Abstract
The nuclear receptor steroidogenic factor 1 (SF-1) is essential for adrenal development and steroidogenesis. The atypical orphan nuclear receptor Dax-1 binds to SF-1 and represses SF-1 target genes. Paradoxically, however, loss-of-function mutations of Dax-1 also cause adrenal hypoplasia, suggesting that Dax-1 may function as an SF-1 coactivator under some circumstances. Indeed, we found that Dax-1 can function as a dosage-dependent SF-1 coactivator. Both SF-1 and Dax-1 bind to steroid receptor RNA activator (SRA), a coactivator that functions as an RNA. The coactivator TIF2 also associates with Dax-1 and synergistically coactivates SF-1 target gene transcription. A naturally occurring Dax-1 mutation inhibits this transactivation, and the mutant Dax-1-TIF2 complex mislocalizes in living cells. Coactivation by Dax-1 is abolished by SRA knockdown. The expression of the steroidogenic gene products steroidogenic acute regulatory protein (StAR) and melanocortin 2 receptor is reduced in adrenal Y1 cells following the knockdown of endogenous SRA. Similarly, the knockdown of endogenous Dax-1 downregulates the expression of the steroidogenic gene products CYP11A1 and StAR in both H295R adrenal and MA-10 Leydig cells. These findings reveal novel functions of SRA and Dax-1 in steroidogenesis and adrenal biology.
Nuclear hormone receptors (NRs) mediate the transcriptional responses to a wide variety of physiological stimuli and thus function as important regulators of development, metabolism, and reproduction. By binding to specific DNA sequences, NRs serve as platforms for the recruitment of various coregulatory factors that effect gene regulation. Transcriptional coactivators often function either through their enzymatic activities (as in the examples of acetyl and methyl transferases) or through the formation of productive complexes with the basal transcription machinery. In contrast, corepressors often have enzymatic activities opposite those of coactivators, such as those of deactylases and demethylases. Thus, coregulators, by functioning as coactivators or corepressors of NR activity, play pivotal roles in mediating hormone action (reviewed in references 13 and 42).
The best-characterized coactivators are the p160 family proteins SRC-1 (NCoA1), TIF2 (GRIP1/NCoA2/SRC-2), and AIB1 (pCIP/ACTR/NCoA3/SRC-3) (3, 51, 62, 66). These coactivators harbor autonomous activation domains and NR interaction domains (28, 65). Recently, the steroid receptor RNA activator (SRA) has been characterized as the only known coregulator that can function as an RNA (36). SRA was shown to coactivate glucocorticoid receptors without direct physical interaction, as part of a ribonucleoprotein complex with p160 coactivators. In addition, SRA coactivates retinoic acid receptors, and this function is dependent upon SRA pseudouridinylation (78). SRA also functions as a thyroid hormone receptor (TR) coactivator by direct physical interaction (72). The TR SRA binding domain is a 41-amino-acid region located between the second zinc finger and the ligand binding domain. Although SRA-protein interactions play important roles in NR activity, the molecular mechanisms and biological functions of these interactions remain largely unknown.
Steroidogenic factor 1 (SF-1/NR5A1/Ad4BP) belongs to the NR5A subfamily of orphan NRs that bind DNA with high affinity as monomers. SF-1 plays critical roles in the regulation of sex determination, adrenal and gonadal development, reproductive function, and steroidogenesis (16, 40, 41, 52, 63). SF-1 interacts with several transcriptional coactivators, such as SRC-1 (11, 25), TIF2 (17), and p300 (9), resulting in the induction of a large number of genes including those for the adrenocorticotropin hormone (ACTH) receptor/melanocortin 2 receptor (Mc2R) and steroidogenic acute regulatory protein (StAR) (58, 70). We and others have shown that sumoylation inhibits and phosphorylation activates SF-1 (73), while recent structural analyses have revealed that phospholipids can serve as activating SF-1 ligands (33, 37).
Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on X chromosome gene 1; NR0B1) is an unusual member of the nuclear receptor superfamily. Although the carboxyl-terminal region of Dax-1 is homologous to the ligand binding domains of other NRs, Dax-1 lacks the typical zinc finger DNA binding domain (75). Instead, the amino terminus of Dax-1 consists of three and a half repeats of a 65- to 67-amino-acid motif that has been proposed to serve as a DNA binding domain. Dax-1 is expressed primarily in the developing urogenital ridge, ovary, testis, and adrenal, hypothalamus, and anterior pituitary glands, and it colocalizes with SF-1 (23). SF-1 activates the expression of Dax-1 and physically interacts with it (48). Dax-1 classically has been thought to function as a repressor of SF-1 target genes (24, 34, 76), probably by interaction with corepressors such as NCoR (10) and Alien (2). In addition, Dax-1 has been reported to inhibit ligand-dependent transactivation by other NRs, including estrogen receptors α and β (77), androgen receptor (20), and progesterone receptor (1). Although the significance of this characteristic is unclear, Dax-1 is an RNA binding protein (35), and unlike many NRs, it localizes to both the nucleus and the cytoplasm (20, 35).
Naturally occurring loss-of-function mutations of the Dax-1 gene NR0B1 cause the human disorder X-linked adrenal hypoplasia congenita (AHC), which is also associated with hypogonadotropic hypogonadism (47, 75). Given that Dax-1 is considered to be a repressor of SF-1 function, it seems paradoxical that SF-1 loss-of-function mutations also result in adrenal hypoplasia (40, 56). These observations suggest that Dax-1 may be capable of enhancing rather than repressing SF-1 function and that Dax-1 may be able to interact with coactivators as well as corepressors, depending upon the cellular and promoter context.
In this study, we report that SF-1 is an RNA binding protein and that both SF-1 and Dax-1 bind to the noncoding RNA SRA. Surprisingly, we have demonstrated that Dax-1 also physically interacts with the p160 coactivator TIF2 in vitro and in living cells. SF-1 recruits Dax-1 to the promoter of the ACTH receptor (Mc2R) gene, and SF-1, Dax-1 and TIF2 synergistically induce Mc2R promoter activity. The knockdown of endogenous Dax-1 downregulates the expression of Mc2R, CYP11A1, and StAR. Furthermore, the knockdown of endogenous SRA in JEG-3 cells reveals that transactivation by Dax-1 is SRA dependent, and SRA knockdown in Y1 adrenocortical cells reveals that SRA plays an important role in the expression of StAR and Mc2R.
MATERIALS AND METHODS
Cell culture, transfection, and reporter gene assays.
JEG-3 cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum with penicillin-streptomycin. COS-1 cells were maintained in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin. Y1 mouse adrenocortical cells were maintained in DMEM supplemented with 7.5% horse serum, 2.5% fetal bovine serum, and penicillin-streptomycin. The human adrenocortical cell line H295R was grown in DMEM-F-12 medium containing 15 mM HEPES and 2.5 mM l-glutamine supplemented with 1% insulin-transferrin-selenium-X (Gibco; catalog no. 51500), 10% fetal bovine serum, and penicillin-streptomycin. The mouse Leydig tumor cell line MA-10 was generously provided by Mario Ascoli (University of Iowa, Iowa City) and was maintained in DMEM-F-12 medium supplemented with 15% horse serum and 10 μg/ml gentamicin, pH 7.7. MA-10 cell culture dishes were precoated with 0.1% gelatin. All cells were incubated at 37°C under a humidified atmosphere of 5% CO2. Transient transfections were carried out using either Lipofectamine Plus (Invitrogen) or FuGene 6 (Roche) reagent. For all luciferase reporter gene assays, JEG-3 cell samples were divided among the wells of 24-well plates, and the cells were cotransfected with pCDNA3 SF-1, pCDNA3 Dax-1, pMc2R-luciferase, and coactivator vector pSCT-SRA, pSG5-TIF2, or pCDNA1 SRC-1, along with pRL-TK Renilla luciferase as a control. The effects of Dax-1 on TR function were tested in cotransfections with pCDM-TRα1 and the T3-responsive luciferase reporter construct 8DR4-Luc (32, 55). Cell lysates were harvested 48 h posttransfection for analyses of firefly and Renilla luciferases with the Promega dual luciferase reporter assay system. For immunoprecipitation experiments, cell samples were divided in 100-mm dishes and the cells were transfected with the plasmids of interest and harvested 48 h later.
Plasmids.
The vector pGEX-KG (14) was used to express wild-type and mutant mouse SF-1 proteins as glutathione S-transferase (GST) fusions in Escherichia coli. To maximize the recovery of full-length protein, the constructs were further tagged with six histidines at their C termini. For simplicity, we hereinafter refer to these proteins as GST-SF-1, etc. GST-SF-1 deletion mutants were constructed by inverse PCR and are depicted in Fig. 1D. GST-tagged wild-type mouse Dax-1 and its mutant versions were constructed in a similar way and are depicted in Fig. 1F. Dax-1 mutation and deletion constructs in the mammalian expression vector pCDNA3 were also generated by inverse PCR, and the products included LBD (the Dax-1 ligand binding domain, amino acids 207 to 472); N3R (the N-terminal three-and-a-half repeat region, amino acids 1 to 206); ΔAF2 (Dax-1 with the activation function 2 [AF2] domain, amino acids 463 to 468, deleted); and the mutant proteins R269P, ΔV271, and N442I, which are homologs of human Dax-1 proteins with naturally occurring AHC mutations. The human SRA expression vector pSCT-SRA was kindly provided by R. Lanz (Baylor College of Medicine, Houston, TX), and pSG5-TIF2 was provided by P. Chambon (IGBMC, Strasbourg, France). pCMV-3Tag-4A (Stratagene) was used to express Dax-1 with three C-terminal Myc tags (from a construct designated pDax-1-Myc) in mammalian cells. The bimolecular fluorescence complementation (BiFC) cloning vectors pFlag-VN173 and pHA-VC155, encoding N-terminal residues 1 to 172 (VN173) and C-terminal residues 155 to 238 (VC155) of the optimized yellow fluorescent protein (YFP) Venus, were described previously (57). For BiFC analysis, cDNAs encoding mouse Dax-1 and its mutant forms were cloned into pHA-VC155, while the cDNA encoding human TIF2 was cloned into pFlag-VN173. The Dax-1 constructs used in the BiFC study encode Dax-1 (wild type), N3R, LBD, and mutant R269P. All constructs were verified by sequencing. The mouse Mc2R-luciferase reporter plasmid containing 1 kb of the mouse Mc2R promoter was a kind gift of F. Beuschlein (University of Freiburg, Freiburg, Germany) (80).
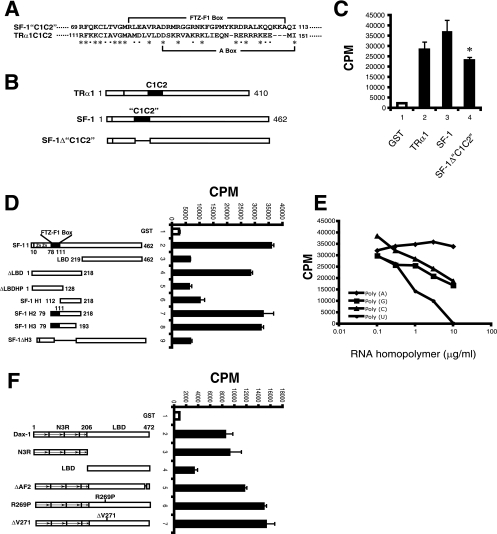
FIG. 1.
SF-1 and Dax-1 bind SRA in vitro. (A) Sequence alignment of mouse SF-1 and the TRα1 RNA binding domain, designated C1C2, showing the region of homology. Asterisks and filled squares indicate identical and similar amino acids, respectively. (B) Schematic presentation of full-length TRα1, SF-1, and SF-1 “C1C2” deletion mutant (SF-1Δ“C1C2”) proteins used for in vitro RNA binding assays. (C) SRA binding to SF-1. Purified full-length GST, GST-TRα1, GST-SF-1, or deletion mutant GST-SF-1Δ“C1C2” bound to glutathione agarose beads was incubated with a 32P-labeled SRA (RNA) probe. GST and TRα1 served as negative and positive controls, respectively. After extensive washing, the counts for the bound proteins were determined. The assay was performed in triplicate, and the results are representative of those from three experiments. Error bars indicate standard deviations. All proteins contained GST at their N termini, although this tag is not indicated in the x-axis labels for simplicity. (*, P = 0.014 for bar 4 versus bar 3 by Student's t test.) (D) Identification of the SRA binding domain of SF-1. Left panel, schematic presentation of full-length SF-1 and truncation and deletion mutants used for in vitro RNA binding assays. Right panel, in vitro [32P]SRA binding was performed as described in the legend to panel C. (E) RNA binding specificity of SF-1. SRA binding to SF-1 was performed as described in the legend to panel C, except that graded doses of nonradiolabeled RNA homopolymer poly(A), poly(G), poly(C), or poly(U) were added as competitors. (F) Identification of the SRA binding domain of Dax-1. Left panel, schematic presentation of full-length Dax-1 and truncated and AHC mutant proteins used for in vitro RNA binding assays. Right panel, in vitro [32P]SRA binding was performed as described in the legend to panel C.
GST fusion protein purification, in vitro RNA binding, and GST pull down.
The purification of GST-six-histidine fusion proteins from E. coli strain BL21 by sequential cobalt and glutathione agarose column chromatography, as well as in vitro RNA binding, was performed essentially as described previously (72). The 32P-labeled RNA probe was made by in vitro transcription using T7 RNA polymerase and [α-32P]UTP from pSCT-SRA that had been linearized by digestion with PvuII. To make 35S-TIF2 protein, in vitro translation was performed using the TNT T7 coupled reticulocyte lysate system kit (Promega). The GST pull-down reactions were performed using 2 μg of purified proteins, 10 μl of glutathione agarose beads, 5 μl of in vitro-translated 35S-TIF2, and 85 μl of 1× binding buffer containing 20 mM Tris (pH 7.9), 170 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.05% Nonidet P-40, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 4 mg/ml bovine serum albumin (BSA). After incubation for 1 h at 4°C with gentle rocking, the beads were washed five times with 500 μl of 1× binding buffer without BSA. The resulting beads were boiled for 5 min in sodium dodecyl sulfate (SDS) loading buffer containing 2-mercaptoethanol, and the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Afterwards, the gels were fixed, dried, and analyzed with a Bio-Rad phosphorimager.
BiFC and confocal microscopy.
BiFC was performed essentially as described previously (57). COS-1 cells were transfected with Venus N terminus (VN) and Venus C terminus (VC) fusion constructs alone or in combination and incubated for 24 h at 37°C. To examine the localization of the wild-type and mutant Dax-1-VC or TIF2-VN fusions, the transfected cells were fixed for 1 min in fresh 1:1 methanol-acetone, blocked for 1 h at room temperature in blocking buffer (2% goat serum, 1% ovalbumin, and 1% BSA), probed with antihemagglutinin (anti-HA) mouse immunoglobulin G (IgG)-Alexa Fluor 488 (Invitrogen) at a 1:1,000 dilution in blocking buffer or anti-Flag M2 (Sigma), and then incubated with Alexa Fluor red 594-goat anti-mouse IgG (Invitrogen) as described previously (71). The cell nuclei were stained with Hoechst 33342. Microscopy was carried out using a Zeiss LSM510 laser-scanning confocal microscope at a magnification of ×63.
Cell lysis, immunoprecipitations, and immunoblotting.
Cells were lysed in M-PER (Pierce) or immunoprecipitation lysis buffer (40 mM HEPES, 120 nM sodium chloride, 10 mM sodium pyrophosphate, 10 mM sodium glycerophosphate, 1 mM EDTA, 50 mM sodium fluoride, 0.5 mM sodium orthovanadate, 1% Triton X-100) containing protease inhibitor cocktail (Roche). Immunoprecipitations were performed by incubating cell lysates with antibody-precoated agarose beads in lysis buffer overnight at 4°C. After extensive washing, the coimmunoprecipitated proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Immunoblotting was performed using the following antibodies: anti-Myc (catalog no. RMYC-45A from Immunology Consultants Laboratory, Inc.; 1:2,000), anti-TIF2 (catalog no. 610985 from BD Transduction Laboratories; 1:250), anti-StAR (a generous gift from Douglas Stocco, Texas Tech University; 1:5,000), anti-Mc2R (catalog no. sc-6876 from Santa Cruz; 1:1,000), anti-Dax-1 (catalog no. sc-13064 from Santa Cruz [for Dax-1 expression in JEG-3 and H295R cells] or catalog no. PP-H7431 from R&D Systems [for Dax-1 expression in MA-10 cells]; 1:1,000), and anti-CYP11A1 (catalog no. sc-18043 from Santa Cruz; 1:500). For the coimmunoprecipitation of SRA, Y1 cells were transiently transfected with pDax-1-Myc or empty pCMV-3Tag-4A vector. The subsequent immunoprecipitation procedures were modified from those described previously (29, 50). Briefly, cells were washed with phosphate-buffered saline and treated with 0.1% formaldehyde for 10 min at room temperature. After the addition of 0.25 M glycine for 5 min, cells were harvested and lysed by exposure to radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton, 0.5% Na deoxycholate, 0.1% SDS containing a protease inhibitor cocktail tablet [Roche], and 40 U of RNase inhibitor) and sonication. After centrifugation, the lysate supernatants were adjusted to the same concentrations and precleared with protein A beads (Sigma) for 30 min at 4°C. For the preparation of antibody-coated beads, 10 μg of anti-Myc antibody was added to 50 μl of protein A beads and blocked with 10 μg of tRNA and 5% BSA overnight at 4°C. The beads were then washed twice with RIPA buffer. For immunoprecipitation, the precleared supernatants were applied to the Myc antibody-precoated protein A beads. After incubation with gentle rotation for 3 h at 4°C, the beads were washed five times with high-stringency RIPA buffer containing 1 M NaCl, 1 mM EDTA, and 1 M urea. The reversal of cross-links was done by adding 100 μl of reversal buffer (100 mM Tris [pH 6.8], 5 mM EDTA, 1% SDS, and 10 mM dithiothreitol) and heating for 45 min at 70°C. The immunoprecipitated beads were treated with 10 U of DNase I (Ambion) for 30 min at 37°C, and then 20 mM EDTA was added to stop the enzyme activity. RNA was extracted with Trizol, precipitated with isopropanol containing glycogen, and used for first-strand cDNA synthesis with SuperScript III (Invitrogen). Real-time PCR was done using 2 μl of the resulting cDNA and the following primers that amplify mouse SRA: forward, 5′-GGCTGGAGGGAAGTTGTCAATAC, and reverse, 5′-CCACTGGTGATGTAAAAGTTCTTG. Data were normalized to the signal obtained using primers that amplify β-actin RNA. Y1 cell immunoprecipitations were also performed using the SF-1-specific antibody (catalog no. 07-618 from Upstate) and normal rabbit IgG as a control.
Silencing by siRNA and shRNA.
JEG-3 human choriocarcinoma cells seeded onto 24-well plates at a confluence of 60 to 70% were transfected with ON-TARGETplus SMARTpool small interfering RNA (siRNA) duplexes for human SRA1 (Dharmacon; catalog no. L-027192-00-0010) by using Dharmacon Duo transfection reagent. Cells were also transfected with the ON-TARGETplus siCONTROL nontargeting siRNA (Dharmacon; catalog no. D-001810-01-05) as a control. The efficiency of siRNA knockdown of endogenous SRA was confirmed by real-time reverse transcription-PCR (RT-PCR) using specific human SRA primers 5′-TTGGAACAGGCATTGGAAGAC and 5′-ACAACTTTCCTCCAGCCCAC. To stably knock down endogenous SRA in Y1 mouse adrenocortical cells, we used a short hairpin RNA (shRNA) construct targeting mouse SRA1 (forward, 5′-GATCCCGACCACTGCAAGATTATTTGTCTTCCTGTCAACAAGTGATCTTGCAGTGGTCTTTTTA, and reverse, 5′-AGCTTAAAAAGACCACTGCAAGATCACTTGTTGACAGGAAGACAAATAATCTTGC AGTGGTCGG; the underlined letters correspond to nucleotides complementary to the SRA sequence). The shRNA was expressed from the retroviral vector pSuperior.retro.puro (OligoEngine) that utilizes the H1 RNA polymerase III promoter. A scrambled-sequence shRNA (forward, 5′-GATCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTT, and reverse, 5′-AGCTTAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGAC ACGTTCGGAGA) in the same vector served as a control. The retroviruses were grown in and harvested from phoenix cells (kindly provided by G. Bommer, University of Michigan) and then used to infect Y1 cells. The Y1 cells were selected with 4 μg/ml of puromycin. The SRA-silencing effects of shRNAs were confirmed by real-time RT-PCR using the mouse SRA-specific primers. To knock down endogenous Dax-1 in H295R and MA-10 cells, pGIPZ lentiviral shRNAmir vectors directing the expression of shRNAs specific to human Dax-1 (Open Biosystems; catalog no. RHS4430-98894425) or mouse Dax-1 (catalog no. RMM4431-99337199) and a nontargeting shRNA control (catalog no. RHS 4346) were obtained from the University of Michigan shRNA library core facility. The human and mouse Dax-1 shRNA targeting sequences (sense) were AGCACAGTCAGCATGGATGATA and AGCTAACAAGCTAATTTCATAA, respectively. 293T cells were cotransfected with the shRNA plasmids and the packaging plasmids (psPAX2 and pMD2.G) by using the calcium phosphate method to produce the virus. Viral supernatants were collected, filtered, and used to infect H295R or MA-10 cells. The infections were repeated three times at intervals of 8 to 12 h. The infected H295R or MA-10 cell samples were then split, and cells were selected with puromycin at 2.5 μg/ml.
ChIP and real-time PCR.
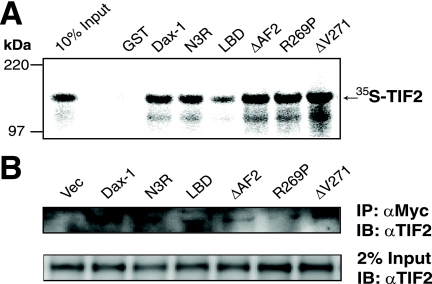
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (70). Y1 cells were transfected with vectors expressing Myc-tagged wild-type Dax-1, deletion constructs (N3R or LBD), or the AHC mutant R269P. Forty-eight hours later, the cells were lysed and ChIP was performed using the Myc antibody. The immunoprecipitated DNA fragments were quantified by real-time PCR using primers that cover the SF-1 response regions within the mouse Mc2R and StAR proximal promoters. The primers used for PCR were as follows: Mc2R promoter forward primer, 5′-GCTATGGACAACGTGGTCAGAA, and reverse primer, 5′-CAGGAAAGGCCGGAACATATAC, and StAR promoter forward primer, 5′AATGACTGATGACTTTTTTATCTCAAGTG, and reverse primer, 5′-AAGTGCGCTGCCTTAAATGC. Exonic primers for the Mc2R gene (positions +21689 to +21791; forward, 5′-GTGCCATGACACTAACCAT, and reverse, 5′-CAGTAAGGGTTATTTGGGC) and the StAR gene (positions +2441 to +2545; forward, 5′-GGACGAAGTGCTAAGTAAG, and reverse, 5′-CGGTCCACAAGTTCTTCAT) served as negative controls for the ChIP studies.
For quantitative real-time PCR analysis of mRNA transcript abundance, total RNA from Y1 cells was isolated by using Trizol reagent (Invitrogen). Four micrograms of total RNA was reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen), and real-time PCR was performed using Power SYBR green (Applied Biosystems) and a 7500 real-time PCR system (Applied Biosystems). All real-time PCRs were done with the following conditions: 10 min at 95°C and then 40 cycles of 15 s at 95°C, 30 s at 54°C, and 1 min at 72°C. All data were analyzed for relative gene expression by using the 2−ΔΔCT method (39) and were normalized to the data for β-actin. Primer sequences for each gene were as follows: mouse StAR gene forward primer, 5′-GTGGTGTCATCAGAGCTGAACACGGCCCCAC, and reverse primer, 5′-CTGCGATAGGACCTGGTTGATGATTGTC; mouse Mc2R gene forward primer, 5′-GTGCCATGACACTAACCATC, and reverse primer, 5′-CAGTAAGGGTTATTTGGGCAG; mouse Dax-1 gene forward primer, 5′-ATTGACACCAAAGAGTATGCC, and reverse primer, 5′-GTTCTCCACTGAAGACCCTC; mouse CYP11A1 gene forward primer, 5′-CGCATCAAGCAGCAAAATTC, and reverse primer, 5′-ATGCGCTCCCCAAATATAAC; mouse CYP17A1 gene forward primer, 5′-ACTAGCTCTGTGCTGAACTG, and reverse primer, 5′-GTTCGACTGAAGCCTACATAC; mouse β-actin gene forward primer, 5′-TATTGGCAACGAGCGGTTCC, and reverse primer, 5′-GGCATAGAGGTCTTTACGGATGTC; human Dax-1 gene forward primer, 5′-CCAAATGCTGGAGTCTGAAC, and reverse primer, 5′-TGAATGTACTTCACGCACTG; human CYP11A1 gene forward primer, 5′-AGCTAGAGATGACCATCTTCC, and reverse primer, 5′-GGCATCAGAATGAGGTTGAATG; human StAR gene forward primer, 5′-AAGACCAAACTTACGTGGC, and reverse primer, 5′-GTGGTTGGCAAAATCCACC; human Mc2R gene forward primer, 5′-AGCCTGTCTGTGATTGCTG, and reverse primer, 5′-AGATGACCGTAAGCACCACC; human CYP17A1 gene forward primer, 5′-TGTGGACAAGGGCACAGAAG, and reverse primer, 5′-GGATTCAAGAAACGCTCAGGC; and human β-actin gene forward primer, 5′-TCACCATTGGCAATGAGCG, and reverse primer, 5′-TGGAGTTGAAGGTAGTTTCGTG.
Isolation of RNA from mouse livers, adrenal glands, and testes.
RNA from the livers, testes, and adrenal glands of 18-week-old wild-type C57BL/6 male mice (n = 4) was isolated using Trizol reagent.
RESULTS
SF-1 and Dax-1 bind to SRA.
TRα1 binds to SRA via a novel RNA binding domain, designated C1C2 (72), distal from the zinc fingers. A sequence alignment revealed modest similarity between TRα1C1C2 and SF-1 amino acids 69 to 113 (“C1C2”), which lie in a region distal from the SF-1 zinc fingers and include the so-called FTZ-F1 box (Fig. 1A and B). Due to this similarity, we tested whether SF-1 also binds to SRA. Indeed, we found that GST-SF-1 binds [32P]SRA in vitro at least as well as TRα1 does (Fig. 1C). However, the deletion of the putative “C1C2” region of SF-1 (generating SF-1Δ“C1C2”) reduced SRA binding by only 30%, indicating that this domain does not fully account for the SF-1 interaction with SRA. We therefore used a series of SF-1 deletion and truncation mutants to map the SF-1 RNA binding domain. As shown in Fig. 1D, the SF-1 ligand binding domain (amino acids 219 to 462) is not involved in SRA binding because this domain by itself has almost no binding activity (Fig. 1D, bar 3) and the deletion of this domain from SF-1 (generating ΔLBD) does not impair SRA binding (Fig. 1D, bar 4). A further C-terminal truncation of the ΔLBD construct so that it extends only through the DNA binding domain and FTZ-F1 box (amino acids 1 to 128, in construct ΔLBDHP) eliminated SRA binding (Fig. 1D, bar 5), suggesting that the RNA binding domain may lie between the FTZ-F1 box and the ligand binding domain (amino acids 129 to 218). However, this possibility was not confirmed, as amino acids 112 to 218, designated SF-1 H1, showed very limited SRA binding (Fig. 1D, bar 6). Extending this construct in the N-terminal direction to include the FTZ-F1 box (producing the SF-1 H2 construct, amino acids 79 to 218) fully restored SRA binding (Fig. 1D, bar 7), and further truncation at the C terminus to produce the SF-1 H3 construct, amino acids 79 to 193, also preserved SRA binding (Fig. 1D, bar 8). In addition, the deletion of the H3 domain (generating SF-1ΔH3) eliminated SRA binding (Fig. 1D, bar 9). Thus, the SF-1 SRA binding domain encompasses residues 79 to 193 and includes the FTZ-F1 box. This domain overlaps with but is larger than the homologous 41-amino-acid RNA binding domain of TRα1.
RNA binding proteins usually bind different RNA homopolymers with different affinities (61). We therefore tested the ability of nonradiolabeled poly(A), poly(C), poly(G), or poly(U) to compete with radiolabeled SRA for binding to SF-1 (Fig. 1E). The data indicate that poly(U) competed most strongly and that poly(A) showed essentially no competition, suggesting that SF-1 may have a preference for U-rich sequences and no binding to A-rich sequences within a target RNA.
Dax-1 is also an RNA binding protein (35), and given that its RNA targets are not well defined, we investigated whether Dax-1 might bind to SRA. Indeed, GST-Dax-1 binds [32P]SRA (Fig. 1F, bar 2), and the N-terminal three-and-a-half repeat region (designated N3R) (Fig. 1F, bar 3) is largely responsible for its RNA binding since the ligand binding domain has virtually no RNA binding activity (Fig. 1F, bar 4). Recent data show that Dax-1 N3R contains LXXLL motifs that bind to SF-1 (59). To test if the LXXLL motifs also play a role in binding to SRA, in vitro RNA binding assays were performed with GST-Dax-1 proteins harboring deletions of these motifs. The specific deletion of any of the three N3R LXXLL motifs or the deletion of all three (without altering the other N3R amino acids) did not diminish the interaction of Dax-1 with SRA, indicating that the LXXLL sequences are not required for the Dax-1-SRA interaction (data not shown). In addition, the deletion of the Dax-1 AF2 domain amino acids 463 to 468 (generating Dax-1 ΔAF2) and the naturally occurring AHC mutations R269P and ΔV271 did not impair SRA binding (Fig. 1F, bars 5 to 7), indicating that these mutations within the Dax-1 ligand binding domain do not have secondary effects on the interaction of N3R with SRA.
SF-1 coactivation properties of SRA, Dax-1, and Dax-1 mutant proteins.
SRA was initially identified as an RNA coactivator for steroid receptor transactivation and was shown to function in a p160 family coactivator complex to enhance target gene transcription (36). These observations and the findings that SF-1 and Dax-1 bind to SRA in vitro prompted us to investigate the functional relevance of SF-1-SRA-Dax-1 interactions in steroidogenic gene transcription. To this end, JEG-3 cells, which do not express endogenous SF-1, were transfected with the SF-1-targeted promoter of the ACTH receptor (Mc2R) linked to a luciferase gene (the Mc2R-luc construct). Transfection was performed either with or without SF-1, SRA, and increasing doses of wild-type or mutant Dax-1. As shown in Fig. 2A, bars 1 to 8, neither SRA nor Dax-1 affected luciferase expression in the absence of cotransfection with SF-1. SF-1 induced luciferase activity ∼2-fold (Fig. 2A, bar 9 versus bar 1), an effect that was marginally inhibited by low-dose Dax-1 (from 10 ng of the Dax-1 expression plasmid) (Fig. 2A, bar 10 versus bar 9). Surprisingly, high-dose Dax-1 (from 100 ng of plasmid) significantly increased luciferase activity above the level seen with SF-1 alone (Fig. 2A, bar 12 versus bar 9). The deletion mutant N3R and the AHC mutants R269P and N422I displayed severely reduced coactivation (Fig. 2A, bar 12 versus bars 25, 31, and 43), although Dax-1 LBD and the AHC mutant ΔV271 retained modest coactivation (Fig. 2A, bar 12 versus bars 19 and 37). The cotransfection of cells with SRA and wild-type Dax-1 vectors further stimulated luciferase expression (Fig. 2A, bar 16 versus bar 12). (The effect of SRA overexpression is not as strong as the effect of the knockdown of endogenous SRA, depicted subsequently in Fig. 5 and 8.) However, SRA in combination with the Dax-1 deletion mutant N3R or AHC mutants R269P and N441I was severely defective in reporter gene transactivation (Fig. 2A, bar 16 versus bars 28, 34, and 46), whereas SRA in combination with Dax-1 LBD or ΔV271 achieved modest gene coactivation, albeit less than that achieved with wild-type Dax-1 (Fig. 2A, bar 16 versus bars 22 and 40).
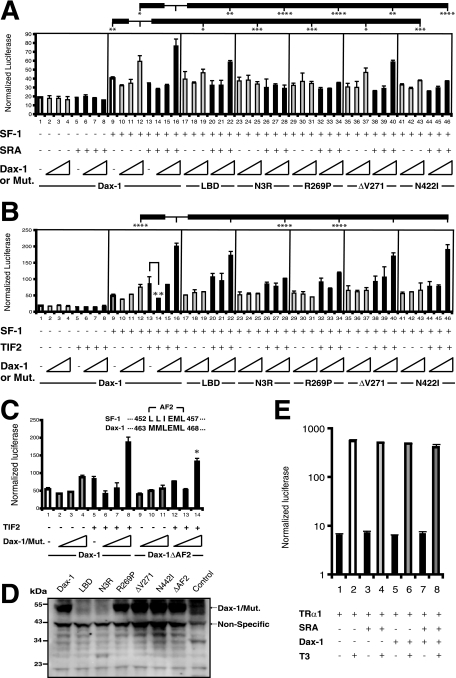
FIG. 2.
Dax-1, the noncoding SRA, and the p160 coactivator TIF2 coactivate SF-1-dependent transcription. (A) JEG-3 cells were transfected with Mc2R-luc and expression vectors for SF-1, SRA, Dax-1, and its deletion or AHC mutant forms (N3R, LBD, R269P, ΔV271, and N422I) in different combinations. Increasing doses (10, 30, and 100 ng) of wild-type or mutant (mut.) Dax-1 plasmids were employed, while cells were transfected with either no SF-1 or constant doses (3 ng) of the SF-1 plasmid, together with constant doses of SRA (30 ng) and the Mc2R-luc reporter plasmid (200 ng). Cells were cotransfected with 10 ng of pRL-TK Renilla luciferase vector as an internal control. Cell lysates were harvested 48 h after transfection. The y axis represents arbitrary firefly luciferase units normalized to Renilla luciferase values. Experiments were performed with triplicate samples and were repeated three times. Error bars indicate standard deviations. The results of statistical analyses by analysis of variance (ANOVA) followed by Scheffe's test, comparing bar 12 with bars 9, 19, 25, 31, 37, and 43 and bar 16 with bars 12, 22, 28, 34, 40, and 46, are shown in the figure. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001). +, present; −, absent. (B) Analyses similar to those described in the legend to panel A, with the SRA plasmid replaced by a TIF2 expression vector (30 ng), were performed. Experiments were done with triplicate samples and were repeated three times. Error bars indicate standard deviations. The results of statistical analyses by ANOVA followed by Scheffe's test, comparing bar 16 with bars 12, 22, 28, 34, and 40 and bar 13 with bars 14, 20, 26, 32, 38, and 44, are shown in the figure. (**, P < 0.01, and ****, P < 0.0001; for all other comparisons, P > 0.05). (C) Role of the Dax-1 AF2 domain in the coactivation of SF-1. The sequence of the mouse SF-1 AF2 hexamer was aligned with that of the putative Dax-1 AF2. Reporter gene activities were analyzed as described in the legend to panel B, except that Dax-1 ΔAF2 was compared with the full-length protein. Experiments were performed with triplicate samples and were repeated three times. Error bars indicate standard deviations. (*, P = 0.002 for bar 14 versus bar 8 by Student's t test.) (D) Expression levels of Dax-1 and its mutant versions in transfected JEG-3 cells. JEG-3 cells were transfected with vectors expressing wild-type Dax-1 or the indicated deletion or AHC mutant. Nontransfected cells served as a negative control. Whole-cell lysates were subjected to anti-Dax-1 immunoblotting. (E) Effect of Dax-1 on the T3-dependent transcriptional activity of TRα1. JEG-3 cells were transfected with 10 ng of pCDM-TRα1 or empty pCDM vector and 30 ng of SRA, 200 ng of the T3-responsive luciferase plasmid 8DR4-Luc, 100 ng of wild-type Dax-1 vector, and 10 ng of pRL-TK Renilla luciferase vector as an internal control. The transfected cells were treated with or without 100 nM T3 for 24 h before being harvested for luciferase assays. The y axis is plotted on a log scale. Error bars indicate standard deviations.
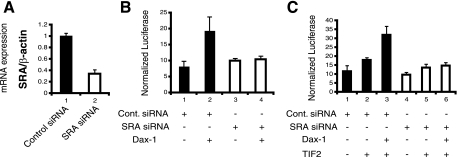
FIG. 5.
The knockdown of endogenous SRA impairs the ability of Dax-1 to function as an SF-1 coactivator. (A) siRNA knockdown of endogenous SRA. JEG-3 cells were transfected with either a nontargeting control siRNA or siRNA directed against SRA. Forty-eight hours after transfection, RNAs were isolated, reverse transcribed, and analyzed by real-time PCR. The relative expression of SRA was normalized to the expression of β-actin and compared to that of the control (set at 1). The experiments were performed in triplicate and repeated three times. Error bars indicate standard deviations. (B) SRA knockdown blocks Dax-1 coactivation of SF-1. JEG-3 cells were cotransfected with either control (cont.) or SRA siRNA, 3 ng of an SF-1 expression plasmid, 200 ng of Mc2R-luc, 10 ng of pRL-TK Renilla luciferase, and 100 ng of a Dax-1 expression plasmid or empty vector. Cells were lysed 48 h later, and luciferase assays were performed. The experiments were performed in triplicate and repeated three times. Error bars indicate standard deviations. +, present; −, absent. (C) SRA knockdown blocks Dax-1-TIF2 coactivation of SF-1. In an analysis similar to that described in the legend to panel B, cells were cotransfected with a TIF2 expression vector (30 ng).
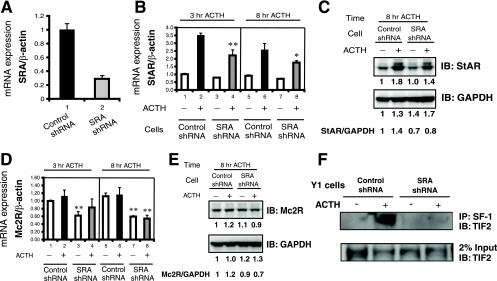
FIG. 8.
The knockdown of endogenous SRA in Y1 cells interferes with the expression of steroidogenic genes and impairs the interaction of SF-1 and TIF2. (A) Knockdown of endogenous SRA in Y1 cells. Y1 cells were infected with a retrovirus expressing an shRNA directed against mouse SRA or a scrambled-sequence shRNA as a negative control. The knockdown of endogenous SRA was assessed by real-time RT-PCR using β-actin as a normalization control. The level of expression of SRA in the control shRNA cells was set at 1. (B) SRA knockdown decreases the ACTH induction of StAR mRNA. shRNA control Y1 cells and shRNA SRA knockdown Y1 cells were treated with (+) or without (−) 10 nM ACTH for 3 and 8 h, and then RNA was isolated and analyzed by real-time RT-PCR for StAR expression, normalized to β-actin expression. Experiments were performed in triplicate and repeated three times. Error bars indicate standard deviations. (**, P = 0.005 for bar 4 versus bar 2, and *, P = 0.034 for bar 8 versus bar 6 by Student's t test.) (C) SRA knockdown decreases the ACTH induction of StAR protein. After 8 h with or without ACTH treatment, extracts from control shRNA or SRA shRNA cells were subjected to immunoblotting (IB) with anti-StAR and then were stripped and reprobed with anti-GAPDH as a loading control. The quantification of each band was performed by using a Bio-Rad Fluor-S MAX MultiImager. For both StAR and GAPDH, the signal of each band was compared to that for control cells without ACTH treatment (for which the level of expression was set at 1). The values are shown below the immunoblots, and the StAR/GAPDH ratios are shown at the bottom. (D) SRA knockdown decreases the expression of Mc2R mRNA. A real-time RT-PCR analysis similar to that described in the legend to panel B was performed for Mc2R rather than StAR (**, P < 0.01 for bar 8 versus bar 6, bar 3 versus bar 1, and bar 7 versus bar 5 [the P value for bar 4 versus bar 2 was not significant] by Student's t test). (E) SRA knockdown decreases the expression of Mc2R protein. Experiments were similar to those described in the legend to panel C but included immunoblotting for Mc2R rather than StAR. (F) SRA knockdown impairs the formation of complexes between SF-1 and TIF2. (Upper panel) Control or SRA knockdown shRNA cells were treated with or without ACTH for 3 h, and then immunoprecipitations (IP) were performed using an anti-SF-1 antibody, followed by immunoblotting using an anti-TIF2 antibody. The lower panel shows anti-TIF2 immunoblotting of 2% of the input for each sample.
Since SRA and SF-1 both are known to form complexes with p160 family coactivators such as TIF2, we also tested whether the coexpression of Dax-1 with TIF2 would have additive or synergistic effects on the SF-1 induction of Mc2R-luc. Neither TIF2 nor Dax-1 influenced Mc2R-luc expression in the absence of SF-1 (Fig. 2B, bars 1 to 8). The inhibition of SF-1 transactivation was again seen with low-dose Dax-1, such that luciferase expression in the presence of TIF2 plus 10 ng of the Dax-1 expression vector was only 50% of that seen with TIF2 alone (Fig. 2B, bar 14 versus bar 13). Conversely, the coexpression of high-dose Dax-1 and TIF2 synergistically enhanced Mc2R-luc expression up to 2.2-fold compared to that in the presence of Dax-1 alone (Fig. 2B, bar 16 versus bar 12). Similar synergistic effects of Dax-1 and SRC-1 were also observed (data not shown). We next studied the same Dax-1 mutants used for the analysis presented in Fig. 2A. Dax-1 LBD was essentially as active as wild-type Dax-1 in coactivation (Fig. 2B, bar 22 versus bar 16), N3R and AHC mutant R269P displayed significantly reduced coactivation (Fig. 2B, bars 28 and 34 versus bar 16), and ΔV271 and N442I behaved similarly to wild-type Dax-1 (Fig. 2B, bars 40 and 46 versus bar 16). Thus, the overall effects of the Dax-1 mutants were qualitatively similar in terms of coactivation with SRA and TIF2, except for Dax-1 N422I, which showed no synergy with SRA but was fully active in conjunction with TIF2.
We also obtained synergistic effects of Dax-1 and TIF2 on the SF-1 induction of StAR-luciferase. Relative to an average baseline luciferase expression level in the presence of SF-1 of 100 ± 5, expression with Dax-1 was 105 ± 3, that with TIF2 was 201 ± 16, and that with Dax-1 plus TIF2 was 281 ± 24.
Although Dax-1 is an atypical orphan nuclear receptor, its putative ligand binding domain does contain a putative AF2 hexamer (residues 463 to 468) homologous to the AF2 domains of conventional NRs such as SF-1 (Fig. 2C). For some NRs such as SF-1 and TRs, the AF2 domain is required for transcriptional activation, but for others such as the androgen receptor, it plays a relatively minor role. To examine the importance of the Dax-1 AF2 domain in the coactivation of SF-1, we deleted it to create Dax-1 ΔAF2. In our standard cotransfection paradigm with SF-1, TIF2, and Mc2R-luc, Dax-1 ΔAF2 retained about 75% of the activity of wild-type Dax-1 (Fig. 2C, bar 14 versus bar 8), indicating that the AF2 domain plays only a minor role in the Dax-1-TIF2 coactivation of SF-1.
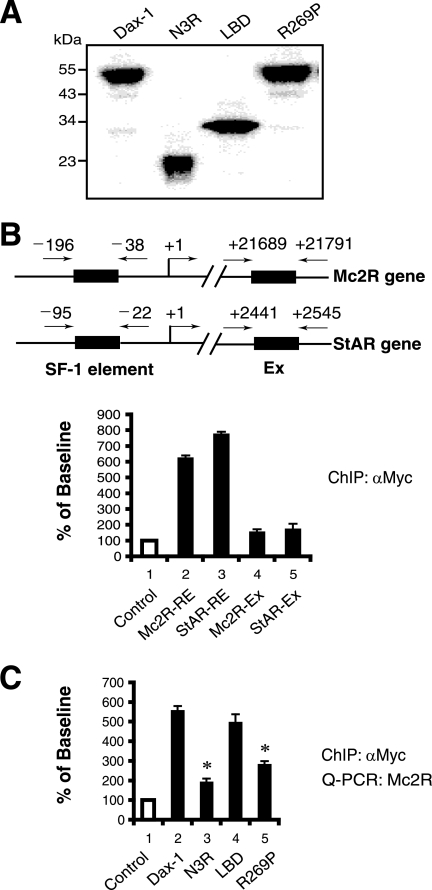
To test whether the Dax-1 mutants were expressed at levels similar to that of wild-type Dax-1 in the above-described transfections, we subjected lysates from transfected JEG-3 cells to immunoblotting using a Dax-1 antibody (Fig. 2D). This study demonstrated that wild-type Dax-1, its AHC mutant forms (R269P, ΔV271, and N442I), and the ΔAF2 mutant are expressed at comparable levels. However, we could not detect the truncated proteins N3R and LBD (calculated sizes, 22 and 30 kDa). This result was likely due to the Dax-1 antibody's not recognizing N3R and LBD, since Myc epitope-tagged versions of N3R and LBD are expressed at levels similar to that of full-length Dax-1 (see Fig. 6A).
FIG. 6.
Dax-1 is recruited to SF-1 target promoters, but the Dax-1 mutants N3R and R269P exhibit decreased promoter occupancy. (A) Expression of Dax-1-Myc and its mutant versions. Y1 cells were transfected with equal amounts of vectors expressing Myc-tagged wild-type Dax-1 and its mutant forms N3R, LBD, and R269P. The cells were lysed 48 h later and subjected to immunoblotting with an anti-Myc antibody. (B) Dax-1-Myc is recruited to the Mc2R and StAR promoters. ChIP assays of transiently expressed Dax-1-Myc in Y1 cells were performed using anti-Myc antibody (αMyc). Immunoprecipitates were analyzed by real-time PCR using primers designed against the SF-1 response elements (RE) in the proximal Mc2R and StAR promoters. Data are normalized to the value obtained using normal IgG as a negative control for immunoprecipitation, set at a baseline of 100. Bars 4 and 5 show the results obtained using PCR primers directed against downstream exons (Ex) to which Dax-1 would not be expected to bind. Experiments were performed in triplicate and repeated three times. Error bars indicate standard deviations. (C) Myc-tagged Dax-1 mutants N3R and R269P are defective in recruitment to the Mc2R promoter. Similar to the experiments described in the legend to panel B, ChIP assays were conducted with transiently expressed Dax-1-Myc wild-type or mutant proteins as indicated (*, P < 0.0001 for bars 3 and 5 versus bar 2 [the P value for bar 4 versus bar 2 was not significant] by ANOVA followed by Scheffe's test). Q-PCR, quantitative PCR.
TRα1 was used to test whether Dax-1 coactivation is specific for SF-1 or occurs with other nuclear receptors. Transfection with high-dose Dax-1 without or with SRA yielded no coactivation activity on T3-induced luciferase but instead showed a slight inhibitory effect (Fig. 2E, bar 6 versus bar 2 and bar 8 versus bar 4). These data suggest that the coactivation properties of Dax-1 cannot be generalized for all nuclear receptors.
Dax-1 binds to TIF2 in vitro and in mammalian cells.
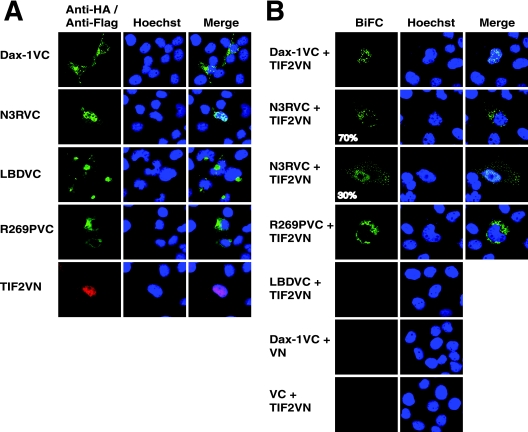
Dax-1 and TIF2 bind to each other in vitro, as shown by GST pull-down assays (Fig. 3A). The N-terminal fragment of Dax-1 (N3R) binds TIF2 as well as wild-type Dax-1 does, whereas Dax-1 LBD binds less well even though LBD has stronger transactivation properties than N3R in the reporter gene assays (Fig. 2A and B). In addition, Dax-1 ΔAF2 and the AHC mutants R269P and ΔV271 bind TIF2 as well as wild-type Dax-1 does.
FIG. 3.
Dax-1 interacts with TIF2 in vitro and in JEG-3 cells. (A) Interaction of GST-Dax-1 and its mutant forms with 35S-TIF2. 35S-labeled in vitro-translated TIF2 was incubated with equal amounts of purified GST-Dax-1 wild-type or mutant proteins adsorbed to glutathione agarose beads. Purified GST protein was the negative control. The bound proteins were subjected to SDS-PAGE, and the signals were analyzed in a Bio-Rad phosphorimager. (B) Dax-1 associates with TIF2 in mammalian cells. JEG-3 cells were cotransfected with a TIF2 expression vector and either an empty Myc vector (Vec) or a vector expressing Dax-1-Myc or its mutant form N3R, LBD, ΔAF2, R268P, or ΔV271. Each cell lysate was immunoprecipitated (IP) with anti-Myc (αMyc) agarose, and the immunoprecipitates were subjected to anti-TIF2 (αTIF2) immunoblotting (IB) as indicated in the upper panel. The lower panel shows anti-TIF2 immunoblotting of 2% of the input lysates.
The Dax-1-TIF2 interaction was substantiated by coimmunoprecipitation from JEG-3 cells transfected with Dax-1-Myc and TIF2 expression vectors (Fig. 3B). The Dax-1 deletion (N3R, LBD, and ΔAF2) and AHC (R269P and ΔV271) mutants also coimmunoprecipitated with TIF2, but the interactions were relatively weak, especially for LBD, ΔAF2, and R269P. The Dax-1-TIF2 interaction in living cells was investigated further, as described below.
Abnormal intracellular localization of Dax-1 mutants interacting with TIF2.
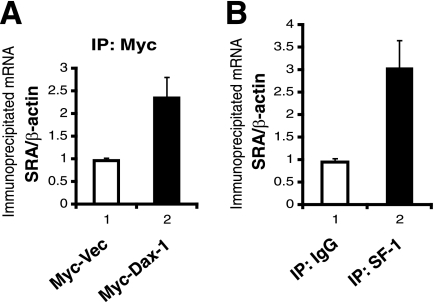
We utilized BiFC (22) in transfected COS-1 cells to confirm the interaction of Dax-1 with TIF2 and to visualize the locations of these proteins in living cells. This technique is based on the reconstitution of YFP from nonfluorescent N-terminal and C-terminal YFP fragments when they are brought together by two interacting proteins fused to the fragments. Recently, this method has been modified to be more specific and sensitive by using two fragments (VN and VC) from the optimized YFP variant Venus (57). Previous studies have reported that Dax-1 localizes both to the nucleus and to the cytoplasm (20, 35) and that TIF2 is almost exclusively nuclear (8, 68). Before the BiFC analysis, we assessed the subcellular localization patterns of the individual wild-type and deletion or AHC mutant Dax-1 proteins expressed from the VC vector (encoding a HA tag) and TIF2 expressed from the VN vector (encoding a Flag tag). Figure 4A shows that Dax-1-VC fluorescence was detected mostly in the cytoplasm, although minor expression in the nucleus was also observed. However, the deletion mutant N3R-VC was localized mainly in the nucleus, LBD-VC was distributed mostly outside of but adjacent to the nucleus, and the AHC mutant R269P-VC was diffusely localized in the cytoplasm. As expected, TIF2-VN was mostly nuclear, although occasionally minor cytoplasmic expression of TIF2-VN was observed (data not shown).
FIG. 4.
Mutant Dax-1-TIF2 complexes mislocalize in the cytoplasm in living cells as shown by BiFC analysis. (A) Subcellular localization patterns of Dax-1 or its mutant forms and TIF2. COS-1 cells were transfected with 100 ng of either a plasmid encoding Dax-1 or one of its mutant versions as a VC fusion or the plasmid encoding TIF2 as a VN fusion. Dax-1 or its mutant fusion proteins were detected with an anti-HA mouse IgG-Alexa Fluor 488 conjugate, while TIF2-VN was detected by exposure to anti-Flag M2 followed by incubation with Alexa Fluor red 594-goat anti-mouse IgG. The cell nuclei were stained with Hoechst 33342. (B) BiFC analysis of Dax-1-TIF2 interactions in living cells. COS-1 cells were cotransfected with combinations of VC and VN fusion constructs as indicated, and the cells were analyzed by laser-scanning confocal microscopy for the reconstitution of Venus protein fluorescence. Hoechst 33342 staining was used to visualize cell nuclei. The cells shown are representative of cells in multiple fields.
The BiFC analysis is shown in Fig. 4B. The coexpression of Dax-1-VC and TIF2-VN resulted in exclusively nuclear fluorescence, indicating that the interaction of Dax-1 and TIF2 occurs in the nucleus and perhaps suggesting that TIF2 recruits Dax-1 to or stabilizes it in this compartment. In contrast, the majority of fluorescence induced by the interaction of N3R-VC and TIF2-VN was cytoplasmic (in 70% of cells), although 30% of the cells showed both nuclear and cytoplasmic fluorescent foci. The interaction of AHC mutant R269P-VC with TIF2-VN was exclusively cytoplasmic, and no or very weak fluorescence complementation was observed in cells coexpressing Dax-1 LBD-VC and TIF2-VN. No fluorescence was observed in control cells cotransfected with Dax-1-VC and empty VN vectors or empty VC and TIF2-VN vectors. Overall, these results suggest that the Dax-1-TIF2 interaction helps maintain Dax-1 in the nucleus and that this localization requires both the N- and C-terminal portions of the Dax-1 protein. Furthermore, the interactions of N3R and the AHC mutant R269P with TIF2 mislocalize to the cytoplasm, consistent with the poor transcriptional activity of these Dax-1 mutants in the luciferase reporter assay (Fig. 2).
Dax-1 transactivation is dependent on SRA.
The observations that Dax-1 binds to SRA and that exogenous SRA enhances the Dax-1 induction of Mc2R-luc raise the question of whether the transactivation properties of Dax-1 depend on endogenous SRA. To test this possibility, we examined the effect of the knockdown of endogenous SRA on Dax-1 induction of Mc2R-luc with or without the cotransfection of cells with p160 coactivators. As shown in Fig. 5A, endogenous SRA was successfully silenced by 70%. The transactivation of Mc2R-luc by Dax-1 was abolished by the knockdown of endogenous SRA (Fig. 5B, bar 4 versus bar 2). Furthermore, the transactivation by coexpressed Dax-1 and TIF2 was nearly abolished after endogenous SRA had been silenced (Fig. 5C, bar 6 versus bar 3). Similar results were obtained when SRC-1 was used in place of TIF2 (data not shown). These observations support the hypothesis that SRA exists in a complex with p160 coactivators and Dax-1 to enhance target gene activation by SF-1.
Dax-1 is recruited to the Mc2R and StAR gene promoters, but the recruitment of N3R and AHC mutant R269P is impaired.
The finding that Dax-1 has the capacity to transactivate Mc2R-luc suggests that Dax-1 may function as a transcriptional activator of genes like the Mc2R gene in steroid hormone metabolism at least in some cellular contexts, which is opposite the conventional model of Dax-1 as a transcriptional repressor (18, 34, 67, 76). To address this possibility, Myc-tagged wild-type Dax-1 and its deletion or AHC mutant forms (N3R, LBD, and R269P) were transiently expressed at similar levels in Y1 adrenocortical cells (Fig. 6A). ChIP experiments were performed, confirming that wild-type Dax-1-Myc is recruited to SF-1 binding sites of the endogenous Mc2R and StAR promoters (Fig. 6B, bars 2 and 3). As negative controls, similar real-time PCRs were performed using exonic primers for Mc2R and StAR genes, neither of which yielded significant amplification (Fig. 6B, bars 4 and 5). Dax-1 mutants N3R and R269P were recruited less well than wild-type Dax-1 to the Mc2R promoter (Fig. 6C), which may explain their deficiencies as SF-1 coactivators (Fig. 2). These ChIP data are also consistent with the extranuclear mislocalization of R269P and N3R complexes with TIF2 observed by BiFC (Fig. 4). Interestingly, the ChIP data also show that Dax-1 LBD was efficiently recruited to the Mc2R promoter (Fig. 6C), consistent with its transactivation properties in the Mc2R-luc reporter gene assay (Fig. 2).
SRA regulates the transcription of endogenous steroidogenic genes in Y1 mouse adrenocortical cells.
The observation that SRA can function as an SF-1 coactivator by directly binding to SF-1 and Dax-1 suggests that SRA may regulate the expression of genes involved in steroidogenesis. To begin to test this hypothesis, we first asked whether endogenous SRA might be associated with SF-1 and Dax-1 in Y1 mouse adrenocortical cells. To examine this possibility, Y1 cells (which do not express endogenous Dax-1) were transfected with an empty Myc vector or a Dax-1-Myc vector and the cell extracts were immunoprecipitated with anti-Myc agarose beads. In parallel, Y1 cell extracts were immunoprecipitated with either anti-SF-1 antibody or normal IgG as a negative control. To eliminate the potential for contaminating genomic DNA, the immunoprecipitated materials were treated with DNase I before being reverse transcribed and analyzed by real-time PCR for SRA (and for β-actin as a negative control). The results indicate that endogenous SRA coimmunoprecipitates with both Dax-1-Myc (Fig. 7A) and endogenous SF-1 (Fig. 7B).
FIG. 7.
Dax-1 and SF-1 associate with endogenous SRA in Y1 cells. (A) Dax-1-Myc associates with endogenous SRA in Y1 cells. Y1 cells were transfected with an empty Myc vector (Myc-Vec) or a Dax-1-Myc expression vector. The cell lysates were immunoprecipitated (IP) using anti-Myc agarose, and the immunoprecipitates were subjected to RT-PCR with primers for mouse SRA or β-actin mRNA (as a nonspecific control). The SRA/β-actin ratio in the cells transfected with an empty Myc vector was set at 1. The experiments were performed in triplicate, and error bars indicate standard deviations. (B) Endogenous SF-1 associates with endogenous SRA in Y1 cells. Extracts from nontransfected Y1 cells were immunoprecipitated with an anti-SF-1 antibody or normal IgG as a negative control. The immunoprecipitates were analyzed by RT-PCR as described in the legend to panel A.
We next asked whether the knockdown of endogenous SRA would interfere with the expression of steroidogenic genes. As shown in Fig. 8A, we efficiently knocked down endogenous SRA in Y1 cells by using a retroviral shRNA, as opposed to a scrambled control shRNA. As shown in Fig. 8B, StAR mRNA expression was induced 3.5-fold by a 3-h exposure to ACTH (Fig. 8B, bar 2 versus bar 1), and this induction was decreased by 37% with SRA knockdown (Fig. 8B, bar 4 versus bar 2). Similar data were obtained after 8 h of exposure to ACTH: ACTH induced StAR mRNA expression 2.7-fold (Fig. 8B, bar 6 versus bar 5), and this induction was decreased by 25% by SRA knockdown (Fig. 8B, bar 8 versus bar 6). StAR protein expression, normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression, also decreased by 40% at 8 h of ACTH treatment (Fig. 8C).
In contrast to the expression of StAR mRNA, the expression of Mc2R mRNA in Y1 cells did not increase with exposure to ACTH (Fig. 8D). However, the knockdown of SRA inhibited Mc2R mRNA expression by about 40%, and a modest inhibition of Mc2R protein expression, normalized to GAPDH expression, was also observed (Fig. 8E). These results indicate that SRA plays a positive role in the expression of the steroidogenic genes for Mc2R and StAR in Y1 adrenocortical cells.
Since the data in Fig. 2 to 5 suggest that SRA forms a complex with p160 coactivators such as TIF2 when inducing the expression of SF-1 target promoters, we asked whether the physical interaction of TIF2 with SF-1 is dependent on SRA. To investigate this issue, we immunoprecipitated SF-1 from Y1 SRA knockdown and control cells and then immunoblotted the precipitates for endogenous TIF2. TIF2 coimmunoprecipitated with SF-1 from control cells but not from SRA knockdown cells (Fig. 8F, upper panel). Since the knockdown of SRA did not influence TIF2 expression (Fig. 8F, lower panel), the data support the hypothesis that SRA facilitates the interaction of SF-1 with TIF2.
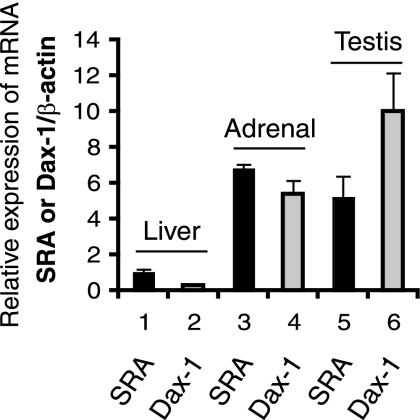
SRA is expressed in mouse adrenal glands and testes.
Although SF-1 and Dax-1 are known to be coexpressed in adrenal and gonadal cells, the expression of SRA in the adrenal gland and gonads has not been reported previously. We found that SRA is expressed at a much higher level in mouse adrenals and testes than in the liver (Fig. 9), which supports the hypothesis that SRA may be a novel regulator of steroidogenic gene expression in vivo, in coordination with SF-1 and Dax-1.
FIG. 9.
Expression levels of SRA and Dax-1 mRNAs in mouse adrenal, testis, and liver tissues. Adrenal, testis, and liver RNAs were isolated from 18-week-old male mice (n = 4). Real-time RT-PCR analysis of SRA, Dax-1, and β-actin for normalization was performed. The level of SRA in the liver tissue normalized to the level of β-actin (the SRA/β-actin ratio) was set at 1, and all other values are expressed relative to that value.
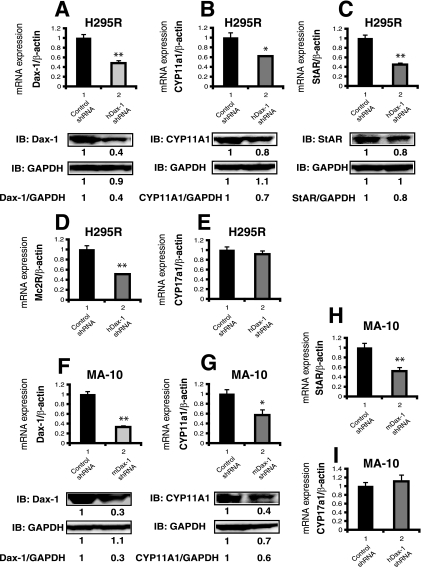
Knockdown of endogenous Dax-1 impairs the expression of a subset of steroidogenic genes.
Since the human adrenocortical cell line H295R (53) and the mouse Leydig tumor cell line MA-10 (4, 46) express Dax-1, SF-1, and steroidogenic enzymes, these cell lines were used in Dax-1 knockdown experiments to test whether endogenous Dax-1 might function as a coactivator for the expression of steroidogenic genes. Endogenous Dax-1 was efficiently knocked down by shRNA at both the mRNA and protein levels in H295R and MA-10 cells (Fig. 10A and F). Although exogenous Dax-1 has been shown to inhibit SF-1-mediated transactivation of steroidogenic genes (24, 59), we found that, in both H295R and MA-10 cells, the knockdown of Dax-1 inhibited the expression of the steroidogenic genes for CYP11A1 (Fig. 10B and 10G) and StAR (Fig. 10C and 10H) at both the mRNA and protein levels (StAR was not detectable in MA-10 cells). In addition, Mc2R mRNA expression was downregulated by Dax-1 silencing in H295R cells (Fig. 10D). However, the knockdown of Dax-1 had no effect on CYP17A1 mRNA expression in either H295R or MA-10 cells (Fig. 10E and I), nor did it affect SF-1 expression (data not shown). These results suggest that endogenous Dax-1 can function as a coactivator to increase the expression of a subset of steroidogenic genes in adrenal and gonadal cells, thus supporting our transfection data that indicate that Dax-1 can function as a coactivator in addition to its previously described role as a corepressor.
FIG. 10.
The knockdown of endogenous Dax-1 in H295R cells (A to E) and MA-10 cells (F to I) inhibits the expression of steroidogenic genes. (A) H295R cells were infected with lentivirus expressing an shRNA directed against human Dax-1 (hDax-1) or a scrambled-sequence shRNA as a negative control. (Upper panel) The knockdown of endogenous Dax-1 was assessed by real-time RT-PCR using β-actin as a normalization control. The level of expression of Dax-1 in the control shRNA cells was set at 1. (Lower panel) The knockdown efficiency was examined by immunoblotting (IB) with anti-Dax-1 antibody. Cell extracts from control shRNA or Dax-1 shRNA cells were subjected to immunoblotting with anti-Dax-1 and then were stripped and reprobed with anti-GAPDH as a loading control. The quantification of each band and the calculation of the relative expression of protein were performed as described in the legend to Fig. 8C. (B) Dax-1 knockdown decreases the expression of CYP11A1 mRNA and protein in H295R cells. (Upper panel) Real-time RT-PCR analysis of CYP11A1 expression in control shRNA or Dax-1 shRNA H295R cells was performed. The level of expression of CYP11A1 mRNA was normalized to that of β-actin mRNA and was set at 1 for control shRNA cells. (Lower panel) Cell extracts from control shRNA or Dax-1 shRNA cells were subjected to immunoblotting with anti-CYP11A1 and then were stripped and reprobed with anti-GAPDH as a loading control. (C, D, and E) Effects of Dax-1 knockdown on the expression of StAR, Mc2R, and CYP11A1 mRNA or protein in H295R cells. A real-time RT-PCR analysis similar to that described in the legend to panel B, but for StAR, Mc2R and CYP11A1, was performed, and immunoblotting was carried out with anti-StAR antibody. (F) MA-10 cells were infected with lentivirus expressing an shRNA directed against mouse Dax-1 (mDax-1) or a scrambled-sequence shRNA as a negative control. (Upper panel) A real-time RT-PCR analysis similar to that described in the legend to panel A was performed using specific primers for mouse Dax-1 or β-actin to assess the efficiency of the knockdown of endogenous Dax-1. (Lower panel) The knockdown efficiency was examined by immunoblotting with anti-Dax-1 and anti-GAPDH antibodies. (G, H, and I) Effects of Dax-1 knockdown on the expression of StAR, Mc2R, and CYP11A1 mRNA or protein in MA-10 cells. A real-time RT-PCR analysis of mouse CYP11A1, StAR, and CYP17A1 was performed, and immunoblotting was carried out with anti-CYP11A1. (**, P < 0.001 for bar 2 versus bar 1, and *, P < 0.01 for bar 2 versus bar 1 by Student's t test for panels A, B, C, D, F, G, and H.)
DISCUSSION
We have shown that Dax-1 can form coactivator complexes with SRA and TIF2 to stimulate SF-1 target gene transcription. This transactivation by Dax-1 is dependent on SRA, and importantly, the expression of a subset of steroidogenic genes is impaired by the knockdown of endogenous SRA or Dax-1 in adrenal or gonadal cell lines.
Dax-1 can function as an SF-1 coactivator or corepressor.
Dax-1 was previously considered to function exclusively as a negative regulator of SF-1-mediated transcription, possibly by recruiting corepressors such as NCoR (10) and Alien (2), and similar repressive activities of Dax-1 on several other nuclear receptors have been demonstrated (26). In addition, male mice with a mutant Dax-1 gene on the X chromosome (Dax-1−/Y mice) have elevated corticosterone/ACTH ratios, consistent with hyperresponsive adrenal glands and thus implying an inhibitory role for Dax-1 (5). However, this in vivo phenotype reflects the balance of many complex interactions. Enhanced steroidogenesis may reflect early enhanced differentiation of the adrenal glands of Dax-1−/Y mice, but the aging organs develop histologic adrenal cytomegaly, predicted to be the result of progenitor cell depletion or failure (unpublished observation). Furthermore, Dax-1 is expressed throughout the hypothalamic-pituitary-adrenal axis and hence may play multiple roles in the feedback regulation of steroidogenesis. Thus, putative inductive actions of Dax-1 may be obscured within this complex biology.
Indeed, loss-of-function mutations of SF-1 and Dax-1 result in similar developmental abnormalities in humans, including primary adrenal hypoplasia, suggesting that Dax-1 may function to enhance SF-1 induction of target genes under some circumstances. In support of this hypothesis, we found that Dax-1 can function as an SF-1 coactivator in cells transfected with high doses of Dax-1 vectors (Fig. 2). Furthermore, the Dax-1 AHC mutations R269P and N422I abolished this coactivation, suggesting that Dax-1 coactivation is important in steroidogenesis and adrenal and gonadal development. Importantly, our more physiologically relevant Dax-1 knockdown experiments revealed that the early-step steroidogenic genes for StAR and CYP11A1 were downregulated by Dax-1 silencing (Fig. 10), which suggests that Dax-1 exerts positive effects on the expression of these genes in both human H295R adrenal and murine MA-10 Leydig cells. These results are consistent with the observation that the disruption of dax1 function by morpholino oligonucleotides downregulates the expression of cyp11a1 and star in zebrafish (79).
Dax-1A, or Dax-1α, is a human Dax-1 splice variant in which the C-terminal 81 amino acids are replaced by a unique 12-amino-acid sequence (19, 21). Dax-1α is unable to repress the SF-1-mediated induction of a reporter gene but instead can increase StAR promoter-luciferase gene expression when SF-1 is present in limiting amounts. Our data indicate that Dax-1 itself can have either a negative or a positive effect on SF-1-mediated transcription, and we provide a mechanism for the positive effect by way of interactions with SRA and p160 coactivators. Since Dax-1 can form homodimers as well as heterodimers with Dax-1A (27), complex possibilities for gene regulation exist.
The coactivation function of Dax-1 is dosage sensitive.
It is important that Dax-1 coactivation of SF-1 is observed only at high doses of Dax-1. In contrast, the repression of SF-1 is seen with low doses of Dax-1. Since Dax-1 expression is induced by glucocorticoids (15) and activated β-catenin (45), Dax-1 coactivation may be favored in situations in which either of these factors, along with SRA and p160 coactivators, is abundant. While the role of Dax-1 dosage in the adrenal cortex has not yet been thoroughly examined, it has become increasingly clear that SF-1 and Dax-1 dosages provide critical regulatory influences on gene expression and that the ratio of each protein to the other may define the overall transcriptional output. Such an interplay is most evident in the transcriptional control of sex determination (gonadal differentiation into testes or ovaries). Indeed, while the Dax-1 gene was cloned as the gene responsible for X-linked AHC, it is also one of the genes in the duplicated Xp21 locus associated with dosage-sensitive XY gonadal sex reversal in humans (hence the acronym Dax-1, for dosage-sensitive sex reversal, AHC, on the X chromosome, number 1). Transgenic mice harboring an additional copy of the Dax-1 gene in a weakened Sry allelic background exhibit an overt intersex phenotype (60). The complex role of the Dax-1 dose in the regulation of transcription becomes more evident with the complete gonadal sex reversal in C57BL/6JEi (but not DBA/2J) XY mice carrying a loss-of-function Dax-1 allele (6, 44). Similarly, SF-1 dosage is critically important for proper adrenal development. The increase in SF-1 brought about by WT-1 and Cited2 in the adrenogonadal primordium specifies early adrenocortical versus gonadal fate (64), yet supraphysiologic transgenic overexpression of SF-1 in the adrenal cortex results in enhanced proliferation and the emergence of gonadal gene expression in the subcapsular adrenal cortex (12).
In addition to its role in the adrenal cortex and gonads (43, 74), Dax-1 is critical in early embryogenesis (49), where it participates in the maintenance of embryonic stem (ES) cell pluripotency (31). In ES cells, Dax-1 occupies the promoters of nearly 2,000 genes, many of which are also occupied by five other transcription factors important for pluripotency (Nanog, Sox2, Nac1, Oct4, and Klf4). Interestingly, ES cell genes occupied by Dax-1 along with these other factors tend to be active whereas those occupied by only one of these transcription factors are repressed, suggesting the importance of transcriptional coactivator networks for Dax-1 to induce gene expression. The prominent expression of Dax-1 in the Wnt-responsive adrenocortical subcapsular cells proposed to function as multipotent adrenocortical progenitor cells provides a similar context for potential activating functions of Dax-1 (30).
Novel role of SRA in steroidogenesis.
SRA was initially characterized as a steroid receptor RNA coactivator (36). However, it is now clear that it has broader biological functions, for example, serving as a coactivator for retinoic acid receptors (78) and the muscle differentiation factor MyoD (7). We found that both SF-1 and Dax-1 bind to SRA (Fig. 1). The SRA binding domain of SF-1 is similar to that identified in TRα1 (72). These domains lie immediately to the C-terminal side of the NR zinc fingers and include the TRα1 A box and the SF-1 FTZ-F1 box, which were previously characterized as playing auxiliary roles in DNA binding by contacting the minor groove just to the 5′ side of the core DNA cis element hexamer AGGTCA (38, 54, 69). Nuclear receptors that can bind DNA as monomers generally contain A box/FTZ-F1 box-like elements, and hence, RNA binding may be a common property of this subset of NRs.
The ability of Dax-1 to coactivate the SF-1-dependent expression of Mc2R-luc was enhanced by exogenous SRA (Fig. 2A) and was abolished by the knockdown of endogenous SRA (Fig. 5). Importantly, SRA knockdown in Y1 mouse adrenocortical cells inhibited endogenous Mc2R and StAR expression (Fig. 8), further supporting a role for this RNA coactivator in steroidogenic gene expression.
TIF2 functions as an SF-1 coactivator with Dax-1 and SRA.
Since SF-1 and SRA are both known to form complexes with p160 coactivators, we also evaluated Dax-1 for interactions with this family of proteins. We found that Dax-1 and TIF2 synergistically enhance the SF-1 induction of Mc2R-luc and that the Dax-1 AHC mutant R269P is severely defective (Fig. 2B). Our data also indicate that endogenous SRA is required for the synergistic enhancement of SF-1 transcriptional activity by Dax-1 and TIF2 (Fig. 5C). Interestingly, by coimmunoprecipitation, we identified a physical interaction between SF-1 and TIF2, which was impaired by SRA knockdown (Fig. 8F). Together, the data indicate that SRA regulates SF-1 target gene expression by functioning as a coactivator in association with p160 proteins and Dax-1.
We found that Dax-1 LBD binds TIF2 weakly compared to wild-type Dax-1 in GST pull-down assays (Fig. 3A), while N3R and the AHC mutant R269P bind TIF2 well. However, Dax-1 LBD is an excellent SF-1 coactivator, but N3R and R269P lose this ability. This result is probably because Dax-1 LBD contains the SF-1 interaction domain while N3R and R269P lose the interaction with SF-1 (24). Consistent with this possibility, our ChIP data revealed that Dax-1 LBD is recruited to the SF-1-responsive region of the Mc2R promoter comparably to wild-type Dax-1 but that the recruitment of N3R and R269P is defective (Fig. 6C). Furthermore, the BiFC analysis indicates that, in living cells, Dax-1 associates with TIF2 in subnuclear foci, suggesting that these foci may constitute a special compartment for the exertion of transcriptional activity. In contrast, N3R and the AHC mutant R269P both interact with TIF2 largely in the cytoplasm (Fig. 4B), again consistent with defective coactivation (Fig. 2) and defective recruitment to the Mc2R promoter in the ChIP assay (Fig. 6C). The findings of different subcellular localization patterns of the Dax-1 wild type and deletion and AHC mutants associated with TIF2 (Fig. 4B) may provide a novel mechanism to explain how Dax-1 AHC mutations lead to the AHC syndrome.
Although our inability to detect BiFC of Dax-1 LBD and TIF2 is consistent with the weaker interaction between these two proteins in the GST pull-down assay (Fig. 3A), the result is surprising given that LBD is an excellent SF-1 coactivator (Fig. 2). Perhaps the interaction is too weak to be detected by BiFC yet is sufficient for transcription, or perhaps the geometry of the LBD-TIF2 interaction does not allow a productive interaction between the two halves of Venus to reconstitute fluorescence.
In conclusion, we have defined the dose-sensitive ability of Dax-1 to function as a coactivator for SF-1 target gene transcription. Biochemical characterizations indicate that Dax-1 functions by binding to the RNA coactivator SRA and p160 coactivator proteins and is capable of playing a positive role in regulating steroidogenic gene expression. SRA is important to stabilize complexes of SF-1 and Dax-1 for the recruitment of p160 coactivators to regulate target gene expression in steroidogenesis.
Acknowledgments
We thank Alessia Trovato for technical help in the shRNA knockdown studies and Alex Kim for assistance with adrenal gland dissections.
This work was supported by NIH grants MH079365 (B.X.), DK44155 (R.J.K.), and DK62027 (G.D.H.) and the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center grant DK020572.
Footnotes
Published ahead of print on 2 February 2009.
REFERENCES
- 1.Agoulnik, I. U., W. C. Krause, W. E. Bingman III, H. T. Rahman, M. Amrikachi, G. E. Ayala, and N. L. Weigel. 2003. Repressors of androgen and progesterone receptor action. J. Biol. Chem. 27831136-31148. [DOI] [PubMed] [Google Scholar]
- 2.Altincicek, B., S. P. Tenbaum, U. Dressel, D. Thormeyer, R. Renkawitz, and A. Baniahmad. 2000. Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J. Biol. Chem. 2757662-7667. [DOI] [PubMed] [Google Scholar]
- 3.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277965-968. [DOI] [PubMed] [Google Scholar]
- 4.Ascoli, M. 1981. Characterization of several clonal lines of cultured Leydig tumor cells: gonadotropin receptors and steroidogenic responses. Endocrinology 10888-95. [DOI] [PubMed] [Google Scholar]
- 5.Babu, P. S., D. L. Bavers, F. Beuschlein, S. Shah, B. Jeffs, J. L. Jameson, and G. D. Hammer. 2002. Interaction between Dax-1 and steroidogenic factor-1 in vivo: increased adrenal responsiveness to ACTH in the absence of Dax-1. Endocrinology 143665-673. [DOI] [PubMed] [Google Scholar]
- 6.Bouma, G. J., K. H. Albrecht, L. L. Washburn, A. K. Recknagel, G. A. Churchill, and E. M. Eicher. 2005. Gonadal sex reversal in mutant Dax1 XY mice: a failure to upregulate Sox9 in pre-Sertoli cells. Development 1323045-3054. [DOI] [PubMed] [Google Scholar]
- 7.Caretti, G., R. L. Schiltz, F. J. Dilworth, M. Di Padova, P. Zhao, V. Ogryzko, F. V. Fuller-Pace, E. P. Hoffman, S. J. Tapscott, and V. Sartorelli. 2006. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 11547-560. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. L., S. C. Wang, B. Hosking, and G. E. Muscat. 2001. Subcellular localization of the steroid receptor coactivators (SRCs) and MEF2 in muscle and rhabdomyosarcoma cells. Mol. Endocrinol. 15783-796. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W. Y., L. J. Juan, and B. C. Chung. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol. Cell. Biol. 2510442-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, P. A., C. Dorn, Y. Sadovsky, and J. Milbrandt. 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 182949-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford, P. A., J. A. Polish, G. Ganpule, and Y. Sadovsky. 1997. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol. Endocrinol. 111626-1635. [DOI] [PubMed] [Google Scholar]
- 12.Doghman, M., T. Karpova, G. A. Rodrigues, M. Arhatte, J. De Moura, L. R. Cavalli, V. Virolle, P. Barbry, G. P. Zambetti, B. C. Figueiredo, L. L. Heckert, and E. Lalli. 2007. Increased steroidogenic factor-1 dosage triggers adrenocortical cell proliferation and cancer. Mol. Endocrinol. 212968-2987. [DOI] [PubMed] [Google Scholar]
- 13.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14121-141. [PubMed] [Google Scholar]
- 14.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192262-267. [DOI] [PubMed] [Google Scholar]
- 15.Gummow, B. M., J. O. Scheys, V. R. Cancelli, and G. D. Hammer. 2006. Reciprocal regulation of a glucocorticoid receptor-steroidogenic factor-1 transcription complex on the Dax-1 promoter by glucocorticoids and adrenocorticotropic hormone in the adrenal cortex. Mol. Endocrinol. 202711-2723. [DOI] [PubMed] [Google Scholar]
- 16.Hammer, G. D., and H. A. Ingraham. 1999. Steroidogenic factor-1: its role in endocrine organ development and differentiation. Front. Neuroendocrinol. 20199-223. [DOI] [PubMed] [Google Scholar]
- 17.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3521-526. [DOI] [PubMed] [Google Scholar]
- 18.Hanley, N. A., W. E. Rainey, D. I. Wilson, S. G. Ball, and K. L. Parker. 2001. Expression profiles of SF-1, DAX1, and CYP17 in the human fetal adrenal gland: potential interactions in gene regulation. Mol. Endocrinol. 1557-68. [DOI] [PubMed] [Google Scholar]
- 19.Ho, J., Y. H. Zhang, B. L. Huang, and E. R. McCabe. 2004. NR0B1A: an alternatively spliced form of NR0B1. Mol. Genet. Metab. 83330-336. [DOI] [PubMed] [Google Scholar]
- 20.Holter, E., N. Kotaja, S. Makela, L. Strauss, S. Kietz, O. A. Janne, J. A. Gustafsson, J. J. Palvimo, and E. Treuter. 2002. Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX-1. Mol. Endocrinol. 16515-528. [DOI] [PubMed] [Google Scholar]
- 21.Hossain, A., C. Li, and G. F. Saunders. 2004. Generation of two distinct functional isoforms of dosage-sensitive sex reversal-adrenal hypoplasia congenita-critical region on the X chromosome gene 1 (DAX-1) by alternative splicing. Mol. Endocrinol. 181428-1437. [DOI] [PubMed] [Google Scholar]
- 22.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9789-798. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, Y., A. Swain, T. J. Weber, K. E. Hentges, E. Zanaria, E. Lalli, K. T. Tamai, P. Sassone-Corsi, R. Lovell-Badge, G. Camerino, and K. L. Parker. 1996. Steroidogenic factor 1 and Dax-1 colocalize in multiple cell lineages: potential links in endocrine development. Mol. Endocrinol. 101261-1272. [DOI] [PubMed] [Google Scholar]
- 24.Ito, M., R. Yu, and J. L. Jameson. 1997. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 171476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, M., R. N. Yu, and J. L. Jameson. 1998. Steroidogenic factor-1 contains a carboxy-terminal transcriptional activation domain that interacts with steroid receptor coactivator-1. Mol. Endocrinol. 12290-301. [DOI] [PubMed] [Google Scholar]
- 26.Iyer, A. K., and E. R. McCabe. 2004. Molecular mechanisms of DAX1 action. Mol. Genet. Metab. 8360-73. [DOI] [PubMed] [Google Scholar]
- 27.Iyer, A. K., Y. H. Zhang, and E. R. McCabe. 2007. LXXLL motifs and AF-2 domain mediate SHP (NR0B2) homodimerization and DAX1 (NR0B1)-DAX1A heterodimerization. Mol. Genet. Metab. 92151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko, S., and J. L. Manley. 2005. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol. Cell 2091-103. [DOI] [PubMed] [Google Scholar]
- 30.Kim, A. C., and G. D. Hammer. 2007. Adrenocortical cells with stem/progenitor cell properties: recent advances. Mol. Cell. Endocrinol. 265-26610-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J., J. Chu, X. Shen, J. Wang, and S. H. Orkin. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 1321049-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koenig, R. J., M. A. Lazar, R. A. Hodin, G. A. Brent, P. R. Larsen, W. W. Chin, and D. D. Moore. 1989. Inhibition of thyroid hormone action by a non-hormone binding c-erbA protein generated by alternative mRNA splicing. Nature 337659-661. [DOI] [PubMed] [Google Scholar]
- 33.Krylova, I. N., E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, L. Lebedeva, M. Suzawa, J. D. Williams, S. P. Williams, R. K. Guy, J. W. Thornton, R. J. Fletterick, T. M. Willson, and H. A. Ingraham. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120343-355. [DOI] [PubMed] [Google Scholar]
- 34.Lalli, E., M. H. Melner, D. M. Stocco, and P. Sassone-Corsi. 1998. DAX-1 blocks steroid production at multiple levels. Endocrinology 1394237-4243. [DOI] [PubMed] [Google Scholar]
- 35.Lalli, E., K. Ohe, C. Hindelang, and P. Sassone-Corsi. 2000. Orphan receptor DAX-1 is a shuttling RNA binding protein associated with polyribosomes via mRNA. Mol. Cell. Biol. 204910-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanz, R. B., N. J. McKenna, S. A. Onate, U. Albrecht, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 9717-27. [DOI] [PubMed] [Google Scholar]
- 37.Li, Y., M. Choi, G. Cavey, J. Daugherty, K. Suino, A. Kovach, N. C. Bingham, S. A. Kliewer, and H. E. Xu. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17491-502. [DOI] [PubMed] [Google Scholar]
- 38.Little, T. H., Y. Zhang, C. K. Matulis, J. Weck, Z. Zhang, A. Ramachandran, K. E. Mayo, and I. Radhakrishnan. 2006. Sequence-specific deoxyribonucleic acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability. Mol. Endocrinol. 20831-843. [DOI] [PubMed] [Google Scholar]
- 39.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 40.Luo, X., Y. Ikeda, and K. L. Parker. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77481-490. [DOI] [PubMed] [Google Scholar]
- 41.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108465-474. [DOI] [PubMed] [Google Scholar]
- 43.Meeks, J. J., S. E. Crawford, T. A. Russell, K. Morohashi, J. Weiss, and J. L. Jameson. 2003. Dax1 regulates testis cord organization during gonadal differentiation. Development 1301029-1036. [DOI] [PubMed] [Google Scholar]
- 44.Meeks, J. J., J. Weiss, and J. L. Jameson. 2003. Dax1 is required for testis determination. Nat. Genet. 3432-33. [DOI] [PubMed] [Google Scholar]
- 45.Mizusaki, H., K. Kawabe, T. Mukai, E. Ariyoshi, M. Kasahara, H. Yoshioka, A. Swain, and K. Morohashi. 2003. Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad. Mol. Endocrinol. 17507-519. [DOI] [PubMed] [Google Scholar]
- 46.Mizutani, T., K. Shiraishi, T. Welsh, and M. Ascoli. 2006. Activation of the lutropin/choriogonadotropin receptor in MA-10 cells leads to the tyrosine phosphorylation of the focal adhesion kinase by a pathway that involves Src family kinases. Mol. Endocrinol. 20619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muscatelli, F., T. M. Strom, A. P. Walker, E. Zanaria, D. Recan, A. Meindl, B. Bardoni, S. Guioli, G. Zehetner, W. Rabl, et al. 1994. Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372672-676. [DOI] [PubMed] [Google Scholar]
- 48.Nachtigal, M. W., Y. Hirokawa, D. L. Enyeart-VanHouten, J. N. Flanagan, G. D. Hammer, and H. A. Ingraham. 1998. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93445-454. [DOI] [PubMed] [Google Scholar]
- 49.Niakan, K. K., E. C. Davis, R. C. Clipsham, M. Jiang, D. B. Dehart, K. K. Sulik, and E. R. McCabe. 2006. Novel role for the orphan nuclear receptor Dax1 in embryogenesis, different from steroidogenesis. Mol. Genet. Metab. 88261-271. [DOI] [PubMed] [Google Scholar]
- 50.Niranjanakumari, S., E. Lasda, R. Brazas, and M. A. Garcia-Blanco. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26182-190. [DOI] [PubMed] [Google Scholar]
- 51.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 2701354-1357. [DOI] [PubMed] [Google Scholar]
- 52.Parker, K. L., D. A. Rice, D. S. Lala, Y. Ikeda, X. Luo, M. Wong, M. Bakke, L. Zhao, C. Frigeri, N. A. Hanley, N. Stallings, and B. P. Schimmer. 2002. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog. Horm. Res. 5719-36. [DOI] [PubMed] [Google Scholar]
- 53.Rainey, W. E., I. M. Bird, and J. I. Mason. 1994. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol. Cell. Endocrinol. 10045-50. [DOI] [PubMed] [Google Scholar]
- 54.Rastinejad, F., T. Perlmann, R. M. Evans, and P. B. Sigler. 1995. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 375203-211. [DOI] [PubMed] [Google Scholar]
- 55.Rosen, E. D., A. L. O'Donnell, and R. J. Koenig. 1992. Ligand-dependent synergy of thyroid hormone and retinoid X receptors. J. Biol. Chem. 26722010-22013. [PubMed] [Google Scholar]
- 56.Sadovsky, Y., P. A. Crawford, K. G. Woodson, J. A. Polish, M. A. Clements, L. M. Tourtellotte, K. Simburger, and J. Milbrandt. 1995. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc. Natl. Acad. Sci. USA 9210939-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shyu, Y. J., H. Liu, X. Deng, and C. D. Hu. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 4061-66. [DOI] [PubMed] [Google Scholar]
- 58.Sugawara, T., J. A. Holt, M. Kiriakidou, and J. F. Strauss III. 1996. Steroidogenic factor 1-dependent promoter activity of the human steroidogenic acute regulatory protein (StAR) gene. Biochemistry 359052-9059. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki, T., M. Kasahara, H. Yoshioka, K. Morohashi, and K. Umesono. 2003. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol. Cell. Biol. 23238-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swain, A., V. Narvaez, P. Burgoyne, G. Camerino, and R. Lovell-Badge. 1998. Dax1 antagonizes Sry action in mammalian sex determination. Nature 391761-767. [DOI] [PubMed] [Google Scholar]
- 61.Swanson, M. S., and G. Dreyfuss. 1988. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 82237-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387677-684. [DOI] [PubMed] [Google Scholar]
- 63.Val, P., A. M. Lefrancois-Martinez, G. Veyssiere, and A. Martinez. 2003. SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl. Recept. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Val, P., J. P. Martinez-Barbera, and A. Swain. 2007. Adrenal development is initiated by Cited2 and Wt1 through modulation of Sf-1 dosage. Development 1342349-2358. [DOI] [PubMed] [Google Scholar]
- 65.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 153667-3675. [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Z. J., B. Jeffs, M. Ito, J. C. Achermann, R. N. Yu, D. B. Hales, and J. L. Jameson. 2001. Aromatase (Cyp19) expression is up-regulated by targeted disruption of Dax1. Proc. Natl. Acad. Sci. USA 987988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe, M., J. Yanagisawa, H. Kitagawa, K. Takeyama, S. Ogawa, Y. Arao, M. Suzawa, Y. Kobayashi, T. Yano, H. Yoshikawa, Y. Masuhiro, and S. Kato. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 201341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Wilson, T. E., T. J. Fahrner, and J. Milbrandt. 1993. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol. Cell. Biol. 135794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winnay, J. N., and G. D. Hammer. 2006. Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol. Endocrinol. 20147-166. [DOI] [PubMed] [Google Scholar]
- 71.Xu, B., and R. J. Koenig. 2005. Regulation of thyroid hormone receptor alpha2 RNA binding and subcellular localization by phosphorylation. Mol. Cell. Endocrinol. 245147-157. [DOI] [PubMed] [Google Scholar]
- 72.Xu, B., and R. J. Koenig. 2004. An RNA-binding domain in the thyroid hormone receptor enhances transcriptional activation. J. Biol. Chem. 27933051-33056. [DOI] [PubMed] [Google Scholar]
- 73.Yang, W. H., J. H. Heaton, H. Brevig, S. Mukherjee, J. A. Iniguez-Lluhi, and G. D. Hammer. 2009. SUMOylation inhibits SF-1 activity by reducing CDK7-mediated serine 203 phosphorylation. Mol. Cell. Biol. 29613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, R. N., M. Ito, T. L. Saunders, S. A. Camper, and J. L. Jameson. 1998. Role of Ahch in gonadal development and gametogenesis. Nat. Genet. 20353-357. [DOI] [PubMed] [Google Scholar]
- 75.Zanaria, E., F. Muscatelli, B. Bardoni, T. M. Strom, S. Guioli, W. Guo, E. Lalli, C. Moser, A. P. Walker, E. R. McCabe, et al. 1994. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372635-641. [DOI] [PubMed] [Google Scholar]
- 76.Zazopoulos, E., E. Lalli, D. M. Stocco, and P. Sassone-Corsi. 1997. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature 390311-315. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, H., J. S. Thomsen, L. Johansson, J. A. Gustafsson, and E. Treuter. 2000. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J. Biol. Chem. 27539855-39859. [DOI] [PubMed] [Google Scholar]
- 78.Zhao, X., J. R. Patton, S. L. Davis, B. Florence, S. J. Ames, and R. A. Spanjaard. 2004. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell 15549-558. [DOI] [PubMed] [Google Scholar]
- 79.Zhao, Y., Z. Yang, J. K. Phelan, D. A. Wheeler, S. Lin, and E. R. McCabe. 2006. Zebrafish dax1 is required for development of the interrenal organ, the adrenal cortex equivalent. Mol. Endocrinol. 202630-2640. [DOI] [PubMed] [Google Scholar]
- 80.Zwermann, O., F. Beuschlein, E. Lalli, A. Klink, P. Sassone-Corsi, and M. Reincke. 2005. Clinical and molecular evidence for DAX-1 inhibition of steroidogenic factor-1-dependent ACTH receptor gene expression. Eur. J. Endocrinol. 152769-776. [DOI] [PubMed] [Google Scholar]