Abstract
Slx5 and Slx8 are heterodimeric RING domain-containing proteins that possess SUMO-targeted ubiquitin ligase (STUbL) activity in vitro. Slx5-Slx8 and its orthologs are proposed to target SUMO conjugates for ubiquitin-mediated proteolysis, but the only in vivo substrate identified to date is mammalian PML, and the physiological importance of SUMO-targeted ubiquitylation remains largely unknown. We previously identified mutations in SLX5 and SLX8 by selecting for suppressors of a temperature-sensitive allele of MOT1, which encodes a regulator of TATA-binding protein. Here, we demonstrate that Mot1 is SUMOylated in vivo and that disrupting the Slx5-Slx8 pathway by mutation of the target lysines in Mot1, by deletion of SLX5 or the ubiquitin E2 UBC4, or by inhibition of the proteosome suppresses mot1-301 mutant phenotypes and increases the stability of the Mot1-301 protein. The Mot1-301 mutant protein is targeted for proteolysis by SUMOylation to a much greater extent than wild-type Mot1, suggesting a quality control mechanism. In support of this idea, growth of Saccharomyces cerevisiae in the presence of the arginine analog canavanine results in increased SUMOylation and Slx5-Slx8-mediated degradation of wild-type Mot1. These results therefore demonstrate that Mot1 is an in vivo STUbL target in yeast and suggest a role for SUMO-targeted degradation in protein quality control.
Posttranslational modifications of proteins by ubiquitin and the ubiquitin-like family of proteins, including SUMO, play pivotal roles in diverse cellular processes (25). One role of ubiquitylation is to target substrates for proteasomal degradation. Targeting of substrates for degradation can be part of a programmed regulatory event, as occurs during cell cycle progression (43) or during downregulation of signal transduction (32), or it can serve as part of a quality control system to remove defective proteins that arise through mutation, transcriptional and translational errors, folding defects, chemical damage, and heat-induced denaturation (14). Quality control has been thoroughly documented for the degradation of misfolded proteins that occurs in the cytoplasm or during transit through the endoplasmic reticulum (6, 15, 18, 34, 49), and a quality control system mediated by the San1 ubiquitin E3 ligase is required specifically for the degradation of some mutant proteins but not that of their wild-type counterparts in the nucleus (12). Current challenges include identifying all the components of the known quality control systems, detailing their individual roles, understanding how defective proteins are recognized and targeted, and determining whether additional quality control systems exist.
Like ubiquitin, SUMO is covalently attached to lysine residues of target proteins by a series of enzymatic reactions that require maturation by a protease (30, 31) and E1 (24), E2 (22), and E3 (23, 53, 61) enzymes. Typically, SUMO is attached to the lysine residue in a ΨKxE/D consensus motif (21), although attachment of SUMO at nonconsensus sites also occurs (9, 20). Although the SUMO conjugation pathway is analogous to the ubiquitin pathway, the downstream consequences of these two modifications are typically different and, in some cases, even antagonistic. SUMOylation of IκBα, for example, prevents its ubiquitylation and subsequent degradation (10, 56). However, recent examples in which SUMO and ubiquitin appear to cooperate or function together have emerged. Like ubiquitin, SUMO can promote degradation of some proteins, such as the mammalian PML-RARα fusion protein that causes acute promyelocytic leukemia (29, 62); BMAL1, which is a component of the mammalian circadian clock (3); and the Saccharomyces cerevisiae Flp recombinase (5). The mechanism of SUMO-targeted degradation remained unclear, however, until the recent discovery by several groups of a novel family of RING domain-containing proteins called SUMO-targeted ubiquitin ligases (STUbLs) (36, 42, 54, 60). STUbLs contain RING domains that confer ubiquitin E3 ligase activity but also contain multiple SUMO-interacting motifs that are required for interaction with SUMOylated substrates (16, 35, 44, 50). STUbLs thus are proposed to preferentially target SUMO conjugates for ubiquitylation by recognizing the SUMO moiety on the substrate, thereby recruiting a ubiquitin E3 protein, which in turn stimulates ubiquitylation of the substrate. Moreover, mutations in proteasome subunits or inhibition of the proteosome by MG132 results in an accumulation of high-molecular-mass SUMO species in the cell, further connecting SUMO to proteasome-mediated degradation (36, 57). The only known direct physiological substrate of SUMO-targeted ubiquitylation is mammalian PML (28, 55), but with the identification of STUbLs and proteins that are targeted for destruction by SUMO, additional substrates are sure to emerge.
We previously identified SLX5 and SLX8, the prototypical STUbL complex in budding yeast, along with the majority of the SUMO pathway, in a selection for genomic suppressors of a temperature-sensitive allele of MOT1, which encodes a regulator of TATA-binding protein (TBP) (58). We proposed that SLX5 and SLX8 were components of the SUMO pathway, on the basis of their coisolation with SUMO pathway mutations, synthetic lethal phenotypes of slx5 or slx8 deletions with SUMO pathway mutations, and accumulation of SUMO conjugates in slx5 or slx8 deletion strains. The specific role of Slx5-Slx8 in the pathway, its downstream target, and its physiological role were unknown. Here, we report that Mot1 is an in vivo target for SUMO and Slx5-Slx8 and that Mot1 is subject to SUMO-targeted degradation via the proteosome. Furthermore, both Mot1 mutant proteins and wild-type Mot1 protein from cells that were grown in canavanine are SUMOylated and degraded to a greater extent than wild-type Mot1. On the basis of these results, we propose that the SUMO-targeted Slx5-Slx8-mediated degradation pathway functions as part of a protein quality control system in the cell.
MATERIALS AND METHODS
Yeast strains, plasmids, media, and genetic methods.
The Saccharomyces cerevisiae strains and plasmids used in this study are listed in Tables 1 and 2. All media used, including rich medium (yeast extract-peptone-dextrose), sucrose medium (yeast extract-peptone-Suc), synthetic complete (SC) drop-out medium (for example, SC-Ura), and sporulation medium, were made as described previously (45). SC-galactose plates contained SC medium with 2% galactose and 1 μg/ml antimycin A. Canavanine sensitivity was tested with SC-Arg plates containing 2.5 μg/ml canavanine (no. C9758; Sigma). For experiments testing the effect of canavanine, cells were grown overnight to log phase in SC medium lacking arginine, and canavanine was added to give a final concentration of 30 μg/ml. When the effect of MG132 was tested, the experiments were performed with a pdr5Δ strain transformed with the hemagglutinin (HA)-tagged MOT1 plasmids. Standard genetic methods for mating, sporulation, transformation, and tetrad analysis were used throughout this study (45).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype |
|---|---|
| GY481 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 |
| ZY142 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) leu2Δ1 trp1Δ63 mot1-301 |
| ZY561 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN |
| ZY616 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 mot1-301-3HA::KAN |
| ZY601 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN siz1Δ::TRP1 |
| ZY602 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 trp1Δ63 MOT1-3HA::KAN siz2Δ::KAN |
| ZY604 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN siz1Δ::TRP1 siz2Δ::KAN |
| ZY593 | MATahis4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN smt3Δ::TRP1 [pF396 (CEN LEU2 SMT3)] |
| ZY594 | MATahis4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN smt3Δ::TRP1 [pF397 (CEN LEU2 3x-Myc-SMT3)] |
| ZY360 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 siz1Δ::TRP1 siz2Δ::KAN |
| ZY624 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) leu2Δ1 trp1Δ63 mot1-301-3HA::KAN ubc9-101 |
| ZY618 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 mot1-301-3HA::KAN siz1Δ::TRP1 |
| ZY623 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 mot1-301-3HA::KAN siz1Δ::TRP1 siz2Δ::KAN |
| OY168 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 mot1Δ::KAN [pMR13 (CEN URA3 MOT1)] |
| OY341 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52(orΔ0) mot1-301 ubc4Δ::KAN |
| OY355 | MATα ura3-52(orΔ0) leu2Δ0 trp1Δ63 mot1-301 ubc2Δ::KAN |
| OY357 | MATahis4-912δ lys2-128δ ura3-52(orΔ0) leu2Δ0 trp1Δ63 mot1-301 ubc5Δ::KAN |
| OY358 | MATahis3Δ1 suc2ΔUAS(-1900/-390) ura3-52(orΔ0) leu2Δ0 mot1-301 ubc7Δ::KAN |
| OY359 | MATahis4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52(orΔ0) trp1Δ63 mot1-301 ubc8Δ::KAN |
| OY360 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52(orΔ0) trp1Δ63 mot1-301 ubc10Δ::KAN |
| OY361 | MATα his4-912δ ura3-52(orΔ0) mot1-301 ubc11Δ::KAN |
| OY362 | MATα his4-912δ ura3-52(orΔ0) leu2Δ0 trp1Δ63 mot1-301 ubc13Δ::KAN |
| OY363 | MATα his4-912δ suc2ΔUAS(-1900/-390) ura3-52(orΔ0) mot1-301 mms2Δ::KAN |
| OY332 | MATahis3Δ1 ura3Δ0 leu2Δ0 met15Δ0 ubc4Δ::KAN |
| OY334 | MATahis3Δ1 ura3Δ0 leu2Δ0 met15Δ0 ubc8Δ::KAN |
| OY281 | MATahis3Δ1 ura3Δ0 leu2Δ0 met15Δ0 pdr5Δ::KAN |
| OY304 | MATahis3Δ1 ura3Δ0 leu2Δ0 met15Δ0 san1Δ::KAN |
| ZY586 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN slx5Δ::URA3 |
| ZY596 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN slx8Δ::TRP1 |
| ZY598 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 MOT1-3HA::KAN slx5Δ::URA3 slx8Δ::TRP1 |
| ZY44 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 slx5Δ::URA3 |
| ZY50 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 slx8Δ::TRP1 |
| ZY109 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 siz2Δ::KAN |
| ZY349 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 trp1Δ63 siz1Δ::TRP1 |
| GY350 | MATα his4-912δ lys2-128δ ura3-52 leu2Δ1 trp1Δ63 HA-SPT6 |
| GY854 | MATα his4-912δ lys2-128δ leu2d1 ura3-52 RPB3-HA::LEU2 |
| GY917 | MATα his4-917δ lys2-173R2 ura3Δ0 leu2Δ0 gal4Δ::KANMX 3HA-SPT20 |
| ZY314 | MATα his4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 trp1Δ63 leu2Δ1 YDR1-FLAG::KAN |
| OY231 | MATα his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 3HA-RTF1 |
| OY302 | MATα his3Δ200 leu2Δ1 ura3-52 RCO1-3HA::HIS3MX6 |
| AY639 | MATahis3Δ200 lys2-128δ trp1Δ63 ura3-52 leu2Δ1 SET2-6HA::TRP1 |
| AY880 | MATα his3Δ200 lys2-128δ ura3-52 trp1Δ63 BUR2-3HA::TRP1 |
| AY906 | MATahis3Δ200 lys2-128δ ura3 leu2Δ1 PAF1-3HA SPT16-3MYC |
| ZY684 | MATα his4-917δ (or-912δ) lys2-128δ ura3-52 leu2Δ0 trp1Δ63 gal4Δ::KANMX 3HA-SPT20 slx5Δ::URA3 |
| ZY689 | MATahis4-917δ (or-912δ) lys2-173R2 ura3-52(orΔ0) leu2Δ1(or Δ0) 3HA-SPT20 siz1Δ::TRP1 siz2Δ::KAN |
| OY493 | MATalys2-128δ suc2ΔUAS(-1900/-390) ura3-52 leu2Δ1 RCO1-3HA::HIS3MX6 siz1Δ::TRP1 siz2Δ::KAN |
| OY494 | MATahis4-912δ lys2-128δ suc2ΔUAS(-1900/-390) ura3-52 trp1Δ63 RCO1-3HA::HIS3MX6 slx5Δ::URA3 |
| PJ69-4A | MATahis3Δ200 leu2-3,112 trp1-901 ura3-52 gal4Δ gal80Δ LYS2::GAL1-HIS3 ade2::GAL2-ADE2 met2::GAL7-LacZ |
TABLE 2.
Plasmids
| Plasmid | Genotype |
|---|---|
| pRS415 | AMPR CEN LEU2 |
| pZW81 | AMPRCEN LEU2 MOT1 |
| pZW83 | AMPRCEN LEU2 mot1-301 |
| pZW217 | AMPR 2μm LEU2 mot1-301 |
| pZW210 | AMPRCEN LEU2 MOT1-3HA::KAN |
| pZW212 | AMPRCEN LEU2 mot1-301-3HA::KAN |
| pZW213 | AMPRCEN LEU2 mot1-301-K101R-K109R-3HA::KAN |
| pZW144 | AMPR 2μm LEU2 SMT3 |
| pZW55 | AMPRCEN URA3 MOT1-FLAG::KAN |
| pZW56 | AMPRCEN URA3 mot1Δ1282-1867-FLAG::KAN |
| pZW57 | AMPRCEN URA3 mot1Δ262-1279-FLAG::KAN |
| pZW153 | AMPRCEN URA3 mot1Δ901-1867-FLAG::KAN |
| pZW154 | AMPRCEN URA3 mot1Δ592-1867-FLAG::KAN |
| pZW155 | AMPRCEN URA3 mot1Δ101-174-FLAG::KAN |
| pZW197 | AMPRCEN URA3 mot1Δ101-174-3HA::KAN |
| pZW195 | AMPRCEN URA3 mot1-K101R-K109R-3HA::KAN |
| pZW196 | AMPRCEN URA3 mot1-K159,169,174R-3HA::KAN |
| pZW193 | AMPRCEN URA3 mot1-K101R-3HA::KAN |
| pZW194 | AMPRCEN URA3 mot1-K109R-3HA::KAN |
| pZW164 | AMPRCEN LEU2 mot1-301-K101R-K109R |
| pZW165 | AMPRCEN LEU2 mot1-301-K159,169,174R |
| pZW187 | AMPRCEN LEU2 mot1-301-K101R |
| pZW188 | AMPRCEN LEU2 mot1-301-K109R |
| pZW215 | AMPRCEN LEU2 mot1-24-3HA::KAN |
| pZW216 | AMPRCEN LEU2 mot1-42-3HA::KAN |
| pGP564 | KANR 2μm LEU2 |
| pGP678 | KANR 2μm LEU2 UBC5 |
| pGP679 | KANR 2μm LEU2 MOT1 |
| pGP680 | KANR 2μm LEU2 UBC4 |
| pCS6514 | AMPR 2μm LEU2 GAL4-AD-HA-SLX5 |
| pCS6821 | AMPR 2μm LEU2 GAL4-AD-HA-SLX8 |
| pGP568 | AMPR 2μm LEU2 GAL4-AD-HA-slx5-104 |
| pGP569 | AMPR 2μm LEU2 GAL4-AD-HA-slx8-103 |
| pZW201 | KANR 2μm TRP1 GAL4-BD-MYC-MOT1 |
| pZW203 | KANR 2μm TRP1 GAL4-BD-MYC-mot1Δ14 (1-1387) |
| pGADT7 | AMPR 2μm LEU2 GAL4-AD |
| pGBKT7 | KANR 2μm TRP1 GAL4-BD |
Yeast two-hybrid assays.
pGBKT7- and pGADT7-based plasmids containing Gal4BD or Gal4AD fused to SLX5 and SLX8 were described previously (19). slx5-104, slx8-103, MOT1, and mot1Δ14 were cloned into pGADT7 or pGBKT7 by standard PCR cloning. Combinations of plasmids were transformed into the yeast two-hybrid reporter strain PJ69-4A and selected on SC plates lacking leucine and tryptophan. Interaction was then determined using SC plates lacking leucine, tryptophan, and adenine.
Preparation of protein extracts.
For cycloheximide chase experiments, crude protein extracts were prepared by the post-alkaline extraction method, as described previously (27). Briefly, cells were resuspended in 200 μl 0.1 M NaOH, incubated for 8 min at room temperature, pelleted, resuspended in 40 μl sodium dodecyl sulfate (SDS) sample buffer, boiled for 3 min, and pelleted again. Ten microliters of supernatant was loaded in each lane for SDS-polyacrylamide gel electrophoresis (PAGE).
For all other experiments, crude protein extracts were prepared by glass bead beating. Typically, yeast cells from 50 ml log-phase culture were pelleted; frozen on dry ice briefly; resuspended in 350 μl lysis buffer containing 50 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 10 mM N-ethylmaleimide, 1% Triton X-100, and protease inhibitors (no. 1836170; Roche); and incubated on ice for 30 min before lysis. Cells were disrupted by vortexing them 10 times (60 s each) in a multitube vortexer with 700-μl glass beads at 4°C. The protein extracts were then clarified by centrifugation at 16,000 × g for 15 min.
Assays of protein half-life.
Yeast cultures were grown overnight at 30°C to log phase in selective medium to maintain plasmids carrying the HA-tagged MOT1 alleles. To start the chase, 1 ml culture was first collected at time zero in an Eppendorf tube preloaded with 10 μl 10% sodium azide. Cells were then pelleted and frozen on dry ice. Cycloheximide (50 mg/ml [no. C7698; Sigma]) was added to the remainder of the culture to give a final concentration of 0.5 mg/ml, and 1-ml samples were collected every 10 or 20 min in tubes containing sodium azide as described above and frozen on dry ice. Crude extracts were prepared by the post-alkaline extraction method. Ten microliters of supernatant was loaded for SDS-PAGE, followed by Western analysis using anti-HA antibody (no. SC-7392; Santa Cruz) to detect Mot1 or anti-G6PDH (no. A9521; Sigma) to detect G6PDH as a loading control. Western signals for Mot1 or Spt20 were quantified by ImageJ, with G6PDH as a loading control. The measured data were then analyzed by Origin8 and fit with the exponential decay function y = y0 + Ae−x/t.
In vivo SUMOylation assay.
Crude protein extracts were prepared from 50 ml log-phase culture by the glass bead beating method, as described above. To immunoprecipitate HA-tagged Mot1, 2 to 3 mg crude extract was diluted in 300 μl lysis buffer, diluted with 900 μl immunoprecipitation dilution buffer (lysis buffer without N-ethylmaleimide or Triton), and incubated with 40 μl anti-HA beads (no. SC-7392AC; Santa Cruz) at 4°C for 2 h. After immunoprecipitation, the beads were washed twice with 1 ml washing buffer (9.1 mM Na2HPO4, 1.7 mM NaH2PO4, 150 mM NaCl, 0.01% SDS, pH 7.4), resuspended in 40 μl SDS sample buffer (10% glycerol, 2% SDS, 62.5 mM Tris [pH 6.8], 0.015% bromophenol blue), and boiled for 3 min. Immunoprecipitated samples were then subjected to SDS-PAGE, followed by Western analysis using anti-SUMO antibody (from Steve Brill) to detect SUMOylated Mot1.
RESULTS
The mot1-301 phenotypes are due to reduced protein level.
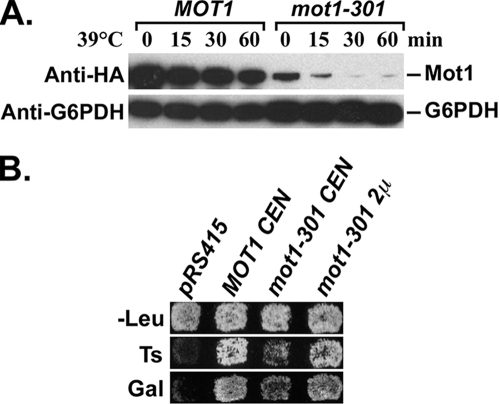
We previously reported that the mot1-301 mutation causes several phenotypes, including Gal− and temperature-sensitive growth, all of which can be suppressed by mutations in genes that encode SUMO pathway components, including SLX5 and SLX8 (58). To understand the mechanism behind these phenotypes, we first determined the defect of the Mot1-301 protein. Strains containing genomic HA-tagged MOT1 or mot1-301 were grown to log phase at the permissive temperature, and Western blots were used to assess protein levels after the cultures were shifted to the nonpermissive temperature. The Mot1-301 protein level was lower than that of wild-type Mot1, even at the permissive temperature, and was dramatically decreased after shifting to 39°C (Fig. 1A). If the major defect of Mot1-301 is protein abundance, then simply increasing the amount of the mutant protein might reverse the mot1-301 phenotype. As predicted, a high-copy-number 2μm mot1-301 plasmid strongly reversed the Ts− and Gal− phenotypes of a strain containing a mot1-301 genomic mutation, and even a low-copy-number CEN mot1-301 plasmid partially complemented mot1-301 (Fig. 1B). Combined, these results indicated that Mot1-301 was present at reduced levels and that simply increasing the Mot1-301 level suppressed mot1-301.
FIG. 1.
The mot1-301 phenotypes are due to reduced protein level. (A) Western blot showing the steady-state level of HA-tagged Mot1 and Mot1-301 during a time course after a temperature shift from 30°C to 39°C. Mot1 was detected by anti-HA antibody. G6PDH served as a loading control. (B) A mot1-301 strain was transformed with the indicated CEN and 2μm plasmids. Transformants were replica plated to SC-Leu plates at 30°C (-Leu) and 39°C (Ts) and to an SC-Leu plate containing galactose (Gal). Photos were taken after 2 days. The mot1-301 Ts− and Gal− phenotypes are reversed by CEN and 2μm mot1-301.
Mot1 is modified by SUMO in vivo.
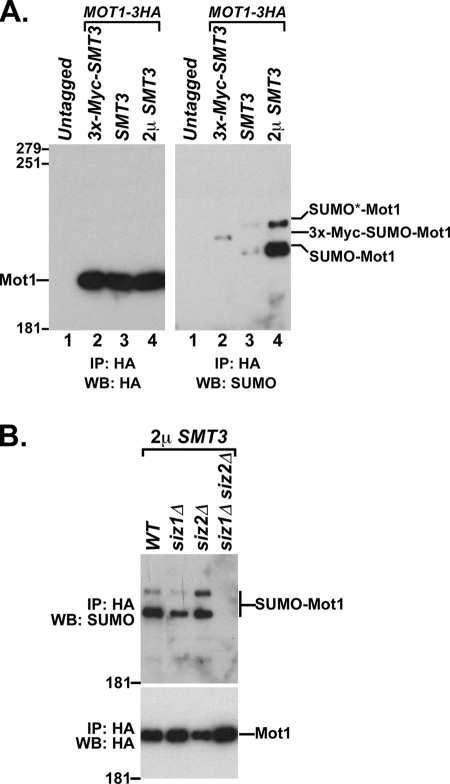
Because mot1-301 can be suppressed by mutations in the SUMO pathway, it seemed likely that Mot1 was the direct target of SUMOylation. To test this possibility, HA-tagged Mot1 was immunoprecipitated from a crude cell lysate and analyzed by Western blotting using an anti-SUMO antibody. The results revealed a weak band with reduced mobility compared to that for unmodified Mot1 and a second band at an even higher position in the gel (Fig. 2A, right, lane 3). Both bands were absent in a Mot1 untagged strain (Fig. 2A, right, lane 1) and were strongly elevated when SMT3, which encodes SUMO in yeast, was overexpressed from a 2μm vector (Fig. 2A, right, lane 4). Replacement of wild-type SMT3 with Myc3-tagged SMT3 resulted in a shifting of the bottom band to a reduced mobility (Fig. 2A, right, lane 2), as expected because the addition of the Myc3 epitope to SUMO generates a ∼20-kDa mobility shift relative to that of SUMO alone (37). The upper band was not detectable in the Myc3-SMT3 strain, perhaps due to the reduced conjugation of epitope-tagged SUMO that has been reported for some proteins (16, 59). To further determine whether the bands recognized by the anti-SUMO antibody are due to SUMOylation of Mot1, we analyzed the effect of deletions of SIZ1 and SIZ2, which encode the major SUMO E3 proteins in yeast (Fig. 2B). The intensities of both bands were reduced by deletion of SIZ1, mostly unaffected by deletion of SIZ2, and reduced to an undetectable level by deletion of both SIZ1 and SIZ2, consistent with previously reported redundancy between SIZ1 and SIZ2 (23) and the abilities of siz1Δ to partially suppress mot1-301 and of siz1Δ siz2Δ to completely suppress mot1-301 (58). Combined, these results demonstrated that Mot1 was modified by SIZ1-SIZ2-dependent SUMOylation in vivo.
FIG. 2.
Mot1 is SUMOylated in vivo. (A) Mot1-HA was immunoprecipitated with anti-HA beads from an untagged strain (lane 1), an smt3Δ strain containing a CEN Myc3-SMT3 plasmid (lane 2), an SMT3+ strain containing empty vector (lane 3), or an SMT3+ strain containing a 2μm SMT3 plasmid (lane 4). The immunoprecipitated (IP) samples were subjected to SDS-PAGE and Western blot (WB) analysis with anti-HA antibody (left) or anti-SUMO antibody (right) to detect total Mot1 and SUMOylated Mot1, respectively. The numbers on the side of the gel are molecular mass markers. (B) Total lysates from the indicated strains overexpressing SMT3 were subjected to the same Mot1 in vivo SUMOylation assay as for panel A. WT, wild type.
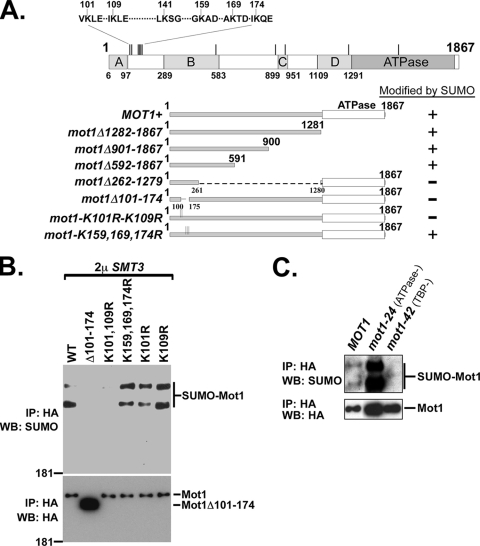
SUMO can be conjugated to lysine residues in consensus (ΨKxE/D) and nonconsensus motifs (9, 20, 21). Mot1 contains 11 lysine residues in SUMO consensus motifs, with a cluster of six consensus motifs located between amino acids 101 and 174 (Fig. 3A). To map the likely SUMO conjugation site(s), deletions of Mot1 were designed and tested for their ability to be SUMOylated in vivo. The mutants that were constructed and their corresponding SUMOylation statuses are summarized in Fig. 3A. The N-terminal 591 amino acids of Mot1 (mot1Δ592-1867) were sufficient to be SUMOylated, and deletion of two nonoverlapping internal regions, comprising amino acids 262 to 1279 or amino acids 101 to 174, abolished SUMOylation. Because the region comprising amino acids 101 to 174 overlapped the cluster of SUMO consensus sites, we next created missense mutations of the lysines within the consensus motifs. A K101R-and-K109R double mutant (mot1-K101R-K109R) abolished Mot1 SUMOylation, while single K101R or K109R mutations or a triple mutant of K159R, K169R, and K174R (mot1-K159,169,174R) had no effect (Fig. 3B). We also assayed SUMOylation of two Mot1 missense mutants, Mot1-24 (51) and Mot1-42 (7), which are defective for ATPase and TBP-binding activities, respectively (Fig. 3C). Interestingly, the ATPase-defective Mot1-24 mutant was much more extensively SUMOylated than wild-type Mot1, while SUMOylation of the TBP-binding defective Mot1-42 mutant was undetectable. We conclude that K101, K109, and the internal region from residue 262 to 591, which is required for binding of Mot1 to TBP (7), are required for SUMOylation of Mot1, with K101 and K109 being likely conjugation sites, and that SUMOylation of Mot1 might require its TBP-binding activity and be inhibited by the ATPase activity.
FIG. 3.
SUMOylation of Mot1. (A) Summary of Mot1 deletions or point mutations and their in vivo SUMOylation statuses. A diagram of Mot1 is presented at the top, showing the locations of the conserved domains (A to D), the C-terminal ATPase domain, and the 11 consensus SUMO motifs (vertical lines). The SUMO consensus motifs within the N-terminal cluster are displayed. (B) Western blot (WB) of immunoprecipitated (IP) samples for assay of in vivo SUMOylation levels of Mot1 mutants within the N-terminal cluster, using anti-SUMO antibody (top) or anti-HA antibody (bottom). WT, wild type. (C) Similar in vivo SUMOylation assay for comparison of the SUMOylation levels of wild-type Mot1, Mot1-24, and Mot1-42.
Mot1 is an authentic in vivo substrate for the SUMO-targeted degradation pathway.
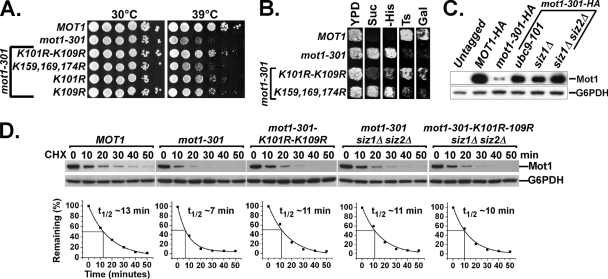
The definitive genetic test for identifying a physiologically relevant posttranslational modification site is to determine whether mutation of the suspected modification site causes the same phenotype as mutations in the modification genes. In the case of Mot1, since mutations in the SUMO pathway suppress mot1-301, we predicted that mutation of the relevant SUMOylation site(s) would suppress the mot1-301 phenotypes. As predicted, and strongly paralleling their effects on SUMOylation, a K101R-K109R double mutation built into mot1-301 strongly reversed the mot1-301 Ts− phenotype, while K101R or K109R single mutations suppressed to a lesser extent, and the K159,169,174R triple mutation had no effect (Fig. 4A). A similar suppression effect was also observed for the other mot1-301 phenotypes, including Gal−, Bur−, and Spt− phenotypes (Fig. 4B). Because mutations in the SUMO pathway, such as ubc9-101 (E2) and siz1Δ siz2Δ (E3 proteins) (58) and introduction of the SUMOylation-defective K101R-K109R mutation into mot1-301, reversed the mutant phenotypes, we tested whether these mutations also reversed the lower protein abundance of Mot1-301. Indeed, the Mot1-301 protein levels at the permissive temperature were restored to near-wild-type levels in ubc9-101 and siz1Δ siz2Δ strains. (Fig. 4C). A cycloheximide chase experiment was performed to determine whether the change in protein level was due to a change in the protein half-life. Indeed, the half-life of Mot1-301 (∼7 min) was much shorter than that of wild-type Mot1 (∼13 min), and a siz1Δ siz2Δ double mutation or introduction of the K101R-K109R mutation into Mot1-301 significantly increased the Mot1-301 half-life (Fig. 4D). The K101R-K109R mutation did not further increase Mot1-301 stability in a siz1Δ siz2Δ strain, suggesting that they functioned in the same linear pathway. The strong correlation between the loss of Mot1 SUMOylation, the increased protein stability of Mot1-301, and the suppression of mot1-301 phenotypes strongly argues that Mot1 is an authentic substrate for SUMO in vivo and that the SUMO-regulated protein instability of Mot1-301 is the mechanism behind the phenotype.
FIG. 4.
Effects of disrupting SUMOylation of Mot1-301. (A) CEN LEU2 plasmids carrying the indicated mot1 alleles were transformed into a mot1Δ strain containing a CEN URA3 MOT1 plasmid. The CEN MOT1 plasmid was shuffled out using 5-FOA selection, and 10-fold serial dilutions of the resulting strains were plated onto yeast extract-peptone-dextrose plates and incubated at 30°C and 39°C for 3 days. (B) A complete test of all four phenotypes (Bur, Spt, Ts, and Gal) of the indicated MOT1 alleles. YPD, yeast extract-peptone-dextrose. (C) Total protein levels of genomic HA-tagged Mot1 and Mot1-301 in the indicated strains were examined by SDS-PAGE followed by anti-HA Western blot analysis. (D) CEN plasmids expressing HA-tagged Mot1, Mot1-301, or Mot1-301-K101R-K109R were transformed into SIZ1+ SIZ2+ or siz1Δ siz2Δ strains, as indicated, and their half-lives assayed during a cycloheximide chase time course. Quantification of Mot1 half-life is shown below the corresponding Western blots.
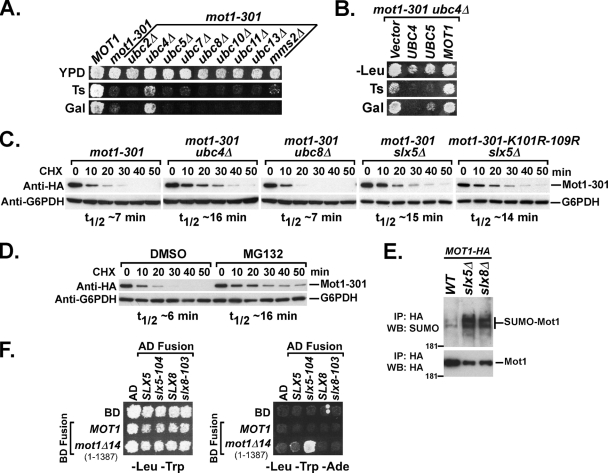
In addition to mutations in the SUMO pathway, we also identified slx5 and slx8 mutations in the mot1 suppressor screen. The recent characterization of Slx5-Slx8 as a STUbL suggests that the ubiquitin-proteasome pathway is also involved. Our mot1 suppressor selection did not identify any mutations in the ubiquitin pathway other than SLX5 and SLX8, however, either because the ubiquitin pathway is not involved, because the selection was not saturated, or due to redundancy within the ubiquitin pathway. To determine whether the ubiquitin-proteosome pathway is involved, we first created double mutants of mot1-301 with deletions of genes that encode nine nonessential ubiquitin E2 proteins. Of the nine ubiquitin E2 deletions tested, only ubc4Δ suppressed mot1-301 for the Ts− and Gal− phenotypes (Fig. 5A), although we note that suppression of mot1-301 by ubc4Δ was not very strong, as the Bur− and Spt− phenotypes were barely affected (data not shown). It was somewhat unexpected that ubc4Δ suppressed mot1-301 at all, since UBC4 and UBC5 encode 92% identical proteins that have overlapping functions (47). The suppression of mot1-301 by ubc4Δ and not by ubc5Δ could be due to the fact that UBC4 is expressed at a much higher level than UBC5 in growing cells (47), or they might have different substrate specificities. To distinguish between these possibilities, we overexpressed UBC5 in a mot1-301 ubc4Δ strain and found that it reversed the suppression by ubc4Δ (Fig. 5B). This result suggests that UBC4 and UBC5 both can contribute to the destabilization of Mot1-301, but the higher expression level of UBC4 effectively masks the contribution of UBC5. We could not test for a combinatorial effect of ubc4Δ ubc5Δ on mot1-301, due to the near lethality of the ubc4Δ ubc5Δ double mutant, even in a MOT1+ background. The specificity of suppression by ubc4Δ was also reflected in assays of Mot1-301 protein half-life, where ubc4Δ and slx5Δ restored Mot1-301 stability, but ubc8Δ, which did not suppress the mutant phenotypes, also had no effect on Mot1-301 stability (Fig. 5C). Mot1-301 was also stabilized in cells treated with the proteasome inhibitor MG132, indicating a direct involvement of the proteosome on the half-life of Mot1-301 (Fig. 5D). Finally, deletion of SLX5 or SLX8 resulted in an accumulation of SUMOylated Mot1 species (Fig. 5E), as expected if Slx5-Slx8 functions downstream of SUMOylation to target Mot1 for degradation. The K101R-K109R mutation did not have a combinatorial effect with deletion of SLX5 on the stability of Mot1-301 (Fig. 5C), suggesting that SUMOylation and SLX5-SLX8 were in the same epistasis pathway. If Slx5-Slx8 directly destabilizes Mot1-301, physical interactions between the proteins should be detectable. Indeed, a yeast two-hybrid interaction was detected between a Mot1 C-terminal truncation, Mot1Δ14 (1-1387), and Slx5-104, a RING finger mutant (58) (Fig. 5F). No interaction was observed, however, with wild-type Mot1 or Slx5, perhaps because wild-type Mot1 is not strongly targeted and because interaction with wild-type Slx5 would result in its degradation. Combined, these results indicated that the Slx5-Slx8 ubiquitin E3 protein functions with the Ubc4-Ubc5 E2 to mediate proteosome-dependent degradation of SUMOylated Mot1-301.
FIG. 5.
The ubiquitin-proteasome pathway is involved. (A) mot1-301 was crossed into strains containing the indicated ubiquitin E2 gene deletions, and the double-mutant phenotype was determined. Only ubc4Δ specifically suppressed the mot1-301 Ts− and Gal− phenotypes. YPD, yeast extract-peptone-dextrose. (B) A mot1-301 ubc4Δ strain was transformed with 2μm LEU2 plasmids carrying the indicated genes. Transformants were patched and replica plated to SC-Leu plates at 30°C (-Leu) and 39°C (Ts) and to an SC plate containing galactose (Gal). Photos were taken after 2 days. 2μm UBC5 reversed the suppression of mot1-301 by ubc4Δ for the Ts+ and Gal+ phenotypes. (C) A CEN plasmid containing mot1-301-HA or mot1-301-K101R-K109R-HA was transformed into strains with the indicated genotypes, and Mot1-301 stability was assayed by Western blotting during a cycloheximide chase. (D) A CEN mot1-301-HA plasmid was transformed into a pdr5Δ strain, and Mot1 stability was assayed upon addition of dimethyl sulfoxide (DMSO) or 50 μM MG132 in DMSO. (E) The levels of Mot1 SUMOylation in wild-type, slx5Δ, and slx8Δ strains were assayed using an anti-SUMO Western blot (WB). IP, immunoprecipitation. (F) Two-hybrid interactions between Slx5-Slx8 and Mot1. Wild-type and mutant alleles were cloned into the pGADT7 (Gal4AD, LEU2+) or pGBKT7 (Gal4BD, TRP1+) vector as indicated. Combinations of plasmids were then transformed into yeast strain PJ69-4A, which contains Gal UAS-driven ADE2 as a reporter gene, and selected on SC plates lacking leucine and tryptophan. Transformants were then patched and replica plated to SC-Leu-Trp (left) and SC-Leu-Trp-Ade (right) plates. Slx5-104 was found to interact with Mot1Δ14.
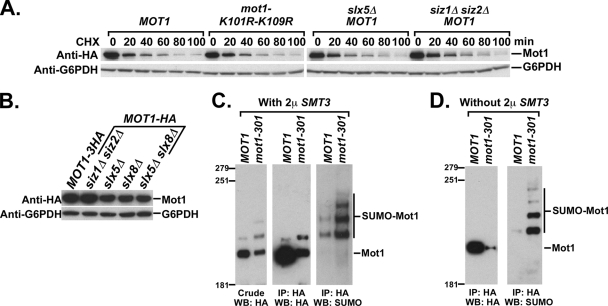
SUMO and Slx5-Slx8 have no detectable effect on the stability of wild-type Mot1.
Because mutations in the SUMO-Slx-ubiquitin pathway affected stability of the mutant Mot1-301 protein, we next determined whether the stability of wild-type Mot1 was also affected. We were unable to detect any increase in either the protein half-life (Fig. 6A) or the steady-state protein level (Fig. 6B) of wild-type Mot1 by deleting SIZ1, SIZ2, SLX5, or SLX8 or by introducing the K101R-K109R mutations into MOT1+. In fact, the Mot1 protein level appeared to decrease slightly in slx5Δ and slx8Δ strains. The lack of any increase of wild-type Mot1 level raised the issue of why SUMOylation strongly destabilized the mutant form of Mot1 but not its wild-type counterpart. To address this issue, we compared the in vivo SUMOylation statuses of Mot1 and Mot1-301 with (Fig. 6C) and without (Fig. 6D) SUMO overexpression. Interestingly, Mot1-301 was SUMOylated to a much greater extent than wild-type Mot1 under both conditions, especially in light of its reduced protein level. The relative proportion of SUMOylated to un-SUMOylated Mot1 could be assessed from the anti-HA Western blot when SUMO is overexpressed. Under this condition, we estimate that ∼34% of Mot1-301 and ∼7% of Mot1 are SUMOylated (Fig. 6C). Thus, the differential stability of Mot1-301 compared to that of wild-type Mot1 likely arises from differential levels of SUMOylation. SUMOylation likely destabilized a subpopulation of wild-type Mot1, but the level of SUMOylation is so low that any effect might be undetectable within the larger un-SUMOylated Mot1 population. The result presented in Fig. 5E is consistent with this conclusion, as SUMOylated Mot1 accumulated in slx5Δ and slx8Δ strains.
FIG. 6.
Differential regulation of Mot1 and Mot1-301 by SUMOylation. (A) Western blot for assaying the half-lives of wild-type Mot1 and Mot1-K101R-K109R during a cycloheximide chase in strains with the indicated genotypes. (B) Western blot showing the steady-state protein levels of wild-type Mot1 in strains with the indicated genotypes. (C) HA-tagged Mot1 and Mot1-301 strains were transformed with a 2μm SMT3 plasmid and assayed for SUMOylation. An anti-HA Western blot (WB) against crude extracts is shown in the left panel. Mot1-HA and Mot1-301-HA were immunoprecipitated (IP) with anti-HA beads and analyzed using anti-HA antibody (center) or anti-SUMO antibody (right). (D) Same as panel C, except in the absence of SUMO overexpression.
SUMO-targeted degradation in response to canavanine.
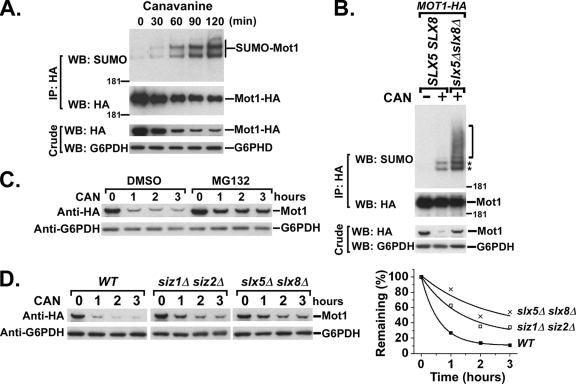
The results presented above show that SUMOylation strongly targets the Mot1-301 mutant protein, perhaps as part of a quality control system, yet also targets a small population of wild-type Mot1 protein. We hypothesized that the population of wild-type Mot1 modified by SUMO might be defective or misfolded proteins analogous to Mot1-301. One prediction of this model is that under conditions where protein misfolding is induced, there should be corresponding increases in SUMOylation and SUMO-dependent degradation of wild-type Mot1. To test this prediction, we grew cells in the presence of canavanine, an arginine analogue. Canavanine can be incorporated into proteins during translation, thus generating abnormal canavanine-containing proteins, which are recognized and degraded by the proteasome through a pathway that requires the Ubc4 and Ubc5 ubiquitin E2 proteins (12, 47).
Cells expressing Mot1-HA were grown to log phase in medium lacking arginine, canavanine was added, cells were harvested during a time course, and extracts were assayed for in vivo SUMOylation of Mot1. Anti-SUMO Western analyses revealed a clear decrease of Mot1 protein that coincided with increase of Mot1 SUMOylation across the time course (Fig. 7A). Canavanine treatment caused even higher levels of SUMOylation in an slx5Δ slx8Δ strain, including the appearance of a ladder of slowly migrating species above the two major SUMO-Mot1 bands, indicative of SUMO-containing chains (Fig. 7B). A longer exposure of this blot revealed that the ladder was also present in the SLX5+ SLX8+ strain, but at greatly reduced levels relative to those for the two main bands (data not shown). A similar experiment performed in the presence of the proteasome inhibitor MG132 revealed that Mot1 was degraded by the proteasome upon canavanine treatment (Fig. 7C). Interestingly, canavanine-induced degradation of Mot1 was impeded by deletions of either the SIZ1 and SIZ2 SUMO E3 proteins or SLX5 and SLX8 (Fig. 7D). The results of these experiments therefore indicated that SUMOylation and Slx5-Slx8 target abnormal Mot1 proteins induced by canavanine treatment and direct them for proteasome-mediated degradation.
FIG. 7.
Canavanine-induced SUMO-targeted degradation of wild-type Mot1 protein. (A) Cells were grown overnight in medium lacking arginine before incubation in 30 μg/ml canavanine. Cultures were harvested every 30 min after canavanine addition for up to 2 h, followed by Western blotting (WB) for detection of in vivo SUMOylation of Mot1-HA. IP, immunoprecipitation. (B) Western blot for assaying the canavanine-induced SUMOylation of Mot1-HA expressed in wild-type or slx5Δ slx8Δ strains. The two major SUMO-Mot1 species are indicated by asterisks, and a ladder of additional SUMO-Mot1 species with reduced mobility are indicated by the bracket. (C) Cells were treated with canavanine, together with DMSO alone or 50 μM MG132 in DMSO, and total Mot1 protein levels were measured by anti-HA Western blot analysis during the time course. (D) Cells with the indicated genotypes were grown and treated with canavanine (CAN) as described for panel A and harvested every hour after canavanine treatment. Total lysates were prepared and probed with anti-HA antibody to detect Mot1 levels. Quantification of the Western blots is shown to the right. G6PDH served as a loading control for all experiments. WT, wild type.
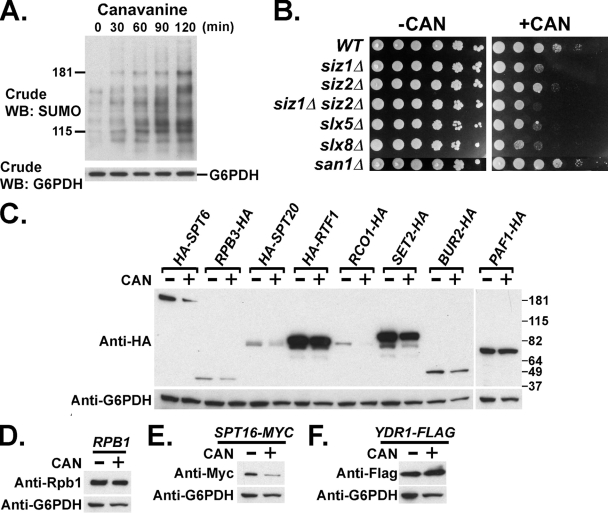
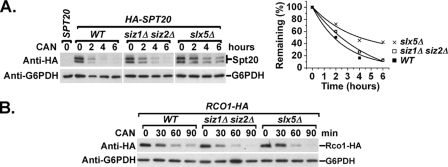
To determine whether the canavanine-stimulated SUMOylation and degradation extend beyond Mot1 to other proteins, Western blot analyses were performed on crude extracts prepared from canavanine-treated cells. Consistent with previous results for Arabidopsis (26), canavanine treatment caused an increase in SUMOylated proteins in yeast crude extracts, suggesting that SUMO targets other proteins in response to canavanine (Fig. 8A). In addition, slx5Δ, slx8Δ, siz1Δ, and siz1Δ siz2Δ double mutants were sensitive to the presence of canavanine in the growth medium, while siz2Δ was moderately sensitive and san1Δ was equivalent to the wild type (Fig. 8B). These results indicated that SUMO and Slx5-Slx8 were required for the cell to survive under this condition and that they and San1 might be in different pathways. To determine if other proteins were destabilized by growth in canavanine-containing medium, the levels of 11 arbitrarily selected transcription factors were tested after growth in the presence of canavanine. The levels of Spt6, Spt16, Spt20, Set2, and Rco1 decreased to various extents upon canavanine treatment (Fig. 8C to F). Interestingly, the decrease in Spt20 level was reproducibly attenuated by siz1Δ siz2Δ and was further impeded in slx5Δ strains (Fig. 9A). The decreased levels of the other four proteins, however, were not dependent on Siz1-Siz2 or Slx5 (Fig. 9B). Taken together, these results suggested that the SUMO-targeted, Slx5-Slx8-mediated degradation pathway might have a more general role in a cellular protein quality control system, targeting some but not all proteins.
FIG. 8.
Canavanine-induced SUMOylation and degradation of other proteins besides Mot1. (A) Crude lysates of cells grown overnight in medium lacking arginine, followed by a time course after addition of 30 μg/ml canavanine. Lysates were subjected to Western blotting (WB) with anti-SUMO antibody to determine the effect of canavanine on SUMOylation level in the crude extract. G6PDH served as a loading control. (B) Tenfold serial dilutions of the indicated strains were spotted onto SC-arginine (-CAN) and SC-arginine plus 2.5 μg/ml canavanine (+CAN) plates, showing the sensitivity to the drug. Photos were taken after incubation at 30°C for 3 days. WT, wild type. (C to F) Cells were grown overnight in medium lacking arginine. Half of the culture was collected (CAN-), and the other half was further incubated in the presence of 30 μg/ml canavanine for 2 h (CAN+). Crude lysates were prepared and analyzed by Western blotting using anti-HA antibody to detect HA-tagged Spt6, Rpb3, Spt20, Rtf1, Rco1, Set2, Bur2, and Paf1; anti-Rbp1 antibody to detect Rpb1; anti-Myc antibody to detect Myc-tagged Spt16; and anti-Flag antibody to detect Flag-tagged yDr1.
FIG. 9.
Siz1-Siz2 and Slx5 effects on canavanine-induced degradation. (A) Wild-type (WT), siz1Δ siz2Δ, and slx5Δ strains expressing HA-tagged Spt20 were grown and treated with 60 μg/ml canavanine (CAN) and harvested every 2 h after canavanine treatment. Crude lysates were prepared and probed with anti-HA antibody to detect Spt20 levels. HA-Spt20 was revealed as a doublet by anti-HA Western blotting, since both bands were absent in the lysates prepared from a SPT20 untagged strain. G6PDH served as a loading control. Quantification of the Western blots is shown to the right. (B) Wild-type, siz1Δ siz2Δ, and slx5Δ strains expressing HA-tagged Rco1 were grown and treated with 30 μg/ml canavanine and harvested every 30 min after canavanine treatment. Crude lysates were prepared and probed with anti-HA antibody to detect Rco1 levels. G6PDH served as a loading control.
DISCUSSION
To understand the functions of SUMO-targeted ubiquitylation, it is important to identify the in vivo targets and consequences of this modification. The only in vivo substrate identified to date for the SUMO-targeted ubiquitylation pathway is mammalian PML (28, 55), where SUMO-targeted ubiquitylation is required for PML degradation. In this study, we identify another in vivo target for SUMO-mediated degradation and, moreover, provide evidence that it has a role in protein quality control. Our results show that Mot1 is a direct target of a pathway that requires the Siz1 and Siz2 SUMO E3 proteins, the Ubc4-Ubc5 ubiquitin E2 protein, the Slx5-Slx8 ubiquitin E3 protein, and the proteasome, resulting in selective degradation. Importantly, a Mot1 mutant is targeted to a much greater extent than wild-type Mot1, but wild-type Mot1 becomes a stronger target when cells are grown in the presence of canavanine, which has previously been used to interfere with protein function, presumably by inducing a misfolded or defective state (13, 47). Increased degradation of a mutant protein relative to the wild-type level, combined with increased degradation in response to canavanine, is a hallmark of a quality control function. The proposed quality control system that we detect likely extends beyond Mot1, since slx5Δ, slx8Δ, and siz1Δ siz2Δ strains are canavanine sensitive, SUMOylation of other proteins increases upon canavanine treatment, and, in a small-scale screen, at least one other protein, Spt20, is degraded upon canavanine treatment in an SLX5 and SIZ1-SIZ2-dependent manner. It should not be too surprising that SUMOylation participates in quality control processes, as the levels of SUMO conjugates are dramatically increased in mammalian cells when cells are exposed to stresses, such as heat, H2O2, and ethanol (33, 46), and in plants that are exposed to heat, ethanol, or canavanine (26). Interestingly, these effects mirror an induction of ubiquitin modification that is observed upon cellular stresses (4, 39, 48). The identification of a quality control function for the Slx5-Slx8 STUbL provides a critical link between SUMO and quality control, mediated through the ubiquitin-proteosome pathway.
The proposed role for Slx5-Slx8 in quality control displays some similarities to and differences from the San1-mediated nuclear quality control system. Like SLX5-SLX8, SAN1 encodes a ubiquitin E3 protein (8) that targets some mutant proteins, but not their wild-type counterparts, for degradation (12). These two pathways appear to have nonoverlapping sets of substrates, as san1Δ suppresses cdc68-1 but does not suppress mot1-301, and reciprocally, slx5Δ and ubc9-101 mutations suppress mot1-301 but do not suppress cdc68-1 (data not shown). These E3 proteins might respond to different defects, since slx5Δ and slx8Δ are canavanine sensitive, while san1Δ is canavanine resistant (Fig. 8B). In addition, San1 and Slx5-Slx8 apparently function with different ubiquitin E2 proteins. Deletion of SAN1, CDC34, or UBC1, but not UBC4, stabilizes Sir4-9, suggesting that Cdc34 and Ubc1 are in vivo ubiquitin E2 proteins for San1. In contrast, deletion of UBC4 specifically suppresses mot1-301 and increases Mot1-301 stability (Fig. 5), suggesting that Ubc4 is an in vivo ubiquitin E2 protein for Slx5-Slx8. Consistent with this conclusion, other links have been established between Slx5-Slx8 and Ubc4: Slx5 interacts physically with Ubc4 by a glutathione S-transferase pulldown assay (57), the Slx5-Slx8 complex stimulates Ubc4 (54, 57, 60) or Ubc5 (36) in vitro, and both ubc4Δ ubc5Δ and slx5Δ are sensitive to canavanine and defective for degradation of canavanine-containing proteins (47). Taken together, these results suggest that the Slx5-Slx8 and San1 pathways are nonredundant quality control systems that target different substrates. Although there is no apparent overlap with the San1 system, we note that destabilization of Mot1 and Spt20 was not completely reversed in our studies, suggesting overlap or redundancy with other currently undefined factors. The basis for the differential and overlapping roles of Slx5-Slx8 with respect to other systems will be an interesting subject for future studies.
For any quality control system, a difficult step is discerning which features of a defective protein are recognized to initiate the process. Our deletion analysis (Fig. 3) indicates that, in addition to the inferred SUMO conjugation sites, a second region of Mot1, located between residues 262 and 591, is required for SUMOylation. This region overlaps a conserved B block that is required for binding of Mot1 to TBP (7). In contrast, deletion of the ATPase domain or conserved blocks C and D, which are not required for binding to TBP, are not required for SUMOylation of Mot1 (1). Interestingly, the mot1-42 allele that reduces Mot1-TBP-DNA complex formation also reduces SUMOylation. In contrast, a missense mutation within the ATPase domain (mot1-24) that abolishes ATPase activity results in increased SUMOylation (Fig. 3), similar to mot1-301 (Fig. 6), which contains a missense mutation just outside the ATPase domain. Although the determinants clearly need to be investigated further, these results suggest that TBP binding might be required for targeting of Mot1 for SUMOylation. SUMOylation of proteins in complexes with other proteins or with DNA appears to be a recurring theme. For example, SUMOylation of mammalian BMAL1 requires its heterodimerization partner CLOCK (3), SUMOylation of thymine DNA glycosylase (TDG) is stimulated by binding to double-stranded DNA (17), and SUMOylation of PCNA by Siz1 is enhanced in vitro in the presence of DNA and replication factor C, which facilitates PCNA loading onto DNA (40). It was proposed that the loading of PCNA onto DNA changes some property, such as its conformation, that makes it a better SUMO substrate. It remains to be seen whether conformational changes in Mot1 affect its modification, but conformational changes have been implied from structural studies of the homologous Snf2/Swi2 family member SsoRad54 (11, 51). SUMOylation of TDG and PCNA does not result in their degradation, but there is no evidence thus far that Slx5-Slx8 is a downstream effector of SUMOylated TDG or PCNA.
Although this study shows that Slx5-Slx8 can participate in quality control, we do not wish to imply that all SUMOylated proteins are targets or that Slx5-Slx8 targets only defective proteins. Different outcomes can result from the same posttranslational modification of different substrates, either due to intrinsic structural changes in the substrate or due to diversity of downstream effectors that recognize the modified substrate, and this is certainly true for SUMOylation. For the proteins targeted by SUMOylation, at least three different outcomes have been documented: SUMOylation can cause intrinsic changes in protein structure, as occurs with TDG (2, 17, 52); SUMOylation can sterically affect protein-protein interactions, as exemplified by altered interactions of SUMOylated PCNA with DNA polymerase subunits (38, 41); and SUMOylation can recruit downstream effectors, such as the STUbLs that trigger degradation, as occurs for PML (28, 55) and Mot1. The recognition of Mot1 as a quality control target is an important step in understanding Slx5-Slx8, but many fundamental questions remain. Future goals will include identifying whether other STUbLs exist, determining whether other STUbLs always target proteins for destruction, defining the contacts within SUMO and the substrate that are recognized by Slx5-Slx8, and identifying additional substrates that are targeted by Slx5-Slx8.
Acknowledgments
This work was supported by grant GM52486 to G.P. from the National Institutes of Health.
We thank Ed Hurt for SUMO plasmids, Steve Brill for anti-SUMO antibody and yeast two-hybrid plasmids, and members of the laboratory for comments on the manuscript.
Footnotes
Published ahead of print on 12 January 2009.
REFERENCES
- 1.Adamkewicz, J. I., K. E. Hansen, W. A. Prud'homme, J. L. Davis, and J. Thorner. 2001. High affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP). J. Biol. Chem. 27611883-11894. [DOI] [PubMed] [Google Scholar]
- 2.Baba, D., N. Maita, J. G. Jee, Y. Uchimura, H. Saitoh, K. Sugasawa, F. Hanaoka, H. Tochio, H. Hiroaki, and M. Shirakawa. 2005. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435979-982. [DOI] [PubMed] [Google Scholar]
- 3.Cardone, L., J. Hirayama, F. Giordano, T. Tamaru, J. J. Palvimo, and P. Sassone-Corsi. 2005. Circadian clock control by SUMOylation of BMAL1. Science 3091390-1394. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, N., S. Rogers, and M. Rechsteiner. 1987. Microinjection of ubiquitin: changes in protein degradation in HeLa cells subjected to heat-shock. J. Cell Biol. 104547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, X. L., A. Reindle, and E. S. Johnson. 2005. Misregulation of 2μm circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 254311-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cyr, D. M., J. Hohfeld, and C. Patterson. 2002. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27368-375. [DOI] [PubMed] [Google Scholar]
- 7.Darst, R. P., A. Dasgupta, C. Zhu, J. Y. Hsu, A. Vroom, T. Muldrow, and D. T. Auble. 2003. Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. J. Biol. Chem. 27813216-13226. [DOI] [PubMed] [Google Scholar]
- 8.Dasgupta, A., K. L. Ramsey, J. S. Smith, and D. T. Auble. 2004. Sir Antagonist 1 (San1) is a ubiquitin ligase. J. Biol. Chem. 27926830-26838. [DOI] [PubMed] [Google Scholar]
- 9.Denison, C., A. D. Rudner, S. A. Gerber, C. E. Bakalarski, D. Moazed, and S. P. Gygi. 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics 4246-254. [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2233-239. [DOI] [PubMed] [Google Scholar]
- 11.Durr, H., C. Korner, M. Muller, V. Hickmann, and K. P. Hopfner. 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121363-373. [DOI] [PubMed] [Google Scholar]
- 12.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Degradation-mediated protein quality control in the nucleus. Cell 120803-815. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, A. L. 1972. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin). Proc. Natl. Acad. Sci. USA 69422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg, A. L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426895-899. [DOI] [PubMed] [Google Scholar]
- 15.Hampton, R. Y. 2002. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14476-482. [DOI] [PubMed] [Google Scholar]
- 16.Hannich, J. T., A. Lewis, M. B. Kroetz, S. J. Li, H. Heide, A. Emili, and M. Hochstrasser. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2804102-4110. [DOI] [PubMed] [Google Scholar]
- 17.Hardeland, U., R. Steinacher, J. Jiricny, and P. Schar. 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 211456-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch, C., E. Jarosch, T. Sommer, and D. H. Wolf. 2004. Endoplasmic reticulum-associated protein degradation—one model fits all? Biochim. Biophys. Acta 1695215-223. [DOI] [PubMed] [Google Scholar]
- 19.Ii, T., J. R. Mullen, C. E. Slagle, and S. J. Brill. 2007. Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair 61679-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73355-382. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, E. S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 27226799-26802. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106735-744. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 165509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22159-180. [DOI] [PubMed] [Google Scholar]
- 26.Kurepa, J., J. M. Walker, J. Smalle, M. M. Gosink, S. J. Davis, T. L. Durham, D. Y. Sung, and R. D. Vierstra. 2003. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 2786862-6872. [DOI] [PubMed] [Google Scholar]
- 27.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16857-860. [DOI] [PubMed] [Google Scholar]
- 28.Lallemand-Breitenbach, V., M. Jeanne, S. Benhenda, R. Nasr, M. Lei, L. Peres, J. Zhou, J. Zhu, B. Raught, and H. de The. 2008. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10547-555. [DOI] [PubMed] [Google Scholar]
- 29.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 1931361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398246-251. [DOI] [PubMed] [Google Scholar]
- 31.Li, S. J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 202367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Y. C., J. Penninger, and M. Karin. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao, Y., S. D. Desai, and L. F. Liu. 2000. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. 27526066-26073. [DOI] [PubMed] [Google Scholar]
- 34.McClellan, A. J., S. Tam, D. Kaganovich, and J. Frydman. 2005. Protein quality control: chaperones culling corrupt conformations. Nat. Cell Biol. 7736-741. [DOI] [PubMed] [Google Scholar]
- 35.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2000. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 27536316-36323. [DOI] [PubMed] [Google Scholar]
- 36.Mullen, J. R., and S. J. Brill. 2008. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J. Biol. Chem. 28319912-19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panse, V. G., U. Hardeland, T. Werner, B. Kuster, and E. Hurt. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 27941346-41351. [DOI] [PubMed] [Google Scholar]
- 38.Papouli, E., S. Chen, A. A. Davies, D. Huttner, L. Krejci, P. Sung, and H. D. Ulrich. 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19123-133. [DOI] [PubMed] [Google Scholar]
- 39.Parag, H. A., B. Raboy, and R. G. Kulka. 1987. Effect of heat shock on protein degradation in mammalian cells: involvement of the ubiquitin system. EMBO J. 655-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker, J. L., A. Bucceri, A. A. Davies, K. Heidrich, H. Windecker, and H. D. Ulrich. 2008. SUMO modification of PCNA is controlled by DNA. EMBO J. 272422-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfander, B., G. L. Moldovan, M. Sacher, C. Hoege, and S. Jentsch. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436428-433. [DOI] [PubMed] [Google Scholar]
- 42.Prudden, J., S. Pebernard, G. Raffa, D. A. Slavin, J. J. Perry, J. A. Tainer, C. H. McGowan, and M. N. Boddy. 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 264089-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed, S. I. 2006. The ubiquitin-proteasome pathway in cell cycle control. Results Probl. Cell Differ. 42147-181. [DOI] [PubMed] [Google Scholar]
- 44.Reverter, D., and C. D. Lima. 2005. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2756252-6258. [DOI] [PubMed] [Google Scholar]
- 47.Seufert, W., and S. Jentsch. 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang, F., X. Gong, and A. Taylor. 1997. Activity of ubiquitin-dependent pathway in response to oxidative stress. Ubiquitin-activating enzyme is transiently up-regulated. J. Biol. Chem. 27223086-23093. [DOI] [PubMed] [Google Scholar]
- 49.Sitia, R., and I. Braakman. 2003. Quality control in the endoplasmic reticulum protein factory. Nature 426891-894. [DOI] [PubMed] [Google Scholar]
- 50.Song, J., L. K. Durrin, T. A. Wilkinson, T. G. Krontiris, and Y. Chen. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 10114373-14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sprouse, R. O., M. Brenowitz, and D. T. Auble. 2006. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J. 251492-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinacher, R., and P. Schar. 2005. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 15616-623. [DOI] [PubMed] [Google Scholar]
- 53.Strunnikov, A. V., L. Aravind, and E. V. Koonin. 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 15895-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun, H., J. D. Leverson, and T. Hunter. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 264102-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatham, M. H., M. C. Geoffroy, L. Shen, A. Plechanovova, N. Hattersley, E. G. Jaffray, J. J. Palvimo, and R. T. Hay. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10538-546. [DOI] [PubMed] [Google Scholar]
- 56.Ulrich, H. D. 2005. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 15525-532. [DOI] [PubMed] [Google Scholar]
- 57.Uzunova, K., K. Gottsche, M. Miteva, S. R. Weisshaar, C. Glanemann, M. Schnellhardt, M. Niessen, H. Scheel, K. Hofmann, E. S. Johnson, G. J. Praefcke, and R. J. Dohmen. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 28234167-34175. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Z., G. M. Jones, and G. Prelich. 2006. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 1721499-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wohlschlegel, J. A., E. S. Johnson, S. I. Reed, and J. R. Yates III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 27945662-45668. [DOI] [PubMed] [Google Scholar]
- 60.Xie, Y., O. Kerscher, M. B. Kroetz, H. F. McConchie, P. Sung, and M. Hochstrasser. 2007. The yeast Hex3·Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 28234176-34184. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, X., and G. Blobel. 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 1024777-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, J., V. Lallemand-Breitenbach, and H. de The. 2001. Pathways of retinoic acid- or arsenic trioxide-induced PML/RARα catabolism, role of oncogene degradation in disease remission. Oncogene 207257-7265. [DOI] [PubMed] [Google Scholar]