Abstract
The mechanisms of aberrant CpG island methylation in oncogenesis are not fully characterized. In particular, little is known about the mechanisms of inhibition of CpG island methylation. Here we show that sal-like 3 (SALL3) is a novel inhibitory factor for DNA methyltransferase 3 alpha (DNMT3A). SALL3 binds to DNMT3A by a direct interaction between the double zinc finger motif of SALL3 and the PWWP domain of DNMT3A. SALL3 expression reduces DNMT3A-mediated CpG island methylation in cell culture and in vitro. CpG island methylation is enhanced in SALL3-depleted cells. Consistently, DNMT3A from SALL3-depleted cells increases methyltransferase activity in vitro. Binding of DNMT3A to chromatin is reduced or increased by SALL3 expression or depletion, respectively, accounting for the mechanism by which SALL3 inhibits DNMT3A-mediated CpG island methylation. We also show that SALL3 is inducible by BMP-4 and silenced by associated DNA methylation in hepatocellular carcinoma (HCC). Our results suggest that silencing of SALL3 results in acceleration of DNA methylation in HCC. This functional characterization of SALL3 sheds light on regulatory mechanisms for DNMT3A and provides new strategies to inhibit aberrant methylation in cancer.
Dnmt3a and -3b are responsible for de novo DNA methylation during embryonic development. Repetitive sequences, including C-type retroviruses, minor satellite, and IAP repeats, were demethylated in Dnmt3a- and -3b-inactivated mouse embryos or embryonic stem cells. Inactivation of Dnmt3a and -3b also resulted in demethylation of a differentially methylated region in Igf2 (34). DNA methylation silences transcription by recruiting methyl-CpG-binding domain proteins, which further interact with histone deacetylase (HDAC) and histone methyltransferase (17, 21, 29). It has been shown that DNA methylation is tightly liked to histone modifications, including deacetylation or methylation involving histone H3 lysines 9 and 27 (2, 9, 11, 31). These reports indicate that DNA methylation and histone modifications act together to establish repressive chromatin structure. DNMT3A and -3B have been shown to interact with components of heterochromatin, including HP1, HDAC, and histone methyltransferase (10, 12), suggesting that DNMT3 also plays a role in the regulation of chromatin structure. It is well known that de novo DNA methylation in CpG islands occurs during cancer development (3, 16). However, the role of DNMT3 in methylation of normally unmethylated CpG islands has not been characterized completely. In DNMT3B and DNMT1 double-knockout colon cancer cells, methylation of histone H3 lysine 9 occurred first and the p16 promoter CpG island was methylated subsequently, after an additional 55 passages (1). Methylation of histone H3 lysine 9 and lysine 27 was suggested to be a prerequisite for de novo DNA methylation in cancer (33, 38). DNMT3 was recruited to some genomic loci depending on the interaction with the PML-RAR fusion protein or EZH2 (7, 42). Upregulation of DNMT3 expression was found in a proportion of cancer samples (37). These reports suggested that de novo DNMT is involved in aberrant CpG island methylation. However, the precise mechanisms of de novo DNA methylation, especially in CpG islands in cancer, remain to be clarified.
sal was originally identified in Drosophila melanogaster as a region-specific homeotic gene (18). sal is thought to be a transcription factor that is involved in specification of terminal patterning (6), positioning of wing veins (40), and differentiation of photoreceptors in embryonic development (28). Decapentaplegic (dpp), a human orthologue of BMP-4, was shown to induce sal expression, by which dpp regulated pattering of the wing disc in Drosophila (6). Mammalian SALL proteins also are involved in embryonic development. SALL1 is mutated in Townes-Brocks syndrome, characterized by a combination of anal, renal, limb, and ear anomalies (24). SALL4 mutations cause Okihiro syndrome, in which hand malformations are associated with impaired eye movement and retracted eye (23). The mouse Sall1, -2, and -4 proteins have been shown to activate or repress transcription (4, 27, 47). Interestingly, SALL1 and Sall4 localized to heterochromatin, showing association with HP-1 (4, 30). Sall1 interacted with components of an HDAC complex, including HDAC, RbAp46/48, and MTA (20). The repression of transcription may be explained by association of SALL with the HDAC complex. However, it is not known whether SALL proteins are involved in regulation of DNA methylation. SALL3 is another human SALL gene for which no germ line mutations have been reported. While Sall3 homozygous mutant mice died after birth with deficiencies in cranial nerves, little is known about functions of mammalian SALL3 (35).
We show that SALL3 interacts strongly with DNMT3A and less readily with DNMT3B. The double zinc finger (DZF) motif of SALL3 and PWWP domain of DNMT3A are necessary for their direct interaction. Expression of DNMT3A promotes CpG island methylation in demethylated cancer cells. Significantly, coexpression of SALL3 with DNMT3A reduces DNMT3A-mediated methylation. Consistent with this, SALL3 depletion increases the methylation levels of CpG islands. In addition, SALL3 inhibits the methyltransferase activity of DNMT3A in vitro. We conclude that SALL3 is a cellular factor that has the ability to inhibit the activity of DNMT3A. SALL3 reduces binding of DNMT3A to chromatin, accounting for the mechanism of SALL3-dependent inhibition of de novo CpG island methylation. We also show that SALL3 is inducible by BMP-4 and silenced in hepatocellular carcinoma (HCC) by the associated methylation of the promoter. We suggest that silencing of SALL3 results in promotion of CpG island methylation in HCC.
MATERIALS AND METHODS
Cell lines and tissue samples.
Human HCC cell lines are as described previously (25). To establish demethylated cells, HuH2 cells were incubated with 1 μM 5-aza-2′-deoxycytidine (5Aza-dC) for 9 days and then cultured with conventional medium for a month. Growing colonies were isolated and expanded. To examine induction of SALL3 by BMP-4, FLC4 cells were seeded in six-well plates. Cells were starved overnight and incubated with or without BMP-4 (R & D systems) for 3 h. Primary HCC samples and adjacent normal liver tissues were resected from patients who provided informed consent for the use of the specimens at The Cancer Institute Hospital, Japanese Foundation for Cancer Research.
Construction of vectors.
Full-length human SALL3c, DNMT3A, DNMT3B, and EZH2 were amplified by PCR. The amplified DNA was inserted into a modified pcDNA3.1 vector (Invitrogen) in which the Xpress tag was replaced with N-terminal hemagglutinin (HA) or a Flag tag. The primer sequences are AGCGCCGCTAGCAGCATGTCT and CGAGTCACTGGCTAGTTGAT for SALL3c, CTCTCGCCTCCAAAGACCACGAT and AACTTTGTGTCGCTACCTCAG for DNMT3A, TCGGCGATCGGCGCCGGAGATTC and GCTGGAACTATTCACATGCAAAG for DNMT3B, and GGACGAAGAATAATCATGGGC and AGCAGATGTCAAGGGATTTCC for EZH2. The primer sequences used in generating SALL3 deletion mutants are ATGTCTCGGCGCAAGCAGGC and ACTTCTCCTTGTGCCTCTGG for SALL3 deletion 1, CCAGAGGCACAAGGAGAAGT and AATGCTGGAGATGACCGAGG for deletion 2, CCTCGGTCATCTCCAGCATT and GCCCGGGCCCATCACTGTCT for deletion 3, and GCCGGGCCTCAGACAGTGATG and TAGAAGGCACAGTCG (pcDNA3.1 reverse primer) for deletion 4. Deletion 5 was an EcoRI-NotI fragment of deletion mutant 1. Deletion 6 was a NotI-XbaI fragment of deletion mutant 1. All SALL3 mutants were inserted into Flag-tagged pcDNA3.1. DNMT3A deletion mutant 1 was the EcoRI-KpnI fragment of full-length DNMT3A. Primers used in amplifying DNMT3A deletion mutant 2 are the T7 primer and CAGGGCCCATTCAATCATGGG, and those for DNMT3A deletion mutant 3 are AAGGCCGTGGAGGTGCAGAAC and pcDNA3.1 reverse primer. The SALL3 CpG island in the promoter region was divided into two fragments (−1157 through −3273 and +20 through −1177) and amplified using primer pairs (CACCGCAGAAATTACTCGTG and GACCGAAAGTTCCAACTCCA for promoter 1 and GTGGAGTTGGAACTTTCGGT and TGCTTGCGCCGAGACATGCT for promoter 2). The products were cloned into pGL3-basic vector (Promega). For expression in Escherichia coli, SALL3 deletion mutant 6 or full-length DNMT3A cDNA in the pcDNA3.1 vector was transferred into the pGEX vector (GE healthcare). Point mutants of SALL3 were generated using the QuikChange site-directed mutagenesis kit (Stratagene).
Reverse transcription (RT)-PCR analysis.
Total RNA of HCC cell lines was prepared using an RNeasy minikit (Qiagen), and normal liver total RNA was obtained from Ambion. cDNA was generated by reverse transcription of 3 μg of total RNA using the Superscript preamplification system (Invitrogen). The primer sequences were AAGCAGCACAACTGCCAGTC and ATGGCGAGCCCGTTAGTGAT for SALL3. RNA from cells treated with 1 μM 5Aza-dC for 3 days was used for the reactivation study.
Promoter assay.
FLC4 cells were cotransfected with 2 μg SALL3 promoter plasmid and 1 ng reference plasmid, pRL-CMV (Promega). After transfection, the cells were starved and incubated with BMP-4 (50 ng/ml). At 48 h posttransfection, luciferase activities were measured using a dual-luciferase reporter assay system (Promega). The values of the SALL3 promoter plasmid were normalized to those of the reference plasmid.
Methylation-specific PCR and bisulfite sequencing analysis.
Bisulfite modification of genomic DNA was performed as described previously (14). Bisulfite-treated DNA was amplified by PCR. For sequencing, the PCR products were cloned and 10 clones selected randomly for each sample were sequenced. The primer pairs used in methylation-specific PCR (MSP) are as follows: TCGGAATTTGGGACGGCGTTTAC and CAAAAATATCGCGCGAATCGCCG for SALL3 methylation, ATTGTTTGGAATTTGGGATGGTGTTTAT and CAAATTACAAAAATATCACACAAATCACCA for SALL3 unmethylation, ATTAGTTGGTGGTGAACGTAGTGC and CTAAACAACAAATTCGCCTCGCCG for SOCS-3 methylation, GAGTATTAGTTGGTGGTGAATGTAGTGT and AACACTAAACAACAAATTCACCTCACCA for SOCS-3 unmethylation, TTTTTATTTTAGCGCGGTCGAGGC and CATAACCTCGAAAAACACGAACTCG for EMX1 methylation, AATTATTTTTATTTTAGTGTGGTTGAGGT and TAATTCATAACCTCAAAAAACACAAACTCA for EMX1 unmethylation, GATTCGCGGTCGTTGGTAGGGC and CTCGCTCACGATCAACCTAAACG for ECEL1 methylation, TTTTGGATTTGTGGTTGTTGGTAGGGT and CTCTTCTCACTCACAATCAACCTAAACA for ECEL1 unmethylation, GGGTTAGATTCGGGGCGTTTTTC and AATCCTCAAAAAAACTCAACGACCG for Hs.670807 gene methylation, and GTTTGGGGTTAGATTTGGGGTGTTTTTT and ACACAAATCCTCAAAAAAACTCAACAACCA for Hs.670807 gene unmethylation. To analyze SALL3 in demethylated clone 12 cells, another MSP primer pair (GGGGTATATTTTGGCGGTCGTTC and CACAAACGCGTACTCCCACCCG for methylation and TTGGGGGTATATTTTGGTGGTTGTTT and ACACACAAACACATACTCCCACCCA for unmethylation) was used, because the region in the SALL3 CpG island where this primer set was designed was more efficiently demethylated by 5Aza-dC treatment.
The primer pairs used in bisulfite sequencing are as follows: GGATTTTTTAAYGAAATTTTAGAGTAGT and ACAACTTACAAAATAAAAATTTCTTACAAA for SALL3, GTGYGTTATGGTTATTTATAGTAAGTT and AAAAACTACCCCCCTCACACTAAAT for SOCS-3, TATTATAGAGTTTTTGGTGGTTAAGGA and CAAAACCCAAAAATAAAAATAAAAAAAATC for EMX1, TGTTTGTTAGAGGGTTGGGAAATTG and ACRCCCCCTAAACRCCCTACCA for ECEL1, and GGGTGTGTGTATTTATATATTTGTAGT and TAACTCTCTCTAACCCCTACCTAAC for Hs.670807. To analyze SALL3 in demethylated clone 12 cells by bisulfite sequencing, a primer pair (GGGAAGTTTTAGGAGGTTATTTGTGT and ACCCCACACACTCRACCCCTAA) was used.
RNA interference.
For SALL3 small interfering RNA (siRNA) experiments, FLC4 cells plated in a 100-mm dish were transfected with 200 nM siRNA using oligofectamine reagent (Invitrogen) according to the manufacturer's instructions. The target sequences used for SALL3 depletion were AGCGAGCTCAGAAACAGCA (SALL3 siRNA1) and TGCTGCGCCGAGTTCTTCA (SALL3 siRNA2). The control siRNA was obtained from Qiagen. The target sequence used for DNMT3A depletion was CTACTACATCAGCAAGCGCAA.
Colony formation assay.
Cells were transfected with a SALL3 expression or backbone plasmid as described previously (25). Transfected HuH2 or Hep3B cells were selected for 4 weeks with 1 mg/ml or 500 μg/ml G418, respectively.
Retroviral transduction.
A retroviral vector with SALL3, SALL3-mut1, or DNMT3A was constructed in pLNCX2 (Takara). Retroviruses were produced according to the company's protocol. Briefly, 2 μg of either SALL3, SALL3-mut1, DNMT3A, or backbone pLNCX2 was cotransfected with 2 μg of pAmpho into GP2-293 cells plated on a 10-cm plate. At 48 h posttransfection, the viral supernatant was collected and added to clone 12 cells plated on a 35-mm dish. For a vector control, 1.2 ml of the viral supernatant from backbone pLNCX2-transfected cells was used. For SALL3 or DNMT3A transduction, 0.6 ml of the viral supernatant from SALL3- or DNMT3A-transfected cells was mixed with 0.6 ml of that from backbone vector-transfected cells. For cotransduction of DNMT3A with SALL3 or SALL3-mut1, 0.6 ml of the viral supernatant from DNMT3A was mixed with 0.6 ml of that from SALL3 or SALL3-mut1 and then added to clone 12 cells. After 8 h of the first infection, the second infection was carried out.
Immunoblotting and immunoprecipitation.
Immunoblotting and immunoprecipitation analyses were carried out essentially as described previously (32). Anti-Flag and anti-HA antibodies were obtained from Sigma and Roche, respectively. Anti-SALL3, anti-DNMT3A, and anti-DNMT3B antibodies used in immunoblotting analysis were obtained from Santa Cruz Biotechnology. Anti-DNMT3A antibody and normal rabbit immunoglobulin G (IgG) used in immunoprecipitation analysis were obtained from Novus Biologicals and Santa Cruz Biotechnology, respectively.
GST pull-down assay.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21 and purified on glutathione-Sepharose beads (GE Healthcare). 35S-labeled in vitro-transcribed-translated SALL3 or DNMT3A proteins, including full-length and deletion mutants (generated using the Promega TNT-coupled reticulocyte lysate system), were mixed with full-length GST-DNMT3A or GST-SALL3 deletion 6, respectively, in binding buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X, and 0.2% NP-40) and incubated for 1 h at 4°C with rotation. The beads were pelleted, washed three times with binding buffer, and then resuspended with sample buffer. The specifically bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For the competition assay, a fixed amount of 35S-labeled in vitro-transcribed-translated full-length EZH2 protein relative to increasing amounts of 35S-labeled in vitro transcribed-translated full-length SALL3 (FL-SALL3) protein was premixed. The mixture was then incubated with GST-DNMT3A, and the proteins specifically bound to GST-DNMT3A were analyzed by SDS-PAGE.
Methyltransferase activity assay.
DNMT3A was isolated from Flag-DNMT3A-transfected HEK293 cell lysates or SALL3-specific siRNA-transfected FLC4 cell lysates by immunoprecipitation using anti-Flag antibody (Sigma) or anti-DNMT3A antibody (Novus Biologicals), respectively. The immunoprecipitates were washed twice with immunoprecipitation buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X, and 0.2% NP-40) and three times with wash buffer (20 mM Tris-HCl). The methylation reaction was carried out using the immunoprecipitated DNMT3A and 150 ng SOCS-3 plasmid DNA as a substrate in methylation buffer (20 mM Tris-HCl [pH 7.5] and 25% glycerol) in the presence of S-adenosyl-l-[methyl-3H]methionine (GE Healthcare). After incubation at 37°C for 1 h, DNA was purified using a gel extraction kit (Qiagen) and then absorbed in a Whatman GF/C filter. The filter was washed with 10% trichloroacetic acid and dried. Then, incorporation of radioactivity was measured by liquid scintillation counting.
ChIP.
Formaldehyde cross-linking and chromatin immunoprecipitation (ChIP) were performed using the EZ ChIP chromatin immunoprecipitation kit (Upstate Biotechnology) according to the company's protocol. Briefly, cells were cross-linked in 1% formaldehyde. The fixed chromatin from 5 × 105 cells was sonicated, precleared, and then incubated with anti-Flag antibody (Sigma), anti-DNMT3A antibody (Novus Biologicals), normal mouse IgG, or normal rabbit IgG overnight at 4°C with rotation. After the chromatin was washed and eluted, cross-links were reversed at 65°C. The DNA was purified and used for PCR analysis. Primers used in ChIP analysis were as follows: AAATGCAAAACAGATCGATG and CACCTTTTCTGTTGCGTAAG for SALL3, CGCGGCCAAGCGCGGCTTTA and CCCAGCTGGTGCGCCGGATG for EMX1, ATGGTCACCCACAGCAAGT and GGTCCGAGCTGTCGCGGAT for SOCS-3, CCGGGGCATTCGTAGGT and CAGCCTGGACGACAGGAACT for ECEL1, and ATCTCCCATCTCACAGCCGA and TTGCTTTCTATTTCCAGCGAG for the Hs.670807 gene.
RESULTS
Methylation-associated silencing of SALL3 in HCC.
We have isolated a NotI site that showed a cancer-specific alteration using a chromosome-assigned restriction landmark genomic scanning method (25, 32, 44-46). We found that the NotI site was located in the 5′ region of the SALL3 gene. Cloning of the NotI site-related SALL3 gene using normal liver RNA revealed that the gene was different from a registered SALL3 gene (accession no. NM_171999) by 216 bp spliced out from nucleotide 2917 through nucleotide 3132 (22). This SALL3 gene was also different from another reported form, in which the same region was spliced out but associated with a different 5′ region (accession no. AJ007421). Therefore, we termed this alternatively spliced form, encoding a protein of 1,228 amino acids, SALL3c. SALL3c is composed of eight C2H2-type zinc fingers, among which four fingers make 2 units of a DZF motif (Fig. 1A). SALL3c and SALL3 (accession no. NM_171999) share the same promoter region, while SALL3 (accession no. AJ007421) has another promoter. We analyzed the DNA methylation status of the promoter region for SALL3c and SALL3 (NM_171999) by MSP. We found that SALL3 was methylated in 5 of 10 HCC cell lines while a normal liver sample was unmethylated (Fig. 1B). The promoter region in the sequence under accession no. AJ007421 was fully methylated in normal liver (data not shown). Bisulfite sequencing analysis demonstrated that the promoter region was densely methylated in HuH2 and Hep3B cells while a normal liver and FLC4 cells showed minimum methylation (Fig. 1C). Next, we analyzed SALL3 RNA expression. We designed primers that recognize the three spliced forms of SALL3, including SALL3c. RT-PCR analysis revealed that among the HCC cell lines examined, only FLC4 expressed SALL3. A normal liver sample showed SALL3 expression as described previously (22) (Fig. 1D). This result demonstrates that methylated cell lines do not express SALL3 RNA. To determine whether DNA methylation is associated with a lack of expression in the HCC cell lines, three methylated cell lines in which SALL3 expression was undetectable were incubated with 5Aza-dC. We found that the methylated cell lines reactivated SALL3 (Fig. 1E), supporting that SALL3 is silenced by the associated DNA methylation in these three cell lines. We also examined the DNA methylation status around the translation start site. However, this region was partially methylated in SALL3-expressing FLC4 cells (data not shown), indicating that methylation at around −3 kb from the translation start site where we analyzed the HCC cell lines (Fig. 1B and C) is more strongly associated with expression silencing. In the SALL3-unmethylated cell lines without SALL3 expression, SALL3 might be inactivated by mechanisms other than DNA methylation, such as histone modifications. We then analyzed methylation of primary HCC samples, and 11 of 30 samples (37%) were methylated. In contrast, the nontumorous liver samples did not show detectable methylation (Fig. 1F). Given the methylation-associated silencing of SALL3, we tested whether SALL3 is involved in the regulation of cell growth. Using two cell lines in which SALL3 was silenced by the associated DNA methylation, we performed a colony formation assay. In this study, we used a SALL3c construct as SALL3, because the SALL3c gene that we cloned from normal liver RNA encoded the common C2H2-type zinc fingers. The colony numbers of SALL3-transfected cells were decreased compared with the control transfectants, demonstrating that SALL3 was able to suppress cell growth (Fig. 1G).
FIG. 1.
Methylation-associated silencing of SALL3. (A) Schematic representation of the SALL3c protein. SALL3c contains eight C2H2-type zinc fingers (vertical bars). Four fingers compose two DZF motifs. NP_741996 and Q9BXA9 are coded by NM_19999 and AJ007421, respectively. (B) Upper panel, CpG density in the SALL3 promoter region. Vertical bars indicate CpG sequences. The translation start site is indicated as “1.” Arrows and arrowheads represent primer sets for MSP and bisulfite sequencing, respectively. Bottom panel, MSP analysis of primary HCC cell lines. HuH1 through FLC4 are HCC cell lines. Visible bands in lanes M are methylated products, and those in lanes U are unmethylated products. (C) Bisulfite sequencing analysis. Methylation status of the 5′ region of SALL3 (from −3139 through −2431) was investigated in three HCC cell lines (FLC4, HuH2, and Hep3B) and a normal liver sample. Ten individual clones were sequenced for each sample. Filled and open circles represent methylation and unmethylation, respectively. (D) RT-PCR analysis. Total RNA samples from 10 HCC cell lines and a normal liver were analyzed by RT-PCR with SALL3-specific primers (upper panel). GAPDH amplification verified the consistency of the RT-PCR (bottom panel). (E) Reactivation by a methylation inhibitor. Three methylated cell lines (Hep3B, HuH2, and HuH7) were treated with or without 5Aza-dC, and SALL3 expression was analyzed by RT-PCR. (F) MSP of primary HCC samples. Methylation of SALL3 was analyzed in 30 primary HCC samples by MSP. Representatives from the 30 samples are shown. Tumor samples are methylated in cases 1 through 5 and unmethylated in cases 6 through 10. “T” represents a tumor sample, and “N” represents its nontumorous counterpart. (G) Colony formation assay. Methylation-silenced cells (Hep3B and HuH2) were transfected with either the SALL3 expression vector or the backbone vector and selected for 4 weeks with G418.
Interaction of SALL3 with DNMT3A.
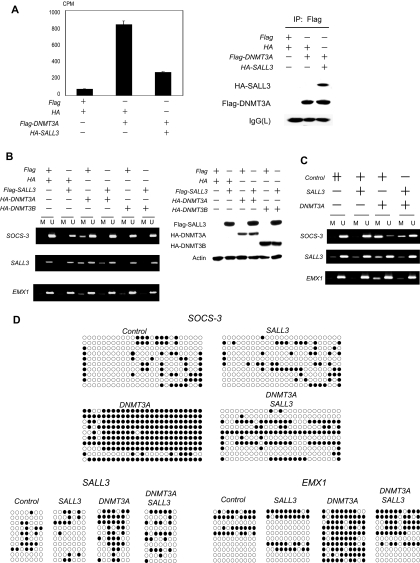
Mouse Sall1 was reported to interact with components of a HDAC complex, including HDAC and MTA, but Sall1 was not shown to interact with Dnmt3 (20). We speculated that SALL3 interacts with a factor that can associate with HDAC complexes. We examined the possibility that SALL3 might interact with DNMT3 by immunoprecipitation analysis. Substantial amounts of DNMT3A precipitated with SALL3 from the cotransfected cell lysate, while DNMT3B was precipitated less readily (Fig. 2A). Because SALL3 interacted with DNMT3A more efficiently than with DNMT3B, we studied further the functions of SALL3 with respect to DNMT3A. Next, we examined the interaction between the endogenous SALL3 and DNMT3A proteins. The endogenous proteins were coprecipitated with anti-DNMT3A antibody but not with control IgG from the nontransfected cell lysate. The amount of precipitated SALL3 was reduced markedly when DNMT3A was depleted by RNA interference (Fig. 2B). Because the anti-SALL3 antibody did not efficiently precipitate SALL3, we utilized HA-SALL3 stably expressing cells. Anti-HA immunoprecipitation demonstrated that endogenous DNMT3A precipitated with HA-SALL3 from the lysate of HA-SALL3-transfected cells but not from that with the HA tag alone (Fig. 2C). These results indicate that the interaction was not an artifact attributable to overexpression. To define the regions that are responsible for the interaction between SALL3 and DNMT3A, we produced deletion mutants of SALL3 and cotransfected them with full-length DNMT3A (FL-DNMT3A). DNMT3A coprecipitated with SALL3 deletion mutants 1, 4, and 6 as well as FL-SALL3 but not with SALL3 deletion mutants 2, 3, and 5. Expression of SALL3 deletion mutants and DNMT3A was at a similar level among the transfectants (Fig. 2D). Interestingly, all SALL3 mutants that interacted with DNMT3A contained the DZF motif, while deletion mutants lacking this motif failed to interact with DNMT3A. The DZF motif appears to be indispensable for the interaction with DNMT3A. To confirm the interaction between the DZF motif of SALL3 with DNMT3A in vitro, we performed a GST pull-down assay. SALL3 deletion mutants were produced by coupled in vitro transcription-translation in the presence of [35S]methionine and used for a pull-down assay with a GST fusion protein corresponding to FL-DNMT3A (GST-FL-DNMT3A). Among the three SALL3 deletion mutants, two mutants containing the DZF motif (deletions 1 and 6) bound selectively to GST-DNMT3A as well as FL-SALL3 while the mutant without the DZF (deletion 3) failed to do so (Fig. 2E). These results argue strongly that the DZF motif of SALL3 is responsible for the direct interaction with DNMT3A. Next, we investigated the domain in DNMT3A that is required for the interaction with SALL3. [35S]methionine-labeled DNMT3A deletion mutants were produced and tested for their ability to bind to SALL3. We used GST-SALL3 deletion mutant 6, which contains the DZF motif, instead of FL-SALL3, because FL-SALL3 could not be produced efficiently in bacteria. FL-DNMT3A bound strongly to SALL3 deletion mutant 6. DNMT3A deletion mutants 1 and 2, which contained the PWWP domain, also bound to SALL3 deletion mutant 6. However, deletion mutant 3, which retained the PHD domain but lacked the PWWP domain, failed to bind to SALL3 deletion mutant 6 (Fig. 2F), demonstrating that the PWWP domain is necessary for direct binding to SALL3. Taken together, we concluded that the DZF motif of SALL3 directly interacts with the PWWP domain of DNMT3A.
FIG. 2.
Interaction of SALL3 with DNMT3. (A) Binding of FL-SALL3 to DNMT3. HA-SALL3 or the backbone vector (HA) was cotransfected with the Flag-DNMT3A or Flag-DNMT3B expression vector. Anti-HA immunoprecipitates were analyzed by immunoblotting with anti-Flag and anti-HA antibodies. (B) Lysate from nontransfected HEK293 cells was immunoprecipitated (IP) with anti-DNMT3A antibody or normal rabbit IgG. The immunoblot was analyzed with anti-SALL3 and anti-DNMT3A antibodies. IVT is in vitro-transcribed-translated SALL3. IgG was used as a control to show that the anti-DNMT3A antibody does not precipitate SALL3 nonspecifically (left panel). Anti-DNMT3A immunoprecipitation was performed using HEK293 cells transfected with DNMT3A-specific siRNA or control siRNA (right panel). (C) HA-tagged SALL3 or HA-stably expressing cells were generated by transfection of HA-SALL3 or HA tag alone, respectively, and subsequent selection with G418. The cell lysates were immunoprecipitated with anti-HA antibody and analyzed by immunoblotting with anti-DNMT3A and anti-HA antibodies. (D) Interaction of SALL3 deletion mutants containing the DZF motif with DNMT3A. Various Flag-tagged SALL3 deletion mutants were cotransfected with FL-DNMT3A. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-DNMT3A antibody. Deletions 1 through 6 are shown schematically (left panel). The cell lysates were analyzed by immunoblotting with anti-DNMT3A and anti-Flag antibodies (right panel). Vector is the vector control. FL is FL-SALL3. (E) Direct binding of SALL3 deletion mutants containing the DZF motif to DNMT3A in vitro. Immobilized GST-FL-DNMT3A or GST alone was mixed with in vitro-translated 35S-labeled FL-SALL3 or SALL3 deletion mutants (1, 3, or 6). Beads were washed, and bound proteins were analyzed by SDS-PAGE and autoradiography. (F) Direct binding in vitro of DNMT3A deletion mutants containing the PWWP domain to the SALL3 deletion mutant containing the DZF motif. A GST pull-down assay was performed using immobilized GST-SALL3 deletion mutant 6 and in vitro-translated 35S-labeled FL-DNMT3A or DNMT3A deletion mutants (schematically shown in the upper panel).
Inhibition of DNMT3A-mediated CpG island methylation by SALL3.
Given the physical interaction between SALL3 and DNMT3A in cells and in vitro, we determined next whether SALL3 regulates the methyltransferase activity of DNMT3A in vitro. We measured incorporation of methyl groups from S-adenosyl-l-[methyl-3H]methionine into a SOCS-3 plasmid DNA using immunoprecipitated DNMT3A. We took advantage of SOCS-3 as the substrate, because aberrant methylation of SOCS-3 has been found in cancer (32). The immunocomplex from DNMT3A-transfected cells demonstrated a marked increase in methyltransferase activity compared with that from the control cells. Interestingly, coexpression of SALL3 with DNMT3A reduced DNMT3A-mediated methyltransferase activity. Immunoprecipitated DNMT3A was at a similar level in cells expressing exogenous DNMT3A alone and cells coexpressing DNMT3A and SALL3 (Fig. 3A). DNMT3A and -3B are believed to mediate de novo DNA methylation at several genomic loci (8, 34, 36). However, the precise mechanisms of de novo DNA methylation are not fully characterized. In particular, a cellular inhibitor of de novo DNA methyltransferases has not been identified. Because we found that SALL3 inhibited the methyltransferase activity of DNMT3A in vitro, we sought a role for SALL3 in inhibition of de novo DNA methylation. By incubation of HuH2 cells with 5Aza-dC for 9 days and subsequent cloning of surviving cells, we developed cancer cell clones in which aberrant DNA methylation was inducible. Using some of these clones, we examined the methylation status of CpG islands that are preferentially methylated in HCC (44). We found that most loci examined were demethylated or hypomethylated (data not shown). This enabled us to investigate efficiently changes in the CpG island methylation status in the cells. We transiently transfected DNMT3A into one of those clones (clone 12) and examined the methylation status of 10 CpG islands in which we had found cancer-specific DNA methylation in parental HuH2 cells. SOCS-3, SALL3, and EMX1 CpG islands became hypermethylated in DNMT3A-transfected cells. In contrast, those islands did not show detectable changes in DNMT3B-transfected cells or control cells transfected with backbone plasmids. Significantly, cotransfection of SALL3 with DNMT3A reduced DNMT3A-mediated methylation of the three CpG islands. DNMT3A expression was equivalent in cells with DNMT3A expression alone and in those with coexpression of DNMT3A and SALL3 (Fig. 3B). Other demethylated clones showed similar results to those for clone 12 (data not shown). We then developed stably transduced clone 12 cells using retrovirus vectors carrying DNMT3A or SALL3. In cells expressing DNMT3A stably, MSP analysis again demonstrated that SOCS-3, SALL3, and EMX1 CpG islands had increased levels of methylation. In particular, substantial methylation was present in the SOCS-3 CpG island. Consistent with the results from transient cotransfection, coexpression of SALL3 with DNMT3A reduced DNMT3A-mediated methylation (Fig. 3C). We then analyzed the stably transduced cells using bisulfite sequencing. DNMT3A-transduced cells acquired profound methylation compared with control cells, while cotransduction of SALL3 with DNMT3A reduced DNMT3A-mediated methylation substantially in the three CpG islands (Fig. 3D). These results indicate that DNMT3A promotes methylation in a fraction of the CpG islands that are sensitive to aberrant methylation in HCC. More importantly, SALL3 is able to inhibit DNMT3A-mediated methylation at the specific genomic loci. This inhibition of genomic methylation by SALL3 is quite consistent with SALL3-dependent inhibition of methyltransferase activity in vitro. Although we found promotion of methylation by DNMT3A in the three CpG islands, seven other islands examined were insensitive to DNMT3A as determined by MSP analysis (data not shown). We produced SALL3 mutants harboring point mutations by substituting key histidine and cysteine residues in the DZF motifs and used them in binding and DNA methylation analyses to confirm inhibition of DNMT3A-mediated CpG island methylation by SALL3. H437, C449, and H1058 were replaced with alanine residues in the mutants. In addition, the C1070A or H1086A mutation was generated in SALL3 mutant 1 (SALL3-mut1) or SALL3 mutant 2 (SALL3-mut2), respectively. Expression of SALL3-mut1 and -mut2 was equivalent to that of wild-type (WT) SALL3 in the transfected cells. Immunoprecipitation analysis revealed that the mutations reduced substantially the ability of SALL3 to interact with DNMT3A. Particularly, the interaction was undetectable in SALL3-mut1 (Fig. 3E, upper panel). Therefore, we used SALL3-mut1 in a methyltransferase activity assay. SALL3-mut1 failed to suppress the methyltransferase activity of DNMT3A in vitro, while WT SALL3 did so efficiently (Fig. 3E, upper panel). We then tested whether SALL3-mut1 is defective in inhibiting CpG island methylation mediated by DNMT3A in clone 12 cells. We coinfected WT SALL3 or SALL3-mut1 with DNMT3A using the viruses and examined the stably transduced cells by bisulfite sequencing. SALL3-mut1 did not suppress methylation of the SOCS-3, EMX1, and SALL3 CpG islands that was induced by DNMT3A infection, while WT SALL3 did reduce the DNMT3A-mediated hypermethylation (Fig. 3E, middle panel). These results strongly support that SALL3 negatively regulates activity of DNMT3A through DZF-dependent interaction between SALL3 and DNMT3A. We further tested whether SALL3-mut1 suppresses growth of HCC cells. A colony formation assay demonstrated that cells transfected with SALL3-mut1 reduced the colony numbers compared with those with the control vector. The suppression of colony formation by SALL3-mut1 was at a similar level to that by WT SALL3 (Fig. 3E, bottom panel).
FIG. 3.
DNMT3A-mediated CpG island methylation and its inhibition by SALL3. (A) In vitro methyltransferase activity assay. DNMT3A was isolated by anti-Flag immunoprecipitations from cells transfected with the plasmids indicated. A methyltransferase reaction was carried out using isolated DNMT3A, S-adenosyl-l-[methyl-3H]methionine, and SOCS-3-plasmid DNA. The values are given as means ± standard deviations of results for the three replicates. The difference in DNMT3A activity between cells transfected with DNMT3A and those transfected with DNMT3A and SALL3 is statistically significant (P = 2.0E−05) by a Student t test (left panel). The anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-Flag and anti-HA antibodies (right panel). (B) MSP analysis of transfected clone 12 cells in SOCS-3, SALL3, and EMX1 CpG islands. The cells were transiently transfected with the plasmids indicated. The genomic DNA was analyzed for DNA methylation using MSP (left panel). Expression of SALL3, DNMT3A, and DNMT3B in transfected clone 12 cells was analyzed by immunoblotting with anti-HA, anti-Flag, and antiactin antibodies (right panel). (C) MSP analysis of stably transduced cells. Cells were infected with retroviruses (SALL3 or DNMT3A alone or SALL3 plus DNMT3A). Control is the retrovirus from the backbone plasmid. The amount of the viral supernatant used in infection of SALL3 or DNMT3A alone was adjusted to that used in coinfection of SALL3 with DNMT3A by adding the viral supernatant from the control. After selection of drug-resistant cells for 2 weeks, the DNA methylation status was examined by MSP. (D) Bisulfite sequencing analysis of the stably transduced cells. The methylation status of SOCS-3, SALL3, and EMX1 CpG islands was examined. Ten individual clones were sequenced for each sample. Filled and open circles represent methylation and unmethylation, respectively. (E) Abrogation of SALL3-dependent inhibition of DNMT3A activity by mutations in the DZF motifs of SALL3. The two SALL3 mutants are shown schematically. A cysteine or histidine residue in each single zinc finger unit in the two DZF motifs of SALL3 was replaced with alanine. Flag-SALL3-mut1 or -mut2 or Flag-WT SALL3 was cotransfected with DNMT3A. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-Flag and anti-DNMT3A antibodies. A methyltransferase activity assay was performed using HA immunoprecipitates, S-adenosyl-l-[methyl-3H]methionine, and SOCS-3 plasmid DNA. The difference in DNMT3A activity between cells transfected with DNMT3A alone and those cotransfected with DNMT3A and SALL3 is statistically significant (P = 2.0E−05) by a Student t test. The difference between cells transfected with DNMT3A alone and those cotransfected with DNMT3A and SALL3-mut1 is not statistically significant (upper panel). Clone 12 cells were infected with retroviruses (DNMT3A alone, WT SALL3 plus DNMT3A, or SALL3-mut1 plus DNMT3A) as in panel C. After selection of drug-resistant cells, the DNA methylation status was examined by bisulfite sequencing (middle panel). Colony formation assay was performed as for Fig. 1G using WT SALL3, SALL3-mut1, or the backbone vector (bottom panel).
Enhancement of CpG island methylation by depletion of SALL3.
Next, we determined whether inhibition of SALL3 expression facilitates CpG island methylation in HCC cells. Among 10 HCC cell lines, endogenous SALL3 expression was found only in FLC4 cells, in which DNMT3A expression was also detectable (Fig. 1D and 4A). Therefore, we depleted SALL3 expression in FLC4 cells by RNA interference. While two siRNAs reduced SALL3 expression, siRNA1 was more effective at depleting SALL3 at the RNA and protein levels and was used in further experiments (Fig. 4B). We performed methyltransferase activity assay using endogenous DNMT3A in SALL3-depleted FLC4 cells. The immunocomplex from SALL3-depleted cells demonstrated greater activity than that from control siRNA-transfected cells. Immunoprecipitated DNMT3A was at similar levels in SALL3-depleted cell lysate and in the lysate of control siRNA-transfected cells (Fig. 4C). Next, we evaluated the methylation status of three CpG islands in SALL3-depleted cells. Because SOCS-3 and EMX1 CpG islands, where we had analyzed DNA methylation in clone 12 cells, were fully methylated in FLC4 cells (data not shown), we examined SALL3, ECEL1, and Hs.670807 CpG islands by bisulfite sequencing. We found that the three CpG islands had increased methylation levels in SALL3-depleted FLC4 cells compared with levels in control siRNA-transfected cells (Fig. 4D). We also determined whether depletion of SALL3 enhances CpG island methylation in another cancer cell line. We detected SALL3 expression in a prostate cancer cell line, PC3. Using SALL3-depleted PC3 cells, we examined the methylation status in SALL3 and Hs.670807 gene CpG islands by bisulfite sequencing. Levels of DNA methylation increased once more in SALL3-depleted cells (see Fig. S1 in the supplemental material). Based on the results from SALL3 expression-and-depletion experiments, we suggest strongly that SALL3 is a novel inhibitor of de novo DNA methylation in a fraction of CpG islands.
FIG. 4.
Hypermethylation by SALL3 depletion. (A) Endogenous DNMT3A expression in FLC4 cells. Cell lysates were analyzed by immunoblotting with anti-DNMT3A antibody. (B) Depletion of SALL3 by RNA interference. FLC4 cells were transfected with SALL3-specific or control siRNA. Cell lysates were analyzed by immunoblotting with anti-SALL3 antibody (left panel). RNA expression was analyzed by RT-PCR as for Fig. 1D (right panel). (C) In vitro methyltransferase activity assay. Endogenous DNMT3A was isolated by anti-DNMT3A immunoprecipitations from lysates of FLC4 cells transfected with SALL3 siRNA1 or control siRNA. A methyltransferase assay was carried out using immunoprecipitated DNMT3A, S-adenosyl-l-[methyl-3H]methionine, and SOCS-3 plasmid DNA. The values are given as means ± standard deviations for the three replicates. The difference in DNMT3A activity between cells transfected with SALL3 siRNA1 and those transfected with control siRNA is statistically significant (P = 0.0031) by a Student t test (left panel). The anti-DNMT3A immunoprecipitates were analyzed by immunoblotting with anti-SALL3 and anti-DNMT3A antibodies (right panel). (D) Bisulfite sequencing analysis of SALL3-depleted FLC4 cells. The methylation status of SALL3, ECEL1, and Hs.670807 CpG islands was examined. Hs.670807 is a registered gene in the UniGene system.
Inhibition of DNMT3A binding to chromatin by SALL3.
To better understand the mechanisms by which SALL3 inhibits DNMT3A-mediated CpG island methylation, we performed ChIP assays using transfected clone 12 cells either with DNMT3A or SALL3 alone or a combination of the two genes. In DNMT3A-transfected cells, DNMT3A bound to chromatin of the three CpG islands (SOCS-3, SALL3, and EMX1) (Fig. 5A) in which DNMT3A promoted DNA methylation (Fig. 3B to E). Significantly, cotransfection of SALL3 with DNMT3A reduced binding of DNMT3A to the three chromatin regions, while binding of DNMT3A to chromatin was at similar levels between cells transfected with DNMT3A alone and those cotransfected with DNMT3A and SALL3-mut1. DNMT3A expression was equivalent among these transfected cells (Fig. 5B). To confirm binding of DNMT3A to the specific chromatin regions and SALL3-dependent inhibition of the association between DNMT3A and chromatin, we performed ChIP assay using SALL3-depleted FLC4 cells. Immunoprecipitated chromatin that associated with endogenous DNMT3A was analyzed for SALL3, ECEL1, and Hs.670807 CpG islands where SALL3 depletion enhanced methylation in the corresponding CpG islands (Fig. 4D). We found that binding of DNMT3A to the three chromatin regions increased in SALL3-depleted cells compared with levels in control siRNA-transfected cells (Fig. 5C). Taken together, these results indicate that SALL3 is able to inhibit binding of DNMT3A to the specific chromatin regions, accounting for SALL3-dependent inhibition of CpG island methylation. We found negative regulation of DNMT3A by SALL3, which was mediated by the interaction of the DZF motif of SALL3 with the PWWP domain of DNMT3A. Histone methyltransferases, including SUV39H1 and EZH2, have been reported to interact with DNMT3 (12). The PHD domain of DNMT3 is the binding module for EZH2 (42). We speculated that SALL3 might have the ability to inhibit EZH2 binding to DNMT3A. To test this, we performed a competitive binding assay in vitro. GST pull-down using GST-DNMT3A, fixed amounts of EZH2, and increasing amounts of SALL3 demonstrated that SALL3 reduced binding of EZH2 to DNMT3A in a dose-dependent manner (Fig. 5D, upper panel). To examine SALL3-dependent inhibition of EZH2 binding to DNMT3A in cells, we cotransfected DNMT3A and EZH2 with or without SALL3 and analyzed the interaction by immunoprecipitation. EZH2 precipitated with DNMT3A from the cotransfected cell lysates. When SALL3 was cotransfected with EZH2 and DNMT3A, EZH2 precipitated less with DNMT3A (Fig. 5D, middle panel). We next examined the relationship between SALL3 and EZH2 in the methyltransferase activity of DNMT3A in vitro. The immunocomplex from EZH2- and DNMT3A-cotransfected cells increased activity of DNMT3A over that from cells transfected with DNMT3A alone. However, this increase was reduced by the additional transfection with SALL3 (Fig. 5D, middle panel). We then depleted SALL3 expression in FLC4 cells by RNA interference and examined the interaction between EZH2 and DNMT3A. The amount of precipitated EZH2 increased in SALL3-depleted cells compared with that in control siRNA-transfected cells (Fig. 5D, bottom panel). These results indicate that SALL3 is able to inhibit the physical interaction of EZH2 with DNMT3A, leading to abrogation of EZH2-mediated enhancement of DNMT3A activity. While we examined levels of CpG island methylation using clone 12 cells, the levels were similar in the cells transfected with DNMT3A alone and those cotransfected with EZH2 and DNMT3A by MSP (data not shown).
FIG. 5.
Inhibition of binding of DNMT3A to chromatin by SALL3. (A) Association of DNMT3A with chromatin. Clone 12 cells were transfected with either SALL3, DNMT3A, or the backbone plasmid. ChIP experiments were performed using anti-Flag antibody or control IgG. Associated DNA was analyzed by PCR with primers amplifying the CpG islands indicated (left panel). Cell lysates were analyzed by immunoblotting with anti-Flag antibody (right panel). (B) Reduced binding of DNMT3A to chromatin by SALL3. Cells were cotransfected with the plasmids indicated. ChIP experiments were performed as for panel A (left panel). Cell lysates were analyzed by immunoblotting with anti-Flag and anti-HA antibodies (right panel). (C) Enhancement of DNMT3A binding to chromatin by SALL3 depletion. FLC4 cells were transfected with SALL3 siRNA1 or control siRNA as for Fig. 4B. ChIP experiments were performed using anti-DNMT3A antibody or control IgG. Associated DNA was analyzed by PCR with primer pairs amplifying the CpG islands indicated. (D) Inhibition of the interaction between EZH2 and DNMT3A by SALL3. The effect of increasing amounts of in vitro-translated 35S-labeled SALL3 (0, 5, 10, and 20 μl) on the binding of a fixed amount of in vitro-translated 35S-labeled EZH2 (20 μl) to GST-DNMT3A was examined by in vitro competition assay (upper panel). Cells were transfected with the indicated expression vectors. The total amount of the vectors used in each transfection was adjusted using the backbone vector (HA). Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-Flag and anti-HA antibodies. The levels of HA-EZH2 or Flag-DNMT3A expression were similar in the cells transfected with HA-EZH2 or Flag-DNMT3A. A methyltransferase activity assay was performed using the immunoprecipitated DNMT3A, S-adenosyl-l-[methyl-3H]methionine, and SOCS-3 plasmid DNA. The difference in DNMT3A activity between cells transfected with Flag-DNMT3A alone and those cotransfected with Flag-DNMT3A and HA-EZH2 is statistically significant (P = 4.8E−06) by a Student t test. The difference between cells cotransfected with Flag-DNMT3A and HA-EZH2 and those cotransfected with Flag-DNMT3A, HA-EZH2, and HA-SALL3 is also statistically significant (P = 5.9E−05) (middle panel). FLC4 cells were transfected with SALL3 siRNA1 or control siRNA. Anti-DNMT3A immunoprecipitates were analyzed by immunoblotting with anti-SALL3, anti-EZH2, and anti-DNMT3A antibodies (bottom panel).
Induction of SALL3 expression by BMP-4.
dpp is known to induce sal expression in Drosophila (6). We determined whether BMP-4, the human orthologue of dpp, induces SALL3 expression by using FLC4 cells. BMP-4 increased the level of SALL3 protein in a dose-dependent manner. This induction by BMP-4 was also detected at the RNA level by RT-PCR analysis (Fig. 6A). The BMP-4-treated cells exhibited increased phosphorylation of Smad1 (Fig. 6B), showing that BMP-4 signaling was intact in FLC4 cells. We also examined induction of SALL3 by BMP-4 using a luciferase assay. The SALL3 CpG island, which extends 3 kb upstream to the translation start site, was divided and cloned into two reporter constructs. Both fragments increased promoter activities upon treatment with BMP-4. Interestingly, the segment proximal to the translation start site showed greater activity (Fig. 6C). Taken together, these results demonstrate that BMP-4 is able to transactivate SALL3.
FIG. 6.
Induction of SALL3 by BMP-4 signaling. (A) Induction of SALL3 expression. FLC4 cells were starved and incubated with the concentrations of BMP-4 indicated. Cell lysates were analyzed by immunoblotting with anti-SALL3 antibody (upper panel). For RT-PCR analysis, cells were incubated with or without BMP-4 (50 ng/ml). Total RNA was analyzed as for Fig. 1D (bottom panel). (B) Induction of Smad1 phosphorylation. Lysates from BMP-4 (50 ng/ml)-treated or nontreated cells were analyzed by immunoblotting with anti-phospho-Smad1 antibody (P-Smad1) and antibody recognizing Smad1, -5, and -8. (C) Promoter assay. The SALL3 CpG island upstream of the translation start site was divided into two fragments and used for a luciferase assay (left panel). Either promoter construct was cotransfected with a reference plasmid into FLC4 cells. Cells were starved and incubated with or without BMP-4 (50 ng/ml). Luciferase activities were measured at 48 h posttransfection. The values of SALL3 reporter activities were normalized to those of the reference reporter and are the means for three replicates. A Student t test was used to generate the P values. The differences between BMP-4-treated and nontreated cells are statistically significant (P = 4.4E−05 and 0.0016 for promoters 1 and 2, respectively) (right panel).
DISCUSSION
We found that SALL3 bound directly to DNMT3A. Endogenous SALL3 and DNMT3A, as well as the exogenously expressed proteins, interacted in cells. The recombinant proteins also interacted in vitro. The sal and SALL proteins are characterized by a DZF motif that is composed of paired C2H2 zinc fingers, although the functions of the motif have been characterized poorly (41). The DZF motif of SALL3 interacted directly with the PWWP domain of DNMT3A. Therefore, the DZF motif appears to be a binding module for the PWWP domain, at least in part. Because SALL2 was shown to bind to DNA through its triple zinc finger motif (27), SALL3 also might interact with DNA through this triple zinc finger, to which DNMT3A failed to bind. The PWWP domain has been suggested to be involved in protein-protein interaction based on the similarity to other protein-binding modules and the location in the proteins (39). As expected, we found that the PWWP domain of DNMT3A bound directly to the DZF motif of SALL3. DNMT3B interacted less readily with SALL3 than did DNMT3A. Therefore, the PWWP domains of DNMT3A and DNMT3B appear to have different activities in the interactions. This is also suggested by the finding that the PWWP domain of DNMT3B bound to DNA more efficiently than that of DNMT3A (5).
We identified SALL3 as a novel inhibitory protein for DNMT3A. DNMT3A-mediated DNA methylation at CpG islands that are preferentially methylated in HCC was substantially reduced by transient and stable expression of SALL3. SALL3 depletion by RNA interference resulted in hypermethylation of CpG islands. Consistently, expression or depletion of SALL3 reduced or enhanced, respectively, in vitro methyltransferase activity of DNMT3A. In addition, substitutions of key residues in the DZF motifs of SALL3 disrupt the interaction of SALL3 with DNMT3A and impair the ability of SALL3 to inhibit the methyltransferase activity of DNMT3A in vitro and DNMT3A-mediated CpG island methylation in cells. To our knowledge, this is the first demonstration that a cellular protein inhibits activity of de novo DNMT. In the SALL3 depletion experiments, we used the FLC4 and PC3 cell lines. Some CpG islands in these cells already were methylated despite the expression of SALL3. We suggest several possibilities for aberrant CpG island methylation found in SALL3-expressing cells. The first possibility is that activity of DNMT3A is regulated by a balance between DNMT3A and its cofactors. We found that coexpression of SALL3 with DNMT3A reduced levels of CpG island methylation that were increased by DNMT3A expression. Even in cells with preexisting CpG island methylation, depletion of SALL3 enhanced aberrant CpG island methylation. SALL3 expression or depletion reduced or enhanced, respectively, the association of DNMT3A with chromatin. In addition, SALL3 competed with EZH2 for binding to DNMT3A. When the amount of DNMT3A or a stimulatory factor exceeds that of SALL3, we suggest that DNMT3A is able to associate with chromatin, resulting in a condition in which DNMT3A potentiates de novo DNA methylation. Along this line, if cells with unmethylated SALL3 express insufficient SALL3 due to a lack of induction by BMP-4, DNMT3A might have the ability to methylate DNA. Notably, methylation-associated silencing of SALL3 is a critical event by which the cells structurally lose the negative regulatory factor, resulting in enhancement of DNMT3A activity. Sall3 knockout mice die soon after birth with deficiencies in cranial nerves, showing the critical involvement of Sall3 in embryonic development (35). Embryonic fibroblasts from sall3-deficient animals may be useful for promoting aberrant DNA methylation in normal cells. The second possibility is that other enzymes might methylate CpG islands in SALL3-expressing cells. The role of SALL3 in DNMT3B activity remains to be elucidated. The third possibility is that SALL3 collaborates with other proteins to inhibit DNMT3A, although these others have not been identified yet. Defects in the accessory proteins might reduce the activity of SALL3. It will be interesting to determine the functions of other SALL family proteins in relation with SALL3.
Our ChIP assays revealed that binding of DNMT3A to chromatin regions at the specific CpG islands where DNMT3A promoted DNA methylation was reduced by wild-type SALL3 but not by mutant SALL3 that is defective in binding to DNMT3A. Consistent with this, SALL3 depletion by RNA interference resulted in increased binding of DNMT3A to chromatin regions in which the corresponding CpG islands were hypermethylated by SALL3 depletion. These findings demonstrate that SALL3 is able to inhibit the association of DNMT3A with chromatin. Thus, SALL3 has the ability to inhibit DNMT3A-mediated DNA methylation. We postulate two possible mechanisms by which SALL3 inhibits the association of DNMT3A with chromatin. We found that SALL3 interacted directly with DNMT3A through binding of the DZF motif of SALL3 to the PWWP domain of DNMT3A. Because the PWWP domain of DNMT3 is thought to interact directly with DNA (5), binding of SALL3 to the PWWP domain may inhibit the ability of DNMT3A to interact with DNA. Secondly, DNMT3A no longer associates with its cofactors for methylation of DNA when SALL3 binds to the PWWP domain of DNMT3A. SALL3 competed with EZH2, which has been reported to bind to the PHD domain of DNMT3A (42). This may suggest that the interaction of DNMT3A with another protein of chromatin components is inhibited by SALL3 binding. We do not exclude another possibility, that SALL3 directly binds to promoter regions, preventing DNMT3A from accessing the chromatin. Because Drosophila sal has been reported to be a transcription factor (6, 40), SALL3 may occlude promoter regions by binding directly. However, we did not detect binding of SALL3 to chromatin regions where DNMT3A promoted DNA methylation.
Our findings identify a novel function of the PWWP domain through which SALL3 is able to inhibit DNMT3A activity. This striking function of the PWWP domain will be useful for manipulation of DNMT3A activity. In particular, targeting of the PWWP domain is one of the promising strategies for protecting the genome from de novo DNA methylation by DNMT3A. Another possibility for inhibiting de novo DNA methylation may be to interfere with the PHD domain-mediated interaction of DNMT3A. Histone methyltransferases, such as SUV39H1 and EZH2, have been reported to form complexes with DNMT3. The PHD domain of DNMT3 is the functional domain for this interaction (10, 12, 42). Inactivation of histone methyltransferases in embryonic stem cells reduced DNA methylation levels at major satellites or several imprinted genes (26, 43). These data suggest a significant relationship between DNA methylation and histone methylation. We found that SALL3 reduced EZH2 binding to DNMT3A and EZH2-mediated enhancement of DNMT3A activity. Therefore, abrogation of DNMT3A binding to other chromatin components, mediated by the PHD domain of DNMT3A, may be effective in inhibiting de novo DNA methylation. Small molecules that are functionally similar to SALL3, if developed, would be expected to inactivate DNMT3A regardless of the SALL3 expression status. The interaction between the DZF motif and the PWWP domain provides significant insight into the regulatory mechanisms of DNMT3A activity. Our data suggest that the internal cysteine residue between the two zinc finger units in a DZF motif is critically involved in the interaction with DNMT3A. Precise analyses of the interaction, such as the determination of tertiary structure, will be helpful in understanding the mechanisms of DNMT3A-mediated DNA methylation and its inhibition in more detail.
We found aberrant methylation of SALL3 in HCC. SALL3 expression was undetectable in methylated HCC cell lines in which SALL3 was reactivated by 5Aza-dC treatment. These results suggest strongly that SALL3 is silenced by associated DNA methylation. Wild-type SALL3 but not mutant SALL3 in the DZF motifs suppressed cell growth when introduced into cells lacking SALL3, suggesting that SALL3 is involved in the regulation of cell growth. This growth suppression is likely to be independent of the DZF-mediated interaction. We also found that SALL3 expression is inducible by BMP-4 signaling. BMP-4, a member of TGF family proteins, is known to play a significant role in cell growth (13). BMP-4 signaling may potentiate aberrant DNA methylation when SALL3 is silenced in the cell. We suggest that methylation silencing of SALL3 facilitates the association of DNMT3A with chromatin and enhances activity of DNMT3A, resulting in a condition in which DNMT3A potentiates de novo DNA methylation. Therefore, inactivation of SALL3 by DNA methylation accelerates aberrant DNA methylation in HCC.
Supplementary Material
Acknowledgments
This research was supported by a Grant-in-Aid for Scientific Research (S) from Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 12 January 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bachman, K. E., B. H. Park, I. Rhee, H. Rajagopalan, J. G. Herman, S. B. Baylin, K. W. Kinzler, and B. Vogelstein. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 389-95. [DOI] [PubMed] [Google Scholar]
- 2.Ballestar, E., M. F. Paz, L. Valle, S. Wei, M. F. Fraga, J. Espada, J. C. Cigudosa, T. H. Huang, and M. Esteller. 2003. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 226335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird, A. 1992. The essentials of DNA methylation. Cell 705-8. [DOI] [PubMed] [Google Scholar]
- 4.Bohm, J., F. J. Kaiser, W. Borozdin, R. Depping, and J. Kohlhase. 2007. Synergistic cooperation of Sall4 and Cyclin D1 in transcriptional repression. Biochem. Biophys. Res. Commun. 356773-779. [DOI] [PubMed] [Google Scholar]
- 5.Chen, T., N. Tsujimoto, and E. Li. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 249048-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Celis, J. F., R. Barrio, and F. C. Kafatos. 1996. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381421-424. [DOI] [PubMed] [Google Scholar]
- 7.Di Croce, L., V. A. Raker, M. Corsaro, F. Fazi, M. Fanelli, M. Faretta, F. Fuks, F. Lo Coco, T. Kouzarides, C. Nervi, S. Minucci, and P. G. Pelicci. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 2951079-1082. [DOI] [PubMed] [Google Scholar]
- 8.Dodge, J. E., M. Okano, F. Dick, N. Tsujimoto, T. Chen, S. Wang, Y. Ueda, N. Dyson, and E. Li. 2005. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 28017986-17991. [DOI] [PubMed] [Google Scholar]
- 9.Fahrner, J. A., S. Eguchi, J. G. Herman, and S. B. Baylin. 2002. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 627213-7218. [PubMed] [Google Scholar]
- 10.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 312305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuks, F., P. J. Hurd, D. Wolf, X. Nan, A. P. Bird, and T. Kouzarides. 2003. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2784035-4040. [DOI] [PubMed] [Google Scholar]
- 12.Geiman, T. M., U. T. Sankpal, A. K. Robertson, Y. Zhao, and K. D. Robertson. 2004. DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochem. Biophys. Res. Commun. 318544-555. [DOI] [PubMed] [Google Scholar]
- 13.Heldin, C. H., K. Miyazono, and P. ten Dijke. 1997. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390465-471. [DOI] [PubMed] [Google Scholar]
- 14.Herman, J. G., J. R. Graff, S. Myohanen, B. D. Nelkin, and S. B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 939821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Jones, P. A., and S. B. Baylin. 2007. The epigenomics of cancer. Cell 128683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19187-191. [DOI] [PubMed] [Google Scholar]
- 18.Jurgens, G. 1988. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 7189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Kiefer, S. M., B. W. McDill, J. Yang, and M. Rauchman. 2002. Murine Sall1 represses transcription by recruiting a histone deacetylase complex. J. Biol. Chem. 27714869-14876. [DOI] [PubMed] [Google Scholar]
- 21.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 3189-97. [DOI] [PubMed] [Google Scholar]
- 22.Kohlhase, J., S. Hausmann, G. Stojmenovic, C. Dixkens, K. Bink, W. Schulz-Schaeffer, M. Altmann, and W. Engel. 1999. SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics 62216-222. [DOI] [PubMed] [Google Scholar]
- 23.Kohlhase, J., M. Heinrich, L. Schubert, M. Liebers, A. Kispert, F. Laccone, P. Turnpenny, R. M. Winter, and W. Reardon. 2002. Okihiro syndrome is caused by SALL4 mutations. Hum. Mol. Genet. 112979-2987. [DOI] [PubMed] [Google Scholar]
- 24.Kohlhase, J., A. Wischermann, H. Reichenbach, U. Froster, and W. Engel. 1998. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat. Genet. 1881-83. [DOI] [PubMed] [Google Scholar]
- 25.Kubo, T., J. Yamamoto, Y. Shikauchi, Y. Niwa, K. Matsubara, and H. Yoshikawa. 2004. Apoptotic speck protein-like, a highly homologous protein to apoptotic speck protein in the pyrin domain, is silenced by DNA methylation and induces apoptosis in human hepatocellular carcinoma. Cancer Res. 645172-5177. [DOI] [PubMed] [Google Scholar]
- 26.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 131192-1200. [DOI] [PubMed] [Google Scholar]
- 27.Li, D., Y. Tian, Y. Ma, and T. Benjamin. 2004. p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP). Mol. Cell. Biol. 243885-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mollereau, B., M. Dominguez, R. Webel, N. J. Colley, B. Keung, J. F. de Celis, and C. Desplan. 2001. Two-step process for photoreceptor formation in Drosophila. Nature 412:911-913. [DOI] [PubMed] [Google Scholar]
- 29.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393386-389. [DOI] [PubMed] [Google Scholar]
- 30.Netzer, C., L. Rieger, A. Brero, C. D. Zhang, M. Hinzke, J. Kohlhase, and S. K. Bohlander. 2001. SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/PIN2 and localizes to pericentromeric heterochromatin. Hum. Mol. Genet. 103017-3024. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen, C. T., D. J. Weisenberger, M. Velicescu, F. A. Gonzales, J. C. Lin, G. Liang, and P. A. Jones. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 626456-6461. [PubMed] [Google Scholar]
- 32.Niwa, Y., H. Kanda, Y. Shikauchi, A. Saiura, K. Matsubara, T. Kitagawa, J. Yamamoto, T. Kubo, and H. Yoshikawa. 2005. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 246406-6417. [DOI] [PubMed] [Google Scholar]
- 33.Ohm, J. E., K. M. McGarvey, X. Yu, L. Cheng, K. E. Schuebel, L. Cope, H. P. Mohammad, W. Chen, V. C. Daniel, W. Yu, D. M. Berman, T. Jenuwein, K. Pruitt, S. J. Sharkis, D. N. Watkins, J. G. Herman, and S. B. Baylin. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99247-257. [DOI] [PubMed] [Google Scholar]
- 35.Parrish, M., T. Ott, C. Lance-Jones, G. Schuetz, A. Schwaeger-Nickolenko, and A. P. Monaghan. 2004. Loss of the Sall3 gene leads to palate deficiency, abnormalities in cranial nerves, and perinatal lethality. Mol. Cell. Biol. 247102-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416552-556. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 272291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlesinger, Y., R. Straussman, I. Keshet, S. Farkash, M. Hecht, J. Zimmerman, E. Eden, Z. Yakhini, E. Ben-Shushan, B. E. Reubinoff, Y. Bergman, I. Simon, and H. Cedar. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39232-236. [DOI] [PubMed] [Google Scholar]
- 39.Stec, I., S. B. Nagl, G. J. van Ommen, and J. T. den Dunnen. 2000. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473:1-5. [DOI] [PubMed] [Google Scholar]
- 40.Sturtevant, M. A., B. Biehs, E. Marin, and E. Bier. 1997. The spalt gene links the A/P compartment boundary to a linear adult structure in the Drosophila wing. Development 12421-32. [DOI] [PubMed] [Google Scholar]
- 41.Sweetman, D., and A. Munsterberg. 2006. The vertebrate spalt genes in development and disease. Dev. Biol. 293285-293. [DOI] [PubMed] [Google Scholar]
- 42.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J. M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871-874. [DOI] [PubMed] [Google Scholar]
- 43.Xin, Z., M. Tachibana, M. Guggiari, E. Heard, Y. Shinkai, and J. Wagstaff. 2003. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 27814996-15000. [DOI] [PubMed] [Google Scholar]
- 44.Yoshikawa, H., S. de la Monte, H. Nagai, J. R. Wands, K. Matsubara, and A. Fujiyama. 1996. Chromosomal assignment of human genomic NotI restriction fragments in a two-dimensional electrophoresis profile. Genomics 3128-35. [DOI] [PubMed] [Google Scholar]
- 45.Yoshikawa, H., K. Matsubara, G. S. Qian, P. Jackson, J. D. Groopman, J. E. Manning, C. C. Harris, and J. G. Herman. 2001. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat. Genet. 2829-35. [DOI] [PubMed] [Google Scholar]
- 46.Yoshikawa, H., K. Matsubara, X. Zhou, S. Okamura, T. Kubo, Y. Murase, Y. Shikauchi, M. Esteller, J. G. Herman, X. Wei Wang, and C. C. Harris. 2007. WNT10B functional dualism: beta-catenin/Tcf-dependent growth promotion or independent suppression with deregulated expression in cancer. Mol. Biol. Cell 184292-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, J., W. L. Tam, G. Q. Tong, Q. Wu, H. Y. Chan, B. S. Soh, Y. Lou, J. Yang, Y. Ma, L. Chai, H. H. Ng, T. Lufkin, P. Robson, and B. Lim. 2006. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 81114-1123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.