Abstract
The regulation of gene expression by nuclear receptors controls the phenotypic properties and diverse biologies of target cells. In breast cancer cells, estrogen receptor alpha (ERα) is a master regulator of transcriptional stimulation and repression, yet the mechanisms by which agonist-bound ERα elicits repression are poorly understood. We analyzed early estrogen-repressed genes and found that ERα is recruited to ERα binding sites of these genes, albeit more transiently and less efficiently than for estrogen-stimulated genes. Of multiple cofactors studied, only p300 was recruited to ERα binding sites of repressed genes, and its knockdown prevented estrogen-mediated gene repression. Because p300 is involved in transcription initiation, we tested whether ERα might be trying to stimulate transcription at repressed genes, with ultimately failure and a shift to a repressive program. We found that estrogen increases transcription in a rapid but transient manner at early estrogen-repressed genes but that this is followed by recruitment of the corepressor CtBP1, a p300-interacting partner that plays an essential role in the repressive process. Thus, at early estrogen-repressed genes, ERα initiates transient stimulation of transcription but fails to maintain the transcriptional process observed at estrogen-stimulated genes; rather, it uses p300 to recruit CtBP1-containing complexes, eliciting chromatin modifications that lead to transcriptional repression.
The regulation of gene expression by nuclear receptors is critical in controlling the phenotypic properties and diverse biologies of their target cells (4). Estrogen receptor alpha (ERα) and ERβ, which are members of the nuclear receptor superfamily of ligand-activated transcription factors, play crucial roles in mammary gland development and also in breast cancer etiology, progression, and treatment (2, 21, 23). From genome-wide transcriptome studies of breast cancer cells, it is clear that hormone-bound ERα is a pivotal regulator that can both stimulate and repress gene transcription and influence, over time, a vast number of target gene mRNAs and proteins, thus creating a well-integrated hormonal response that affects numerous cell processes (6, 14, 42). Global gene expression profiling by microarray analysis has revealed that estradiol (E2), acting through ERα, upregulates the expression of genes encoding positive proliferation regulators, including multiple growth factors, growth factor receptors, and proteins involved in cell cycle progression, and downregulates transcriptional repressors and antiproliferative and proapoptotic genes, these together contributing to the stimulation of proliferation and suppression of apoptosis by estradiol in breast cancer cells (14, 15, 23).
Most of the studies thus far have elucidated the mechanisms by which ERα stimulates gene transcription, often focusing on the TFF1/pS2 gene as a model of ERα action (18). At the TFF1/pS2 enhancer, ERα works in a highly dynamic fashion as a nucleation factor for a cohort of coregulators (e.g., p160 family members and others) and enzymatic complexes (e.g., histone acetyltransferases [HATs] and SWI/SNF) that relax chromatin structure, allowing the basal transcriptional machinery to increase the transcriptional output (31, 38).
In contrast to all that is known about the molecular mechanisms by which the estrogen-occupied ERα stimulates gene expression, the mechanisms that this receptor utilizes to repress gene transcription are only starting to be elucidated. In our laboratory and elsewhere, a direct role of ERα and of corepressor/histone deacetylase (HDAC)-containing complexes has been demonstrated in the regulation of some estrogen (E2)-repressed target genes (32, 40). Carroll et al. (5) have highlighted a role for the coregulator NRIP1/RIP140 in the regulation of late E2-repressed target genes, while other mechanisms proposed to be involved in ERα-mediated transcriptional repression include physiological squelching of coactivator proteins and involvement of components of the basal transcriptional machinery (7, 19).
We sought to investigate the mechanisms involved in ERα-mediated transcriptional repression of endogenous genes downregulated at early times, in contrast to most previous studies of nuclear receptor-mediated gene expression that have used artificial gene constructs or addressed repression events at late time points of E2 treatment (≥8 to 24 h). Herein, we have characterized a group of E2 target genes whose downregulation by ERα is rapid and direct. These estradiol-repressed genes include the cyclin G2 (CCNG2) gene, which encodes a negative regulator of the cell cycle; SMAD6, important in modulating transforming growth factor β signaling pathways; and monocyte-to-macrophage differentiation-associated protein (MMD), highly expressed in the placenta and some cancer cell lines and believed to play critical roles in extracellular matrix remodeling and wound healing.
Our studies reveal that ERα is transiently recruited to the ER binding sites of these estrogen-repressed genes and that at these repressed genes there is also a transient increase in transcriptional rate. Further, p300 is observed to be recruited upon E2 treatment both at stimulated and repressed gene binding sites, and notably, p300 knockdown blocked both E2-mediated gene stimulation and repression. The repressive action of p300 at estrogen-repressed genes appears to be exerted through its partnering with the repressive CtBP1 complex, and through a variety of studies we confirm the central role of CtBP1 in specifically mediating transcriptional repression by this nuclear receptor.
Our study thus provides evidence that ERα is directly utilizing p300 and CtBP1 in transcriptional repression, and we propose that this is due to a failure in sustaining a stable transcriptionally active complex at these sites that causes p300 to partner with repressive rather than activating coregulator complexes.
MATERIALS AND METHODS
Cell culture, treatments, RNA extraction, and real-time qPCR.
MCF-7 cells were cultured in minimal essential medium (MEM) (Sigma Chemical Co., St. Louis, MO) supplemented with 5% heat-inactivated calf serum (HyClone, Logan, UT) and 1% antibiotics. Before experiments, the cells were maintained in phenol red-free MEM containing 5% charcoal-stripped calf serum (CD-CS) for a minimum of 4 days with the medium changed every other day. Cells were treated with E2 or vehicle control (0.1% ethanol) alone or in combination with other ligands at the concentrations and for various times. Total RNA was harvested and isolated using Trizol reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. Quantitative real-time PCR (qPCR) was performed as previously described (39). All PCR primer sequences are available upon request.
ChIP and sequential ChIP (reChIP) assays.
Chromatin immunoprecipitation assays (ChIPs) were performed with minor modifications as described in reference 31. The antibodies used were purchased from Santa Cruz Biotechnology (ERα [HC-20], NRIP1 [H-300], RNA polymerase II [N-20], CBP [A-22], p300 [N-15], SRC-3 [H-270], CtBP1 [C-1], and rabbit, goat, and mouse immunoglobulin G [IgG]), Millipore-Upstate (histone H3 [no. 07-690]), and Abcam (H3K14ac [ab52946] and H3K9ac [ab10812]). The DNA isolated was subjected to qPCR. Data were normalized to the results for 36B4 used as an internal control (measuring total input DNA in every sample), and a recruitment index was calculated (difference between specific antibody signals over Ig signal). For histone mark experiments, data were also normalized to total histone H3 content. For α-amanitin (Sigma) experiments, MCF-7 cells were kept for 4 days in MEM containing 2% CD-CS and then treated with 5 μM α-amanitin in serum-free medium for 2 h. After treatment, the cells were washed four times and treated with 1 nM E2 for 1 h in 5% CD-CS medium.
ChIP/reChIP experiments were done following the same ChIP protocol. After the first pull-down, immunoprecipitated material was recovered with 10 mM dithiothreitol in IP buffer at 37°C for 30 min, diluted, and submitted to a second round of immunoprecipitation.
RNA interference and Western immunoblotting.
MCF-7 cells were transfected with small interfering RNAs (siRNA) for ERα (3); p300, CBP, CtBP1, or CtBP2 SMARTpool; or GL3 luciferase control (Dharmacon), following the manufacturer's instructions. After 72 h, cells were treated with 1 nM E2 for 4 h. Total RNA was prepared from cells and analyzed as described in the previous section. Western immunoblotting was performed on MCF-7 whole-cell extracts following standard protocols. The antibodies used were the same as for ChIP assays, and antibody for CtBP2 was from BD Biosciences.
Nuclear run-on assay.
Nuclear run-on assays were carried out as described previously (30, 33). Briefly, MCF-7 cells were treated with 10 nM E2 or vehicle for different times, washed with PBS, harvested, and lysed (lysis buffer consisted of 0.5% NP-40, 10 mM KCl, 10 mM MgCl2, 10 mM HEPES [pH 7.9], 0.5 mM β-mercaptoethanol) on ice for 5 min. After centrifugation, nuclei were washed with lysis buffer without NP-40 and resuspended in 100 μl of storage buffer (50 mM Tris-HCl, 5 mM MgCl2, 0.5 mM β-mercaptoethanol, 40% glycerol) before being frozen at −80°C. A 100-μl amount of transcription buffer (10 mM Tris-HCl [pH 8.00], 0.3 M MgCl2, 5 mM dithiothreitol, 40 U RNase inhibitor [Roche], 1× biotin labeling mix [Roche]) was added to the nuclei, and the reaction mixture was incubated at 30°C for 45 min. RNA was isolated by using Trizol reagent (Invitrogen). A 50-μl amount of streptavidin-conjugated magnetic beads (Invitrogen) resuspended in binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl) was added to each sample and incubated at room temperature for 2 h. Beads were washed twice in 500 μl of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 15% formamide for 15 min and once in 500 μl of 2× SSC for 5 min and then dissolved in 12 μl of diethyl pyrocarbonate water. RNA was reverse transcribed, and qPCR reactions were carried out as described above.
RESULTS
Characterization of early estrogen-repressed target genes.
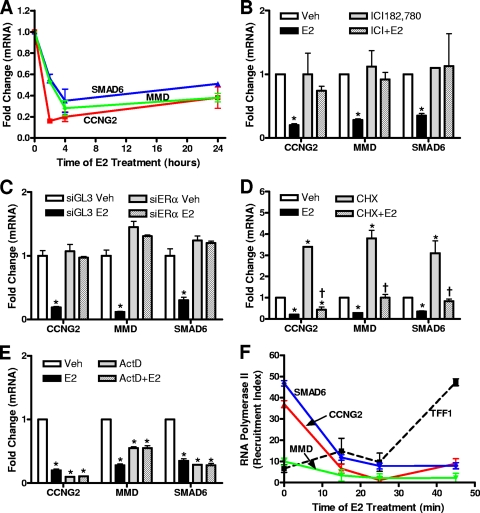
To explore the mechanisms involved in E2-mediated transcriptional repression, we identified a subset of early E2-repressed target genes in MCF-7 breast cancer cells based on previous cDNA microarray studies performed in our laboratory (14, 15). Using qPCR, we examined the effect of E2 on this group of genes and found that their expression was repressed rapidly (by 2 h) and remained repressed in a sustained manner following E2 treatment (Fig. 1A). To determine whether ERα was necessary for the transcriptional repression of these genes, we used both the ERα antagonist ICI182,780 and ERα knockdown with siRNA (Fig. 1B and C). The results of both approaches demonstrated that ERα was required for gene repression, because the E2-elicited repression was completely abrogated by ICI182,780 cotreatment along with the E2 or by ERα knockdown.
FIG. 1.
Characterization of early estradiol (E2)-repressed genes in MCF-7 breast cancer cells. (A) Time course of E2 regulation. MCF-7 cells were treated with 1 nM E2, and RNA was isolated at various time points. cDNA was measured by qPCR using primers for the genes indicated and internal control 36B4. (B) ICI182,780 antagonist experiments. MCF-7 cells were treated with 1 μM ICI182,780 (ICI) with or without 1 nM E2. (C) ERα siRNA experiments. MCF-7 cells were transfected with siRNA for ERα (siERα) or for GL3 control (siGL3) (luciferase) for 48 h prior to treatment with 1 nM E2. (D) Protein translation inhibitor experiments. MCF-7 cells were treated with cycloheximide (10 μg/ml) (CHX) with or without 1 nM E2. (E) Transcriptional inhibitor experiments. MCF-7 cells were treated with actinomycin D (5 μg/ml) (ActD) with or without 1 nM E2. In the experiments whose results are shown in panels B to E, the 1 nM E2 treatment was for 4 h, and total RNA was extracted and quantified by qPCR using 36B4 as the internal control. (F) RNA polymerase II ChIP was performed as described in Materials and Methods after 15, 25, or 45 min of 1 nM E2 treatment. Data are represented as the recruitment index (RNA polymerase II signal/IgG signal ratio). Data shown in all panels are averages ± standard errors of the means of the results of three or more independent experiments. In all panels, an asterisk indicates a P value of <0.05 for results of E2 treatment versus results for vehicle control. In panel D, a dagger indicates a P value of <0.05 for results for cycloheximide with E2 versus results for cycloheximide alone. Veh, vehicle.
To examine whether there was a direct, primary transcriptional mechanism of ERα regulation of these target genes, we treated MCF-7 cells with the protein translation inhibitor cycloheximide or the transcriptional inhibitor actinomycin D (Fig. 1D and E). We found that repression of the expression of these genes did not require protein synthesis, whereas actinomycin D greatly reduced basal gene mRNA and eliminated the E2-induced downregulation. The results of these experiments therefore imply that these genes are primary ERα targets. Further, there was no effect of E2 on the stability of these mRNAs, as assessed by actinomycin D treatment (Fig. 1E) and time course studies (data not shown).
We next examined the status of RNA polymerase II at these genes, in particular whether after E2 treatment RNA polymerase II would be pausing or be displaced from the promoters of these E2-repressed genes, because both mechanisms have been shown to be possible at ERα-regulated genes (25, 40). We performed ChIP time course experiments using a specific antibody directed against RNA polymerase II. As shown in Fig. 1F, RNA polymerase II was displaced from the transcriptional start sites of all E2-repressed genes within 15 min of E2 treatment, indicating that dismissal, rather than stalling or pausing, of RNA polymerase II occurred rapidly at these repressed target genes. In contrast, recruitment of RNA polymerase II continued to increase over time at the E2-stimulated gene TFF1 (Fig. 1F).
Characterization of ERα binding sites for early E2-repressed genes.
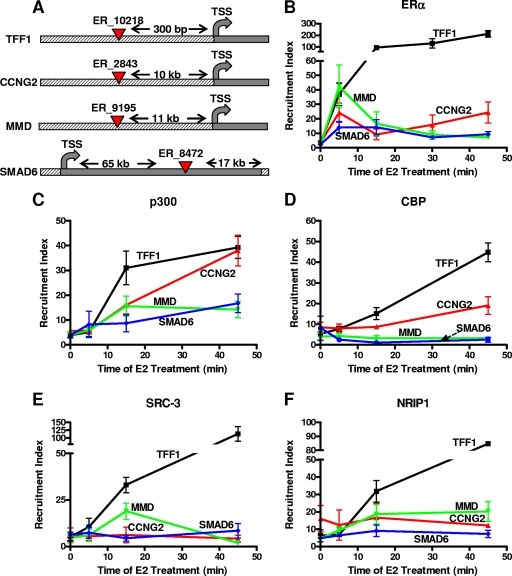
Genome-wide studies using ChIP-on-chip (combined use of ChIP and microarrays) and ChIP-paired-end-tag (ChIP-PET) technologies have identified putative ERα binding sites in MCF-7 breast cancer cells (5, 28). We used this available information to analyze ERα binding sites associated with ERα-repressed genes. The early E2-repressed target genes that we characterized in the experiments whose results are shown in Fig. 1 (CCNG2, MMD, and SMAD6) were chosen because each has a single ERα binding site in the proximity of the gene (Fig. 2A). As a positive control for our experiments, we used the well-characterized enhancer of the E2-stimulated gene TFF1/pS2. To examine the presence of ERα at these putative binding sites after E2 treatment, an ERα time course ChIP assay was performed (Fig. 2B). As expected, at the TFF1/pS2 enhancer region, ERα recruitment was greatly increased, starting after 5 min of E2 treatment and increasing over the 45 min studied. Interestingly, the time pattern of ERα recruitment at the binding sites of E2-repressed genes was different. ERα was recruited after 5 min of E2 treatment, as observed for the E2-stimulated TFF1/pS2 gene, but then ERα occupancy decreased rapidly or remained constant and only mildly elevated over the time analyzed (Fig. 2B). Thus, the magnitudes and time courses of ERα recruitment to the binding sites of repressed and stimulated genes are distinctly different.
FIG. 2.
Characterization of occupancy across time of ERα and selected coregulators at ERα binding sites associated with E2-repressed genes. (A) UCSC Genome Browser schematics of selected ERα binding sites according to Carroll et al. (5). TSS, transcription site. (B to F) ChIP assays were performed at various times of 1 nM E2 treatment, using specific antibodies against ERα (B), p300 (C), CBP (D), SRC-3 (E), NRIP1 (F), or IgG negative control antibody. Data are averages ± standard errors of the means of the results of four or more independent experiments and are represented as the recruitment index (specific antibody signal/IgG signal ratio).
Characterization of the recruitment of coregulators at the ERα binding sites of early E2-repressed genes.
The role of specific ERα coregulatory factors in E2-mediated gene repression is still an open question. Therefore, we performed ChIP assays on MCF-7 cells treated with 1 nM E2 for 5, 15, and 45 min using specific antibodies directed against HATs (CBP and p300), the p160 coactivator SRC-3, and the mixed coactivator/corepressor NRIP1 (Fig. 2C, D, E, and F) to examine their patterns of recruitment over early times of hormone treatment.
Since one of the main features of E2-mediated gene repression appears to be histone deacetylation (25, 40) and because CBP has been found to be squelched away at the GnRH receptor gene (7), we tested whether physiological squelching of HATs was also occurring at early E2-repressed genes. As shown in Fig. 2C, p300 recruitment increased at the binding sites of all E2-repressed genes with a time profile that was similar and at levels that were comparable (CCNG2) or somewhat less (MMD and SMAD6) than that for the E2-stimulated TFF1/pS2 gene. In the case of CBP (Fig. 2D), its occupancy increased at TFF1/pS2 and, more slightly, at the CCNG2 site, while being low at the SMAD6 and MMD binding sites. Hence, although we cannot exclude that squelching of CBP or p300 might occur from other regions of DNA, we find that this does not occur from the ERα binding sites or from regions close to the transcriptional start site (Fig. 2D and data not shown). Regarding p160/SRC coactivators (Fig. 2E), ChIP assays revealed SRC-3, the most abundant SRC in MCF-7 cells, to be either absent or present at very low levels at the ERα binding sites of the examined genes prior to E2 treatment. Upon E2 treatment, SRC-3 was strongly recruited to the TFF1/pS2 enhancer while remaining absent at the binding sites of E2-repressed genes. An intriguing exception was the transient increase in occupancy of SRC-3 at the MMD binding site at the 15-min time point, possibly indicating the formation of a transient ERα-p160 complex at this E2-repressed gene (Fig. 2E).
Since NRIP1 is itself upregulated by the E2-ERα complex and has been proposed to be an important factor mediating ERα repression of late/secondary genes (5), we examined whether this coregulator might also be employed by ERα in the repression of early primary target genes. NRIP1 occupancy increased strongly at the ERα binding site of the TFF1/pS2 gene after E2 treatment, with a profile similar to that observed for SRC-3 (Fig. 2F), whereas at ERα binding sites of E2-repressed genes, NRIP1 was present at generally low levels in control vehicle-treated cells (Fig. 2F, zero time), and its occupancy did not change or increased mildly over time after E2 treatment, especially at the MMD binding site, suggesting that NRIP1 plays only a limited role in early gene repression (Fig. 2F). Likewise, we observed no recruitment of the corepressors NCoR or SMRT to the ERα binding sites of the three E2-repressed genes and no recruitment to the E2-stimulated TFF1 gene (data not shown).
In summary, the results of our ChIP time course experiments reveal that p300 was the only coregulator recruited at the ERα binding sites of E2-repressed, as well as E2-stimulated, genes, whereas other factors, like SRC-3, CBP, and NRIP1 may have more gene-specific roles.
p300 knockdown blocks E2-mediated gene repression.
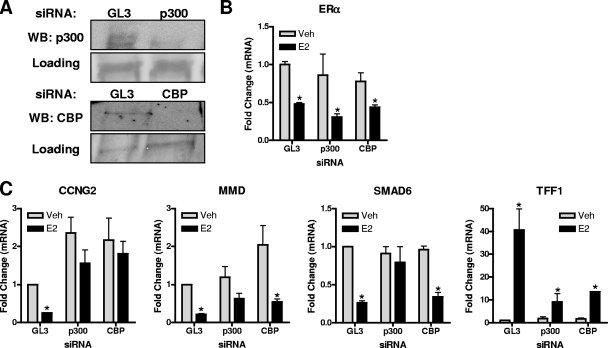
To directly address the roles of p300 and CBP in E2-mediated gene repression, we used RNA interference knockdown (Fig. 3) that selectively reduced p300 or CBP without affecting ERα (Fig. 3A and B). As seen in Fig. 3C, knockdown of p300 resulted in a nearly complete loss of E2-mediated gene repression, indicating a crucial role for p300 in the repression process. We also observed that the E2-stimulated increase in TFF1/pS2 mRNA was also greatly reduced by p300 knockdown (Fig. 3C), confirming the already known role of p300 in E2-mediated stimulation of gene expression. Since p300 and CBP are homologous proteins and their pattern of occupancy at the binding sites of repressed genes was different, we wanted to verify whether their character was retained after CBP-specific knockdown. As shown in Fig. 3C, CBP knockdown blocked the stimulation of TFF1/pS2 and also the repression of CCNG2 by E2, which was the only E2-repressed gene where CBP recruitment at the ERα binding site was detected. In contrast, CBP knockdown had little impact on the repression of MMD or SMAD6 gene expression by E2. These experiments highlight the central role for an ERα-p300-containing complex in the repression of transcription, while CBP appears to have a more gene-specific role.
FIG. 3.
p300 knockdown prevents E2-induced gene repression and E2-induced gene stimulation. (A) MCF-7 cells were transfected with siRNA against p300, CBP, or control GL3 luciferase for 72 h prior to 0.1% control ethanol vehicle or 1 nM E2 treatment for 4 h. p300 and CBP levels were assessed by immunoblotting. WB, Western blotting. (B and C) RNA was extracted after GL3, p300, or CBP siRNA treatment of cells followed by control vehicle or E2 treatment for 4 h, and the levels of E2 target genes were evaluated by qPCR. Data are means ± standard errors of the means of the results of three independent experiments and are expressed relative to results for GL3 siRNA vehicle-treated cells, set at 1. Veh, vehicle; *, P < 0.05 for results of E2 treatment versus results for vehicle control.
ERα transiently activates gene transcription at E2-repressed gene sites.
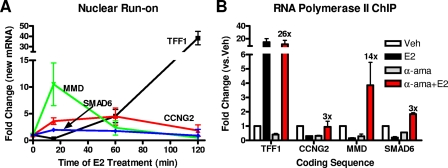
Because we found p300 to be involved in the repression of gene activity by the E2-ERα complex and p300 is known to be an essential factor for ERα-mediated gene activation, especially having a role in the establishment of the transcription initiation complex (1, 27), we explored the possibility that ERα might initially stimulate gene transcription also at early E2-repressed genes while ultimately failing to continue the process, with subsequent inhibition of transcription. To test this possibility, we took two different approaches. In the first, we performed nuclear run-on assays to determine whether there was a transient increase in newly synthesized mRNA at E2-repressed genes. As shown in Fig. 4A, we observed that E2 did transiently increase the production of newly made mRNA at the MMD and also, to a lesser extent, at the CCNG2 and SMAD6 genes. This stimulation was more rapid than that seen for the E2-stimulated gene TFF1/pS2, for which nuclear run-on-assays showed a continued increase over time in TFF1 mRNA production (Fig. 4A).
FIG. 4.
ERα transiently increases gene transcription at E2-repressed genes. (A) MCF-7 cells were treated with 10 nM E2, and nascent mRNA was labeled with biotin-UTP, isolated, and quantified via qPCR using 36B4 as the internal control. Data shown are means ± standard errors of the means of the results of three independent experiments. (B) RNA polymerase II ChIP was performed after 2 h of treatment with 5 μM α-amanitin followed by 1 h of 1 nM E2 treatment (α-ama+E2) or vehicle (α-ama). Change for α-ama+E2 versus α-ama is indicated above the red bars. Data are means ± standard errors of the means of the results of three independent experiments. Veh, vehicle.
In the second approach, we examined the effect of E2 treatment on RNA polymerase II levels at the regulated genes in MCF-7 cells by performing ChIP using an RNA polymerase II-specific antibody. These studies were done either before or after treatment with the RNA polymerase II inhibitor α-amanitin, which is known to block ongoing transcription (31, 38) and to clear the coding sequences from transcribing RNA polymerase II. In cells not treated with α-amanitin, E2 treatment greatly increased RNA polymerase II levels at the TFF1/pS2 gene and lowered RNA polymerase II levels at the repressed genes, consistent with repression of the latter genes (Fig. 4B). In contrast, when MCF-7 cells were first treated with α-amanitin for 2 h, followed by extensive washes, E2 treatment actually increased RNA polymerase II levels in the coding sequence of all three of the early E2-repressed genes, although to a lesser extent (3 times or 14 times or 3 times) (Fig. 4B) than at the TFF1/pS2 gene (26 times) (Fig. 4B). Because ERα is able to stimulate gene transcription after clearance of the transcriptional machinery even at the E2-repressed genes, it suggests that certain “transcriptional barriers” may be playing an important role in deciding the direction of gene regulation by ERα after an initial phase of stimulation.
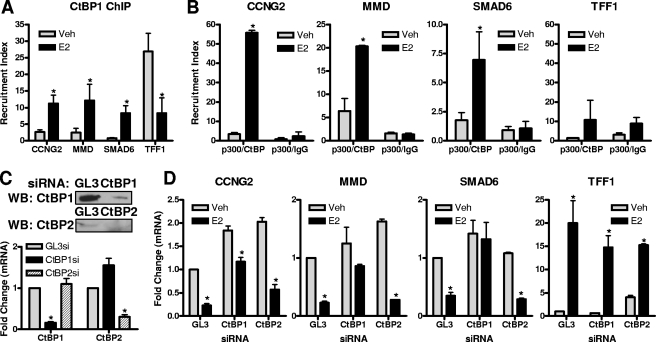
CtBP1 is recruited together with p300 to the binding sites of E2-repressed genes and is essential for E2-mediated gene repression.
To address how p300 might be working in opposite ways at E2-repressed versus E2-stimulated genes, we analyzed the recruitment of the CtBP1 corepressor complex, since it has been shown to directly interact with p300 and to block its HAT activity by binding to its bromodomain (24). As shown in Fig. 5A, CtBP1 was recruited to the binding sites of the three E2-repressed genes, whereas CtBP1 was dismissed from the TFF1/pS2 enhancer. This confirms that its presence in E2-stimulated genes before hormone treatment might be important for maintaining low basal activity and its hormone-regulated clearance might be important for transcriptional activation (35). Consistent with this pattern, at genes repressed by E2, CtBP1 levels were low prior to hormone treatment when basal gene activity is high, and the hormone-regulated increased recruitment of CtBP1 (Fig. 5A) coincides with transcriptional repression.
FIG. 5.
CtBP1, in a complex with p300, is required for E2-mediated gene repression. (A) ChIP assay using CtBP1 antibody was performed on MCF-7 cells after 45 min of control (0.1% ethanol) vehicle or 1 nM E2 treatment. (B) ChIP/reChIP was performed on MCF-7 cells after 45 min of 1 nM E2 treatment, using p300 antibody for the first pull-down and CtBP1 antibody or IgG control for the second pull-down. (C and D) CtBP1, CtBP2, or GL3 control siRNA was transfected into MCF-7 cells for 72 h prior to treatment with 1 nM E2 for 4 h. mRNA levels of target genes were measured by qPCR, and Western immunoblotting (WB) was used to confirm CtBP protein knockdown. Data are means ± standard errors of the means of the results of three independent experiments. In panels A, B, and D, an asterisk indicates a P value of <0.05 for results of E2 treatment versus results for vehicle control. For panel C, an asterisk indicates a P value of <0.05 versus results for GL3 siRNA. Veh, vehicle; si, siRNA.
As seen in Fig. 5B, by performing ChIP/reChIP experiments we observed CtBP1 to be in the same complex with p300 uniquely at E2-repressed target genes, whereas they were not present together at the TFF1/pS2 ERα binding site (Fig. 5B).
To test the functional role of CtBP1, we used siRNA that effectively reduced CtBP1 levels in MCF-7 cells (Fig. 5C). CtBP1 knockdown completely blocked E2-mediated gene repression, whereas it had little impact on TFF1/pS2 gene stimulation, thus implicating CtBP1 as a key factor in E2-mediated gene repression (Fig. 5D).
Because of the effect of CtBP1 depletion in preventing gene repression by estrogen, we also examined the effect of the closely related protein CtBP2. Of interest, depletion of CtBP2 had no effect on gene repression; repression remained as robust as that observed in control siRNA-treated cells (Fig. 5D), suggesting that gene repression uniquely requires CtBP1. Surprisingly, however, CtBP2 knockdown greatly reduced estrogen stimulation of the TFF1 gene, suggesting that CtBP2 is likely to be a factor involved in restricting transcriptional activation.
CtBP1 recruitment requires the presence of p300 and elicits lysine 9 of histone 3 (H3K9) and H3K14 deacetylation.
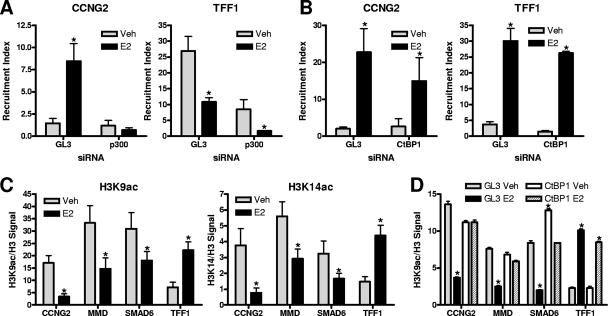
To determine whether p300 recruitment is a pioneer event for CtBP1 action, we reduced cellular p300 levels by using specific siRNA and then examined CtBP1 recruitment. As shown in Fig. 6A, CtBP1 recruitment to the E2-repressed gene CCNG2 was increased by E2 treatment in control (GL3) siRNA-exposed cells and was greatly impaired after p300 knockdown, indicating p300 to be essential for CtBP1-mediated transcriptional repression at E2-inhibited target genes and suggesting that p300 might act as a bridge between ERα and CtBP1. Similar observations were made for the E2-repressed genes MMD and SMAD6 (data not shown). In contrast, at the TFF1 gene, CtBP1 recruitment was similar after p300 or control siRNA knockdowns. On the other hand, p300 recruitment was not impaired after knockdown of CtBP1 at either E2-repressed or E2-stimulated genes (Fig. 6B), indicating that CtBP1 is probably recruited after p300 interaction with the ERα-containing complex. Hence, the presence of CtBP1 requires p300, but the reverse is not the case.
FIG. 6.
p300 is required for CtBP1 recruitment, and CtBP1 is important for changes in histone marks. (A and B) ChIP assays were performed on MCF-7 cells after knockdown of p300 (A), CtBP1 (B), or GL3 control (A and B) with siRNA for 72 h. Antibodies for CtBP1 (A) or p300 (B) were used. (C) ChIP assays for histone marks (H3K14ac and H3K9ac) were performed after 45 min of 1 nM E2 or vehicle treatment, and data were normalized to total histone H3 content. (D) Histone H3K9ac changes were evaluated after CtBP1 or control GL3 (luciferase) knockdown as described for panel A. In panels A, B, and C, an asterisk indicates a P value of <0.05 for results of E2 treatment versus results for vehicle control. In panel D, an asterisk indicates a P value of <0.05 versus results for siGL3 vehicle. Veh, vehicle.
Since CtBP1-containing complexes have been shown to possess multiple histone-modifying activities (8) and transcriptional repression has been correlated with histone deacetylation, we assessed changes in specific histone marks after E2 treatment at the repressed and stimulated genes.
H3K14ac is a major site for p300-mediated histone acetylation and has been correlated with transcriptional activation (10). As shown in Fig. 6C, H3K14 was deacetylated at two out of three ERα binding sites near E2-repressed genes, indicating that p300 does not act as a HAT at these sites and that, after recruitment, its activity may be blocked by CtBP1 binding. The opposite was observed at the TFF1/pS2 enhancer, where H3K14 acetylation was increased after E2 treatment of cells (Fig. 6C).
The second acetylation event that we checked was at the H3K9 mark (H3K9ac) (Fig. 6C). At the ERα binding sites of the three E2-repressed genes, H3K9 was deacetylated, confirming that one of the important features of gene repression is general histone deacetylation, while at the TFF1 binding site we observed strong acetylation (Fig. 6C).
To demonstrate a direct link between the histone deacetylation events and CtBP1-containing complexes, we performed ChIP assays after CtBP1 knockdown and probed for H3K9ac, which showed the strongest deacetylation at estrogen-repressed gene sites (Fig. 6C). As shown in Fig. 6D, after knockdown of CtBP1, the deacetylation of H3K9 observed with E2 in control (GL3) siRNA-treated cells at the CCNG2, MMD, and SMAD6 genes was nearly completely prevented, while acetylation at the TFF1/pS2 enhancer was not affected. Our findings shown in Fig. 6 suggest that the actions of CtBP1 are unique for E2-mediated gene repression and that the histone deacetylation observed is likely due to HDAC activities in the CtBP1 complex.
DISCUSSION
In this study, we have examined the mechanisms involved in E2-ERα mediated transcriptional repression of early primary target genes, and we describe a new mechanism for ER-mediated transcriptional repression that involves the recruitment of a p300-dependent CtBP1 corepressor complex following ERα failure to activate gene transcription. We demonstrate that ERα can be recruited directly, albeit transiently and less strongly, to DNA elements close to E2-repressed genes where ERα recruits p300 and is able to transiently increase the transcriptional output; however, ERα and p300 are unable to become a nucleation site for p160 coactivators and to sustain positive transcriptional regulation. This leads to recruitment of the corepressor CtBP1, via p300, with RNA polymerase II eviction and histone deacetylation that result in transcriptional repression. Of note, CtBP1 was a crucial factor for gene repression, whereas it was irrelevant for gene stimulation, by estrogen. Furthermore, the important effects of CtBP1 in E2-ERα-mediated gene repression were unique to CtBP1 and were not reproduced by the related CtBP2 protein.
Modulation of gene transcription by ERα: stimulation versus repression.
It is now well accepted that the ER is a master regulator of gene transcription, as demonstrated by numerous studies using both cell culture models and whole-animal target tissues (11, 14, 22). From these studies it is evident that, upon E2 treatment, gene transcription is widely impacted, creating highly complex regulatory networks whose ultimate goal is the stimulation or suppression of specific biological processes. In fact, in MCF-7 breast cancer cells, the expression of more genes is repressed than stimulated by the E2-occupied ERα (14, 15). Hence, it was of interest to understand how the ER, being a strong transcriptional activator, can also behave as a transcriptional repressor.
Thus far, several mechanisms have been hypothesized for E2-mediated gene repression, including physiological squelching of cofactors (e.g., p160s and CBP), direct action of corepressors (NCoR, SMRT, and NRIP1) accompanied by histone deacetylation, and participation of elements of the basal transcriptional machinery (e.g., TAFII30). Most of these studies, though, employed exogenous reporter systems or overexpression of selected factors and/or considered events mostly at late time points of E2 treatment (8 to 24 h). What we addressed in this study is the analysis of early, primary ERα-repressed target genes and the mechanisms that occur at their ERα binding sites in the endogenous cell chromatin setting.
From genome-wide studies of ERα binding sites, it appears that ca. 60% of E2-repressed genes at early time points (1 to 4 h) possess at least one ERα binding site in their proximity, indicating that direct effects of ERα are likely and that they may account for at least a portion of the repressive events. In this study, we characterized a group of primary E2-repressed target genes and document that ERα is recruited to these binding sites by E2 treatment but, interestingly, in a manner that is different from recruitment to the enhancer of the TFF1/pS2 gene, which is strongly E2 stimulated. At the ERα binding sites that we studied, ERα occupancy increased comparatively similarly to that at the TFF1/pS2 enhancer in the first 5 to 15 min of E2 treatment, after which ERα occupancy decreased or remained constant at approximately 10 to 15% of the level of TFF1/pS2 enhancer occupancy. This indicates that ERα may interact less efficiently and more transiently with these sites and also points to the fact that the number of binding sites close to E2-repressed genes may have been underestimated by these genome-wide techniques, because only the strongest interactions would be detected and also because, based on our study, a different result might be obtained by using earlier time points (i.e., 15 min) of E2 treatment versus the typical 45-min time point most commonly examined (5, 31). From our bioinformatic analysis of the composition of the ERα binding sites, there appears not to be any preferential factor linked to E2-repressed versus E2-stimulated genes (F. Stossi, unpublished observation), suggesting that it may be difficult to isolate specific transcription factors that are associated only with E2-mediated transcriptional repression.
p300 plays a central role in both gene activation and repression.
Nuclear receptor coregulators encompass a large family of proteins with multiple enzymatic activities that appear to be essential in performing and fine-tuning the actions of the ER at the chromatin level (29). Several studies (16, 31, 34, 38) have highlighted a very dynamic picture of multiple coactivating and corepressing complexes exchanging during the transcriptional process, adding an important level of complexity and control in the regulation and direction (up/down) of the transcriptional output.
Using time-course ChIP assays, we could establish that p300 was the only cofactor that appeared to be recruited at all the sites analyzed, while other factors, like CBP, NRIP1, and p160s, might play more gene-specific roles. Also of note, an important finding in our study was the essential role of p300 in gene repression as well as gene stimulation. p300 was found to be recruited to the binding sites of both E2-stimulated and E2-repressed genes, and p300 knockdown fully prevented E2-mediated gene repression and also markedly reduced E2-mediated gene stimulation.
The role of p300 in ERα-mediated gene stimulation has been extensively characterized where it plays a central role in transcriptional initiation, but not reinitiation (27), and is normally seen before recruitment of SRCs to the TFF1 gene (31, 38). There is evidence that ERα and p300 interact directly (12, 26), although some have suggested that this may involve mediating proteins (e.g., SRC-3).
p300 is also known to elicit negative roles in transcription, as recently shown in a completely purified in vitro transcription system (36) where p300 acted as a negative cofactor whose repressive activity was reversed by the addition of acetyl-coenzyme A. Moreover, p300 contains a strong repressive domain (cell cycle regulatory domain 1, amino acids 1017 to 1029) that functions independently from the HAT domain via sumoylation and HDAC6 recruitment (17).
Since a role for p300 in transcriptional initiation has been extensively characterized, we hypothesized that the p300-ERα-containing complex might be trying to stimulate gene transcription also at E2-repressed targets but ultimately fails to continue the process. To investigate this possibility, we first performed nuclear run-on assays that clearly demonstrated that, at early time points, ERα can transiently stimulate the transcription of E2-repressed genes. Second, after we cleared the coding sequences of the genes from transcribing RNA polymerase II, E2 treatment resulted in the reloading of RNA polymerase II at both E2-repressed and E2-stimulated genes, indicating that ERα is capable of driving positive transcription from binding sites close to E2-repressed genes. The results of these two experiments also lead us to speculate that elements in the basal transcription machinery and/or in the elongation complexes might be important in choosing the direction of regulation of transcription by ERα after this initial phase of stimulation at both types of genes.
The corepressor CtBP1 is utilized by ERα, via p300, to repress gene transcription.
In addressing how ERα and p300 elicit transcriptional repression, we focused on the corepressor CtBP1. Although CtBP1 had not previously been directly linked to ERα action, it had been shown to interact directly with p300 and inhibit the HAT activity of p300 via interaction with its bromodomain, thus impeding p300's recognition and acetylation of histone tails (24, 37). Moreover, CtBP1-containing complexes have been characterized as containing numerous enzymatic activities, including histone deacetylation via multiple HDACs (i.e., HDAC1 and HDAC2). Although the relationships between histone posttranslational modifications and positive or negative gene activities are known to be very complex (9, 20), it was striking that robust H3K14 and H3K9 deacetylation accompanied the gene repression by E2 and that these were prevented by depletion of CtBP1.
We demonstrated that CtBP1, in complex with p300, is recruited to E2-repressed genes and is essential for the repressive process and histone tail deacetylation events, highlighting a central role for this factor in ERα-mediated transcriptional repression. In addition, p300 recruitment appeared to be a prerequisite for CtBP1 recruitment, although there may also be additional mechanisms, because CtBP1 has been shown to interact with the corepressors NRIP1/RIP140 and LCoR, which can directly interact with ERα (13, 41).
A model for E2-mediated gene repression of early target genes.
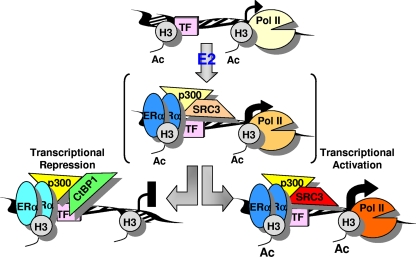
Based on our observations, we present a model for E2-mediated transcriptional repression of early target genes (Fig. 7). In this scenario, ERα would interact either directly, indirectly, or cooperatively with DNA elements in a manner that is comparable for stimulated and repressed target genes in the first 5 to 15 min after E2 treatment. During this first phase, p300 and, in a gene-specific fashion (i.e., MMD), other cofactors (i.e., SRC-3) are being recruited by ERα, causing a spike in transcriptional activation, possibly due to the RNA polymerase II already cycling at these genes. After this first phase, ERα occupancy starts to decrease or remains constant, and this is followed by a loss of capacity for sustaining a steady increase in transcription that causes RNA polymerase II loss, recruitment of CtBP1-containing complexes, and histone deacetylation at early repressed genes.
FIG. 7.
Proposed model for ERα-mediated repression of early target genes. Following E2 treatment, ERα is initially recruited to ERα binding sites of both E2-stimulated and E2-repressed target genes, either via direct DNA binding or tethering to pioneer transcription factors (TF), where it transiently recruits coactivator complexes such as p300 and causes an increase in transcriptional output. After this early transient complex, ERα can maintain transcriptional activation by serving as a more-stable nucleation site for coactivator proteins, leading to histone acetylation (Ac) and engagement of RNA polymerase II (Pol II), or ERα can cause transcriptional repression by recruiting, via p300, CtBP1-containing repressor complexes which lead to RNA polymerase II dismissal and histone deacetylation. The change in color intensity for individual factors indicates the change in occupancy in the transcriptional complex over time.
It is notable that even at E2-repressed genes, one sees some molecular attributes of gene stimulation, albeit transiently: recruitment of ERα, p300, and to some genes, CBP, and a transient increase in nuclear run-on and RNA polymerase II recruitment on α-amanitin-cleared genes. While it might seem paradoxical to see such changes at genes that show a net reduction of RNA levels, it is clear that elevated RNA production from repressed genes is, at most, brief. Also, at the E2-repressed genes, there is a net dismissal of RNA polymerase II upon E2 treatment, RNA polymerase II recruitment by E2 being evident only on the artificially lowered background following α-amanitin treatment.
The most significant and durable differences between the E2-repressed genes and E2-stimulated genes we have studied appear to be the relative instability of the recruitment of ERα and the clear differential recruitment of certain coregulator complexes, e.g., the corepressor CtBP1 to repressed genes versus the coactivator SRC-3 to stimulated genes. It is the consequences of the known differential chromatin-modifying activities of these coregulators that appear to be responsible for the ultimate differential effects on the production of RNA from the repressed versus the stimulated genes.
Our work raises two interesting questions. First, what is responsible for the fact that the E2-ERα-p300 complex, which forms at both stimulated and repressed genes, recruits CtBP1 only to E2-repressed genes? It is possible that differential posttranslational modifications of p300 or other coregulators by specific enzymatic complexes will determine the choice of protein partners for p300. It is indeed known that a “posttranslational code” exists for coactivators like SRC-3 (29), and p300 is known to possess multiple sites of posttranslational modification that influence its activity (17, 43, 44). Second, if E2 treatment results in dismissal of the CtBP1 corepressor system from E2-stimulated genes, by what mechanism is CtBP1 present in the absence of hormone, ERα, and p300? Presumably, in the absence of hormone, CtBP1 is held at stimulated genes through other transcription factors via different coregulatory proteins, like TBL1 (35). It would be interesting to investigate this system, because such transcription factor-CtBP1 corepressor complexes might be responsible for maintaining the low basal activity expected of genes poised to be stimulated by estrogens acting through ERα.
Our studies highlight a new mechanism utilized by the ER to elicit transcriptional repression of target genes. This mechanism includes a new role for p300 as a bridging factor between ERα and coregulator complexes that appears to be crucial in deciding the direction of transcription after ERα activation and binding to DNA. Moreover, we demonstrate for the first time the involvement of the corepressor CtBP1 in estrogen-mediated gene repression. Thus, the cooperation between CtBP1 and p300 appears to be central in discriminating nuclear receptor repression versus stimulation of genes at early times after hormone exposure.
Acknowledgments
This work was supported by NIH grant CA18119 (B.S.K.) and a grant from The Breast Cancer Research Foundation (B.S.K.). Z.M.-E. received partial support from NIH T32 ES07326.
Footnotes
Published ahead of print on 2 February 2009.
REFERENCES
- 1.Acevedo, M. L., and W. L. Kraus. 2003. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates. Mol. Cell. Biol. 23335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, E. 2002. The role of oestrogen and progesterone receptors in human mammary development and tumorigenesis. Breast Cancer Res. 4197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, D. H., S. Sheng, T. H. Charn, A. Waheed, W. S. Sly, C. Y. Lin, E. T. Liu, and B. S. Katzenellenbogen. 2008. Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 683505-3515. [DOI] [PubMed] [Google Scholar]
- 4.Bookout, A. L., Y. Jeong, M. Downes, R. T. Yu, R. M. Evans, and D. J. Mangelsdorf. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, A. H., A. K. Bednarek, R. L. Daniel, K. A. Hawkins, K. J. Laflin, S. Gaddis, M. C. MacLeod, and C. M. Aldaz. 2000. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 605977-5983. [PubMed] [Google Scholar]
- 7.Cheng, C. K., B. K. Chow, and P. C. Leung. 2003. An activator protein 1-like motif mediates 17beta-estradiol repression of gonadotropin-releasing hormone receptor promoter via an estrogen receptor alpha-dependent mechanism in ovarian and breast cancer cells. Mol. Endocrinol. 172613-2629. [DOI] [PubMed] [Google Scholar]
- 8.Chinnadurai, G. 2007. Transcriptional regulation by C-terminal binding proteins. Int. J. Biochem. Cell Biol. 391593-1607. [DOI] [PubMed] [Google Scholar]
- 9.Clayton, A. L., C. A. Hazzalin, and L. C. Mahadevan. 2006. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell 23289-296. [DOI] [PubMed] [Google Scholar]
- 10.Daujat, S., U. M. Bauer, V. Shah, B. Turner, S. Berger, and T. Kouzarides. 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 122090-2097. [DOI] [PubMed] [Google Scholar]
- 11.Denger, S., T. Bahr-Ivacevic, H. Brand, G. Reid, J. Blake, M. Seifert, C. Y. Lin, K. May, V. Benes, E. T. Liu, and F. Gannon. 2008. Transcriptome profiling of estrogen-regulated genes in human primary osteoblasts reveals an osteoblast-specific regulation of the insulin-like growth factor binding protein 4 gene. Mol. Endocrinol. 22361-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, S., Y. X. Ma, C. Wang, R. Q. Yuan, Q. Meng, J. A. Wang, M. Erdos, I. D. Goldberg, P. Webb, P. J. Kushner, R. G. Pestell, and E. M. Rosen. 2002. p300 modulates the BRCA1 inhibition of estrogen receptor activity. Cancer Res. 62141-151. [PubMed] [Google Scholar]
- 13.Fernandes, I., Y. Bastien, T. Wai, K. Nygard, R. Lin, O. Cormier, H. S. Lee, F. Eng, N. R. Bertos, N. Pelletier, S. Mader, V. K. Han, X. J. Yang, and J. H. White. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11139-150. [DOI] [PubMed] [Google Scholar]
- 14.Frasor, J., J. M. Danes, B. Komm, K. C. Chang, C. R. Lyttle, and B. S. Katzenellenbogen. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 1444562-4574. [DOI] [PubMed] [Google Scholar]
- 15.Frasor, J., F. Stossi, J. M. Danes, B. Komm, C. R. Lyttle, and B. S. Katzenellenbogen. 2004. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 641522-1533. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Bassets, I., Y. S. Kwon, F. Telese, G. G. Prefontaine, K. R. Hutt, C. S. Cheng, B. G. Ju, K. A. Ohgi, J. Wang, L. Escoubet-Lozach, D. W. Rose, C. K. Glass, X. D. Fu, and M. G. Rosenfeld. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 111043-1054. [DOI] [PubMed] [Google Scholar]
- 18.Green, K. A., and J. S. Carroll. 2007. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat. Rev. Cancer. 7713-722. [DOI] [PubMed] [Google Scholar]
- 19.Hao, H., M. d'Alincourt-Salazar, K. M. Kelley, A. Shatnawi, S. Mukherjee, Y. M. Shah, and M. Ratnam. 2007. Estrogen-induced and TAFII30-mediated gene repression by direct recruitment of the estrogen receptor and co-repressors to the core promoter and its reversal by tamoxifen. Oncogene 267872-7884. [DOI] [PubMed] [Google Scholar]
- 20.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39311-318. [DOI] [PubMed] [Google Scholar]
- 21.Heldring, N., A. Pike, S. Andersson, J. Matthews, G. Cheng, J. Hartman, M. Tujague, A. Strom, E. Treuter, M. Warner, and J. A. Gustafsson. 2007. Estrogen receptors: how do they signal and what are their targets. Physiol. Rev. 87905-931. [DOI] [PubMed] [Google Scholar]
- 22.Hewitt, S. C., B. J. Deroo, K. Hansen, J. Collins, S. Grissom, C. A. Afshari, and K. S. Korach. 2003. Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol. Endocrinol. 172070-2083. [DOI] [PubMed] [Google Scholar]
- 23.Katzenellenbogen, B. S., and J. Frasor. 2004. Therapeutic targeting in the estrogen receptor hormonal pathway. Semin. Oncol. 3128-38. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. H., E. J. Cho, S. T. Kim, and H. D. Youn. 2005. CtBP represses p300-mediated transcriptional activation by direct association with its bromodomain. Nat. Struct. Mol. Biol. 12423-428. [DOI] [PubMed] [Google Scholar]
- 25.Kininis, M., B. S. Chen, A. G. Diehl, G. D. Isaacs, T. Zhang, A. C. Siepel, A. G. Clark, and W. L. Kraus. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 275090-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, Y., T. Kitamoto, Y. Masuhiro, M. Watanabe, T. Kase, D. Metzger, J. Yanagisawa, and S. Kato. 2000. p300 mediates functional synergism between AF-1 and AF-2 of estrogen receptor alpha and beta by interacting directly with the N-terminal A/B domains. J. Biol. Chem. 27515645-15651. [DOI] [PubMed] [Google Scholar]
- 27.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, C. Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C. L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lonard, D. M., and B. W. O'Malley. 2007. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell 27691-700. [DOI] [PubMed] [Google Scholar]
- 30.Madak-Erdogan, Z., K. J. Kieser, S. H. Kim, B. Komm, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2008. Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol. Endocrinol. 222116-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115751-763. [DOI] [PubMed] [Google Scholar]
- 32.Oesterreich, S., W. Deng, S. Jiang, X. Cui, M. Ivanova, R. Schiff, K. Kang, D. L. Hadsell, J. Behrens, and A. V. Lee. 2003. Estrogen-mediated down-regulation of E-cadherin in breast cancer cells. Cancer Res. 635203-5208. [PubMed] [Google Scholar]
- 33.Patrone, G., F. Puppo, R. Cusano, M. Scaranari, I. Ceccherini, A. Puliti, and R. Ravazzolo. 2000. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. BioTechniques 291012. [DOI] [PubMed] [Google Scholar]
- 34.Perissi, V., and M. G. Rosenfeld. 2005. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 6542-554. [DOI] [PubMed] [Google Scholar]
- 35.Perissi, V., C. Scafoglio, J. Zhang, K. A. Ohgi, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 2008. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol. Cell 29755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santoso, B., and J. T. Kadonaga. 2006. Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nat. Struct. Mol. Biol. 13131-139. [DOI] [PubMed] [Google Scholar]
- 37.Senyuk, V., K. K. Sinha, and G. Nucifora. 2005. Corepressor CtBP1 interacts with and specifically inhibits CBP activity. Arch. Biochem. Biophys. 441168-173. [DOI] [PubMed] [Google Scholar]
- 38.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103843-852. [DOI] [PubMed] [Google Scholar]
- 39.Stossi, F., D. H. Barnett, J. Frasor, B. Komm, C. R. Lyttle, and B. S. Katzenellenbogen. 2004. Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) alpha or ERbeta in human osteosarcoma cells: distinct and common target genes for these receptors. Endocrinology 1453473-3486. [DOI] [PubMed] [Google Scholar]
- 40.Stossi, F., V. S. Likhite, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2006. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J. Biol. Chem. 28116272-16278. [DOI] [PubMed] [Google Scholar]
- 41.Vo, N., C. Fjeld, and R. H. Goodman. 2001. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 216181-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, H., A. Suzuki, T. Mizutani, S. Khono, D. B. Lubahn, H. Handa, and T. Iguchi. 2002. Genome-wide analysis of changes in early gene expression induced by oestrogen. Genes Cells 7497-507. [DOI] [PubMed] [Google Scholar]
- 43.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 2942507-2511. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, L. W., and J. E. Gambee. 2000. Phosphorylation of p300 at serine 89 by protein kinase C. J. Biol. Chem. 27540946-40951. [DOI] [PubMed] [Google Scholar]