that is rapidly induced following T-cell activation, and the heparanase gene, a gene that exhibits basal expression and more modest inducibility. The major finding of this study is that the transcriptional activation of these genes is accompanied by chromatin accessibility that occurs without nucleosome loss on inducible gene promoters. Instead, histone variant exchange takes place in the promoter regions with a loss of histone H3 and a gain of H3.3. In addition, a concomitant decline of H2A.Z levels accompanied gene induction. These events were most dramatic in the 5′-transcribed region of the CD69 gene. Our data lend support to a model whereby histone H2A.Z preferentially coexists with histone H3 on inducible gene promoters in the basal state, whereas it is more likely to be associated with histone variant H3.3 during the establishment of the transcriptionally active phase. Furthermore, H3.3 deposition appeared to be associated with enrichment of the histone PTM K9Ac and concomitant depletion of K4me2 and K9me2.
The CD69 gene represents a classic poised gene with docked Pol II on its promoter in the basal state, and this gene displays rapid induction following activation (16, 28, 45). As with other inducible immune genes, such as the IL-2 gene, the CD69 gene exhibits low levels of chromatin accessibility prior to activation (41). In contrast, the heparanase gene is expressed in resting T cells and can be further induced upon T-cell activation. Consistent with the basal expression of heparanase, the heparanase gene promoter exhibits considerable nuclease accessibility and Pol II occupancy in NS T cells. However, both Pol II occupancy and chromatin accessibility do increase further upon T-cell activation. The transcript induction kinetics and accessibility patterns of heparanase are similar to those of the granulocyte-macrophage colony-stimulating factor gene in T cells (20). Thus, the CD69 and heparanase genes represent different classes of genes in terms of transcript induction kinetics and chromatin accessibility patterns.
Nevertheless, many of the chromatin changes that accompany the transcriptional induction of both of these genes are similar. While chromatin remodeling accompanies gene induction in both cases, these remodeling events are not accompanied by nucleosome loss. This is somewhat surprising, as other studies have demonstrated that nucleosome loss accompanies gene induction in T cells (7, 9). This difference may be because these studies demonstrating nucleosome loss were performed with murine cells and hence may reflect differences between the mouse and human system. Alternatively, it may be due to gene-specific differences in chromatin remodeling mechanisms. Nevertheless, this finding demonstrates that nucleosome loss is not required for chromatin remodeling to occur. Indeed, a recent genome-wide study with human CD4+ T cells demonstrated that transcriptional activation of other genes in T cells occurs without nucleosome loss, although this study did not investigate any changes in chromatin accessibility (45).
The increase in the chromatin accessibility of both genes in activated T cells was accompanied not by nucleosome loss but rather by exchange of H3 histones. With a new H3.3-specific antibody, which we generated, it was found that H3.3 levels are absent or low on both genes in the basal state. However, the induction of both genes was accompanied by H3.3 deposition. Moreover, H3.3 was not restricted to the proximal promoter region, as extensive H3.3 deposition was observed both upstream of the promoter and in the transcribed region of the CD69 gene just downstream of the transcription initiation site. Similar distributions in H3.3 have been observed in genome-wide studies utilizing epitope-tagged H3.3 (32, 52). Our data thus further support the ideas that H3.3 deposition occurs upon transcription and that H3.3 is associated with transcriptionally active regions of the genome.
Chromatin remodeling and induction of both the heparanase and CD69 genes were also associated with the decline of H2A.Z, particularly in the 5′-transcribed region of the CD69 gene. In the resting state, H2A.Z enrichment near the TSS and its subsequent removal upon induction have been observed in other studies (3, 17, 26, 27, 39, 53, 54). Our finding that H2A.Z-containing chromatin is present just downstream of the transcriptional start site on the poised inducible CD69 gene is analogous to the results of a recent genome-wide analysis of the location of H2A.Z-containing nucleosomes in Drosophila (30a). This latter study raised the possibility that this H2A.Z-containing nucleosome is involved in blocking the movement of the poised RNA Pol II complex. Our results support this notion because in response to an activation signal, the biochemical makeup of this downstream nucleosome within the CD69 gene is altered with the loss of H2A.Z. Hence, one could speculate that H2A.Z-containing nucleosomes on poised genes may block elongation by preventing the movement of RNA Pol II in resting T cells (13, 37).
The histone variant composition of a nucleosome was recently demonstrated to influence nucleosome stability. Specifically, H3.3-containing nucleosomes were shown to be less stable than their H3 counterparts, with H3.3/H2A.Z-containing nucleosomes being particularly sensitive to disruption (21). Given that histone exchange accompanies chromatin accessibility across inducible genes in this study, it is therefore tempting to speculate that changes in nucleosome composition may impact upon chromatin accessibility. Thus, the extensive H3.3 deposition we observed upon gene induction will diminish nucleosome stability across the promoter and transcribed regions. Importantly, the H2A.Z present at the time of remodeling (4 h postactivation) on both gene promoters appeared to be predominantly associated with H3.3 rather than H3. Thus, the highly unstable H3.3/H2A.Z nucleosome also increases in frequency upon gene induction. However, while an increase in H3.3/H2A- and H3.3/H2A.Z-containing nucleosomes may facilitate nucleosome remodeling, this does not necessarily mean that H3.3 deposition is sufficient to trigger an increase in chromatin accessibility. While H3.3-containing nucleosomes may be more susceptible to remodeling, the recruitment of the appropriate ATP-dependent remodeling activities to the gene promoter is still likely to be required. Furthermore, it could be reasoned that H3.3 deposition in different gene regions plays different roles. While deposition of H3.3 onto promoters may facilitate an increase in chromatin accessibility, one could speculate that the enrichment of H3.3 in transcribed regions facilitates Pol II elongation.
Another way in which H3.3 deposition may promote chromatin accessibility is through carrying the K9Ac PTM onto its target regions. In fact, we demonstrated that the appearance of K9Ac over the CD69 gene strongly correlated with regions of H3.3 enrichment. This finding is in line with studies showing that K9Ac is strongly associated with H3.3 (18, 30, 31) and represents the first in vivo association between H3.3 and K9Ac at the gene-specific level. As ATP-dependent chromatin remodeling enzymes, such as Brg1, contain an acetylated lysine binding bromodomain (34, 46), an increase in K9Ac may promote remodeler recruitment and subsequent accessibility increases. The observation that unincorporated H3.3 is enriched in K9Ac (30) raises the possibility that H3.3 is “premodified” and carries K9Ac onto the regions of the CD69 gene upon deposition. Nevertheless, we cannot rule out the possibility that H3.3 is preferentially acetylated subsequent to incorporation into the CD69 region. Surprisingly, induction of the heparanase gene is actually accompanied by promoter deacetylation. While it could be argued that the more modest H3 loss and H3.3 deposition on the heparanase gene promoter make it more susceptible to deacetylase activity, our sequential ChIP assays indicate that the H3.3 deposited onto the heparanase gene promoter is also deacetylated. Therefore, opposing enzymatic activities can counterbalance the PTMs deposited on a promoter in association with H3.3. This also means that, despite a strong association (18, 30, 31), H3.3 is not invariably marked with K9Ac. Deacetylation on the heparanase gene promoter clearly dampens transcriptional induction. As the transcript of the heparanase gene is highly stable, a large increase in transcriptional activity is not required for transcript accumulation to occur. Thus, heparanase gene promoter deacetylation upon T-cell activation may represent a biological mechanism for restraining gene induction.
We also found that H3.3 appeared depleted of the PTMs K9me2 and K4me2. The failure to detect any K9me2-modified H3.3 on either gene promoter at any time point was unexpected given previous data showing the presence of this PTM on H3.3 in human cell lines (30). The observation that K9me2 is present on unincorporated H3.3 (30) suggests that active demethylation may be occurring upon deposition onto these gene promoters. However, unincorporated H3.3 is depleted of K4me2 (30), meaning that the loss of K4me2 that parallels initial H3.3 deposition onto the promoters of the CD69 and heparanase genes is not unexpected. The patterns of K4me2 at later time points do not conform to any clear pattern, indicating that whether or not H3.3 K4 dimethylation occurs subsequent to deposition is highly dependent on the gene region examined. Overall, we can conclude that PTM modulation by H3.3 deposition is only one of many factors governing the final pattern of histone PTMs adopted by a gene region. This is consistent with the observation that the PTM pattern associated with H3.3 changes markedly upon H3.3 deposition into chromatin (30) and agrees with a recently proposed model in which chromatin environment controls histone variant PTM patterns (29).
Importantly, histone variant exchange at gene promoters may have an unanticipated relevance to cancer biology. Heparanase is a degradative enzyme thought to play a key role in cancer cell invasion and metastasis (35). The observation that a shift in histone variant usage across the heparanase gene promoter accompanies the acquisition of a metastatic phenotype in mammary carcinoma cells suggests that dysregulation of histone variant distribution may play a role in oncogenesis. This observation, coupled with our other data, further underscores the fundamental role that histone variants are likely to play in gene induction.
Supplementary Material
Acknowledgments
S.R. is supported by an Australian National Health and Medical Research Council (NHMRC) New Investigator Project grant, and C.R.P. is supported by an NHMRC program grant.
We thank Amy Weinmann for helpful advice and suggestions during the compilation of the manuscript.
Footnotes
Published ahead of print on 21 January 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99(Suppl. 4)16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 91191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 111587-1598. [DOI] [PubMed] [Google Scholar]
- 7.Brettingham-Moore, K. H., O. R. Sprod, X. Chen, P. Oakford, M. F. Shannon, and A. F. Holloway. 2008. Determinants of a transcriptionally competent environment at the GM-CSF promoter. Nucleic Acids Res. 362639-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellanos Mdel, C., S. Lopez-Giral, M. Lopez-Cabrera, and M. O. de Landazuri. 2002. Multiple cis-acting elements regulate the expression of the early T cell activation antigen CD69. Eur. J. Immunol. 323108-3117. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., J. Wang, D. Woltring, S. Gerondakis, and M. F. Shannon. 2005. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell. Biol. 253209-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Mestre, A. M., L. M. Khachigian, F. S. Santiago, M. A. Staykova, and M. D. Hulett. 2003. Regulation of inducible heparanase gene transcription in activated T cells by early growth response 1. J. Biol. Chem. 27850377-50385. [DOI] [PubMed] [Google Scholar]
- 11.de Mestre, A. M., M. A. Staykova, J. R. Hornby, D. O. Willenborg, and M. D. Hulett. 2007. Expression of the heparan sulfate-degrading enzyme heparanase is induced in infiltrating CD4+ T cells in experimental autoimmune encephalomyelitis and regulated at the level of transcription by early growth response gene 1. J. Leukoc Biol. 821289-1300. [DOI] [PubMed] [Google Scholar]
- 12.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9172-176. [DOI] [PubMed] [Google Scholar]
- 13.Fan, J. Y., D. Rangasamy, K. Luger, and D. J. Tremethick. 2004. H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16655-661. [DOI] [PubMed] [Google Scholar]
- 14.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14165-173. [DOI] [PubMed] [Google Scholar]
- 15.Geisberg, J. V., and K. Struhl. 2005. Analysis of protein co-occupancy by quantitative sequential chromatin immunoprecipitation. Curr. Protoc. Mol. Biol. 200521.8.1-21.8.7. [DOI] [PubMed] [Google Scholar]
- 16.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette, F. Robert, and L. Gaudreau. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hake, S. B., B. A. Garcia, E. M. Duncan, M. Kauer, G. Dellaire, J. Shabanowitz, D. P. Bazett-Jones, C. D. Allis, and D. F. Hunt. 2006. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281559-568. [DOI] [PubMed] [Google Scholar]
- 19.Hake, S. B., B. A. Garcia, M. Kauer, S. P. Baker, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2005. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl. Acad. Sci. USA 1026344-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holloway, A. F., S. Rao, X. Chen, and M. F. Shannon. 2003. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor κB proteins. J. Exp. Med. 197413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, C., and G. Felsenfeld. 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 211519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamakaka, R. T., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19295-310. [DOI] [PubMed] [Google Scholar]
- 23.Kouskouti, A., and I. Talianidis. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 26.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 27.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 10218385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorincz, M. C., and D. Schubeler. 2007. RNA polymerase II: just stopping by. Cell 13016-18. [DOI] [PubMed] [Google Scholar]
- 29.Loyola, A., and G. Almouzni. 2007. Marking histone H3 variants: how, when and why? Trends Biochem. Sci. 32425-433. [DOI] [PubMed] [Google Scholar]
- 30.Loyola, A., T. Bonaldi, D. Roche, A. Imhof, and G. Almouzni. 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24309-316. [DOI] [PubMed] [Google Scholar]
- 30a.Mavrich, T. N., C. Jiang, I. P. Ioshikhes, X. Li, B. J. Venters, S. J. Zantan, L. P. Tomsho, J. Qi, R. L. Glasser, S. C. Schuster, D. S. Gilmour, I. Albert, and B. F. Pugh. 2008. Nucleosome organization in the Drosophila genome. Nature 453358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 1011525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 371090-1097. [DOI] [PubMed] [Google Scholar]
- 33.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303343-348. [DOI] [PubMed] [Google Scholar]
- 34.Mujtaba, S., L. Zeng, and M. M. Zhou. 2007. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 265521-5527. [DOI] [PubMed] [Google Scholar]
- 35.Parish, C. R., C. Freeman, and M. D. Hulett. 2001. Heparanase: a key enzyme involved in cell invasion. Biochim. Biophys. Acta 1471M99-M108. [DOI] [PubMed] [Google Scholar]
- 36.Park, Y. J., J. V. Chodaparambil, Y. Bao, S. J. McBryant, and K. Luger. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 2801817-1825. [DOI] [PubMed] [Google Scholar]
- 37.Park, Y. J., P. N. Dyer, D. J. Tremethick, and K. Luger. 2004. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 27924274-24282. [DOI] [PubMed] [Google Scholar]
- 38.Pokholok, D. K., J. Zeitlinger, N. M. Hannett, D. B. Reynolds, and R. A. Young. 2006. Activated signal transduction kinases frequently occupy target genes. Science 313533-536. [DOI] [PubMed] [Google Scholar]
- 39.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao, S., S. Gerondakis, D. Woltring, and M. F. Shannon. 2003. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J. Immunol. 1703724-3731. [DOI] [PubMed] [Google Scholar]
- 41.Rao, S., E. Procko, and M. F. Shannon. 2001. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 1674494-4503. [DOI] [PubMed] [Google Scholar]
- 42.Ruthenburg, A. J., H. Li, D. J. Patel, and C. D. Allis. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 44.Sarma, K., and D. Reinberg. 2005. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 6139-149. [DOI] [PubMed] [Google Scholar]
- 45.Schones, D. E., K. Cui, S. Cuddapah, T. Y. Roh, A. Barski, Z. Wang, G. Wei, and K. Zhao. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen, W., C. Xu, W. Huang, J. Zhang, J. E. Carlson, X. Tu, J. Wu, and Y. Shi. 2007. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 462100-2110. [DOI] [PubMed] [Google Scholar]
- 47.Shiow, L. R., D. B. Rosen, N. Brdickova, Y. Xu, J. An, L. L. Lanier, J. G. Cyster, and M. Matloubian. 2006. CD69 acts downstream of interferon-α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440540-544. [DOI] [PubMed] [Google Scholar]
- 48.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311844-847. [DOI] [PubMed] [Google Scholar]
- 49.Shteper, P. J., E. Zcharia, Y. Ashhab, T. Peretz, I. Vlodavsky, and D. Ben-Yehuda. 2003. Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene 227737-7749. [DOI] [PubMed] [Google Scholar]
- 50.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 51.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 11651-61. [DOI] [PubMed] [Google Scholar]
- 52.Wirbelauer, C., O. Bell, and D. Schubeler. 2005. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 191761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 202009-2017. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.