Abstract
IbeR is a regulator present in meningitic Escherichia coli strain E44 that carries a loss-of-function mutation in the stationary-phase (SP) regulatory gene rpoS. In order to determine whether IbeR is an SP regulator in E44, two-dimensional gel electrophoresis and LC-MS were used to compare the proteomes of a noninvasive ibeR deletion mutant BR2 and its parent strain E44 in the SP. Four up-regulated (TufB, GapA, OmpA, AhpC) and three down-regulated (LpdA, TnaA, OpmC) proteins in BR2 were identified when compared to E44. All these proteins contribute to energy metabolism or stress resistance, which is related to SP regulation. One of the down-regulated proteins, tryptophanase (TnaA), which is regulated by RpoS in other E. coli strains, is associated with SP regulation via production of a signal molecule indole. Our studies demonstrated that TnaA was required for E44 invasion, and that indole was able to restore the noninvasive phenotype of the tnaA mutant. The production of indole was significantly reduced in BR2, indicating that ibeR is required for the indole production via tnaA. Survival studies under different stress conditions indicated that IbeR contributed to bacteria stress resistance in the SP. Taken together, IbeR is a novel regulator contributing to the SP regulation.
1. Introduction
Neonatal bacterial meningitis continues to be the most common serious infection of the central nervous system (CNS) in newborns with high morbidity and mortality despite the availability of effective bactericidal antibiotics over the last sixty years [1, 2]. This high morbidity and mortality are due to inadequate knowledge of the pathogenesis of this disease.
E. coli is the most common gram-negative bacterium causing neonatal sepsis and meningitis [1]. Bacterial meningitis in newborns is due to hematogenous spread of the pathogen to the meninges. The most important issue in the pathogenesis of E. coli meningitis is how circulating pathogens cross the blood-brain barrier (BBB), which is mainly composed of brain microvascular endothelial cells (BMECs) [3, 4]. Our previous studies showed that E. coli K1 invasion of human BMEC was significantly greater with stationary-phase (SP) cultures than with exponential-phase cultures, suggesting that expression of E. coli K1 invasion-associated virulence genes is strongly regulated in a growth-phase-dependent manner [5]. A nonsense mutation in the SP regulatory gene rpoS was identified in E. coli K1 strains E44 and IHE3034 [5]. Complementation with the wild type E. coli K12 rpoS gene significantly enhanced IHE3034 invasion of BMEC, but failed to improve the invasion activity of another E. coli K1 strain E44. These studies suggest that the growth-phase-dependent invasion of BMEC by IHE3034 is affected by RpoS and that E44 carries a loss-of-function mutation in the rpoS gene. However, the SP gene regulation in E44 has remained an unanswered question.
Several virulence factors, including Ibe (termed after invasion of brain endothelial cells) proteins [6, 7], OmpA [8], YiijP [9], FimH [13], AslA [14], and TraJ [15], have been identified in various strains of E. coli in the in vitro and in vivo models of the BBB as invasins. Most of those invasion genes are present in the E. coli K-12 genome [4, 16]. However, the ibeA gene encoding a 50 kDa protein has been found to be unique to some pathogenic E. coli K1 strains (e.g., C5 and RS218), while laboratory strains of E. coli K-12 (e.g., DH5 and HB101), as well as noninvasive E. coli (e.g., E412), lack ibeA [4]. Recently, vimentin has been identified as an IbeA-binding protein on the surface of human BMEC [17].
Using the ibeA gene as a probe, we have identified a 20.3 kb genomic locus as a genetic island of meningitic E. coli containing ibeA (GimA) [16]. This locus is situated between yjiD and yjiE, adjacent to the fim operon, and absent in nonpathogenic E. coli K12 strains. GimA consists of 15 genes that form 4 operons. The first three operons (ptnIPKC, cglDTEC, gcxKRCI) may be involved in energy metabolism and the last operon (ibeRAT) contributes to E. coli K1 invasion of BMEC. Our previous work showed that GimA-mediated invasion of human endothelial cells is regulated by carbon source [16]. This is consistent with the observations by others that carbon source modulates expression of virulence factors in several pathogenic bacteria [4]. The ibeRAT operon encodes IbeR, IbeA, and IbeT. IbeA and IbeT contribute to E. coli K1 invasion and adhesion [18, 19]. Our previous studies suggest that IbeR is a novel regulatory protein that is present in pathogenic E. coli K1 [16]. It belongs to the NtrC/NifA family of transcriptional activators, carrying a sigma 54-interaction domain and showing significant sequence homology to various regulatory proteins for glycerol metabolism operon in Citrobacter freundii (P45512), acetoacetate metabolism in E. coli K12, sigma L-dependent transcription in B. subtilis (P54529), NIF-specific regulation in Herbaspirillum seropedicae, dhaR transcription in E. coli K12, and globe signal transduction in Clostridium beijerinckii [4, 20]. However, it is unknown how IbeR contributes to the pathogenesis of meningitic infection by modulating the virulence of the pathogen. As E44 carries a nonsense mutation in the rpoS gene and exhibits strong invasion activity in the SP, there must be alternative regulatory mechanisms responsible for the SP gene expression. We speculated that ibeR is an rpoS-like regulator in SP gene expression in E44. In order to dissect the regulation of SP gene expression in E44 that is associated with the pathogenesis of E. coli meningitis, a comparative proteomic analysis of an ibeR deletion mutant (BR2) and its parent strain E44 was carried out in this study. Our studies suggested that the ibeR gene was involved in regulating SP gene expression related to stress resistance and pathogenesis.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Medium
The bacterial strain and plasmid vectors and their relevant characteristics are shown in Table 1. The mutant strains used in this study were derived from E44, which is a rifampin-resistant strain derived from a neonatal meningitis isolate, E. coli K1 RS218 (O18 : K1 : H7) [4, 21]. E. coli DH5α and pGEM-T easy vector were used for subcloning. SM10 (λpir), DH5α (λpir), and pCVD442 were used for making isogenic deletion mutants of ibeR and tnaA [6, 10, 11]. E. coli K12 strain MC4100 [22] and its rpoS insertion mutation Tn10 mutant RH90 [23] were used as positive and negative controls for RpoS, respectively. Plasmid pStyABB, which carries the gene for monooxygenase, was used for indole assay [12]. E. coli strains were grown at 37°C in Luria broth (LB; 1% tryptone, 0.5% yeast extract, 0.5% NaCl) or brain heart infusion (BHI, Difco Laboratories, Detroit, Mich, USA) broth and were stored at −70°C in LB plus 20% glycerol. When it was necessary, the medium was supplemented with ampicillin (100 μg/mL) and rifampin (100 μg/mL) for the positive selection of plasmids or bacterial strains (Table 1).
Table 1.
E. coli K1 (meningitic) or K12 (nonpathogenic) strains and plasmids used in this study.
| Strain or plasmid | Characteristics | Reference(s) |
|---|---|---|
| Strains | ||
| RS218 | O18 : K1 : H7 (CSF) | [6–9] |
| E44 | RS218, Rifr | [6–9] |
| DH5α (λpir) | K 12 strain | [6, 7] |
| SM10 (λpir) | K 12 strain | [6, 7] |
| MC4100 | K 12 strain | [5] |
| RH90 | K 12 strain, rpoS deletion mutant | [5] |
| BR2 | ibeR deletion mutant of E44 | This study |
| TNA44 | tnaA deletion mutant of E44 | This study |
| Plasmids | ||
| pCVD442 | Ampr, oiRr6K, sacB, mobRP4 | [10, 11] |
| pCBR2 | pCVD442 carrying an ibeR deletion, Ampr | This study |
| pGEM-T | Ampr, lacZ | Promega |
| pCTNA2 | pCVD442 carrying a tnaA deletion, Ampr | This study |
| pGTNA | pGEM-T carrying a 3.7 kb fragment containing tnaA gene | This study |
| pStyABB | containing the gene of monooxygenase for indole assay, Ampr | [12] |
| pWKS30 | Ampr, lacZ | [6] |
| pWKS1030 | pWKS30 carrying an 18 Kb ibeR locus | [6] |
Ampr, ampicillin resistant; lacZ, a partial gene coding for the N-terminal fragment of β-galactosidase; Kanr, kanamycin resistant; Rifr, rifampin resistant.
2.2. Extraction and Manipulation of Plasmids and Subcloning
All genetic manipulations were done by using standard methods, as described elsewhere [24]. Plasmid DNA was extracted by using a plasmid mini kit (Qiagen, Calif, USA). DNA fragments were purified and were extracted from agarose gel slices, using QIAquick Gel Extraction Kit (Qiagen). Competent E. coli cells were made in 10% glycerol and were transformed by electroporation as described previously [6, 7].
2.3. Construction of Isogenic in-Frame Deletion Mutants of ibeR and tnaA
To determine the role of the ibeR gene in the growth-phase-dependent E. coli K1 invasion of BMEC, an isogenic deletion mutant of ibeR was generated as follows. Two PCR DNA fragments, B (1.2 kb) and R (1.0 kb), flanking a 1.8-kb region to be deleted were produced from two pairs of primers (IbeR-S1/IbeR-B1 for B and IbeR-B2/IbeR-X2 for R, see Table 2). The two fragments were ligated to make a 2.2 kb fragment (BR) that carries an ibeR internal deletion. The BR fragment was subcloned between SalI and XbaI sites on pCVD442 [10], and the resulting recombinant plasmid was named pCBR2. The mutants named BR2 were obtained by mating E44 with SM10 λpir that carries pCBR2 as described previously [6]. We used PCR and DNA sequencing to confirm the internal deletion in the ibeR deletion mutant BR2 and the desired chromosomal gene ibeR of the mutant with primers IbeR-S1 and IbeR-X2 (Table 2). Amplification was done by using the following cycle profile: 35 cycles at 94°C for 1 minute, 58°C for 1 minute, and 70°C for 1.5 minutes.
Table 2.
Oligonucleotides used for cloning, sequencing, and making the deletion mutants of ibeR and tnaA genes.
| Primers | Sequences of primers | Retained amino acids |
|---|---|---|
| primers for ibeR deletion | Total 56 residues | |
| IbeR-S1 (S = SalI) | 5′-GATGTCGACGGGCTTTTCGGCGTCA-3′ | 52 N-terminal residues |
| IbeR-B1 (B = BamHI) | 5′–CGGGATCCAGTGGCGAGGGTCACA-3′ | (MDIIIMNKES...) |
| IbeR-B2 (B = BamHI) | 5′-CAGGATCCAAATGTTGAGCATGCAG-3′ | 4 C-terminal residues |
| IbeR-X2 (X = XbaI) | 5′-CGTCTAGATAAGGGCTAAACATATCG-3′ | (...GSKC) |
| primers for tnaA deletion | Total 56 residues | |
| TN-S1 (S = SalI) | 5′-GGGTCGACCAGAGATCTGGCCGGAAT T-3′ | 21 N-terminal residues |
| TN-B1 (B = BamHI) | 5′-ACGGATCCAATAACACGAATGCGGAACGGTTC-3′ | (MKDYVMENFK...) |
| TN-B2 (B = BamHI) | 5′-TTAGATCTTTTAAACATGTGAAAGAGAACGCG-3′ | 35 C-terminal residues |
| TN-X2 (X = XbaI) | 5′-CCTCTAGATTAGCCAAATTTAGGTAACAC G-3′ | (...RHFTAKLKEV) |
For the tnaA in-frame deletion mutant, the same method was used as ibeR deletion. Briefly, 2 PCR DNA fragments, FTN5 (1.0-kb) and FTN3 (1.4-kb), were made to flank a 1.3 kb region containing tnaA to be deleted, by using 2 primer pairs (TN-S1/TN-B1 for FTN5 and TN-B2/TN-X2 for FTN3, see Table 2). Then the two fragments were ligated to make a 2.4 kb fragment (FTN53) that carried a tnaA internal deletion. The FTN53 fragment was subcloned into pCVD442 between SalI and XbaI sites to get the suicide plasmid pCTNA2. To get the tnaA in-frame deletion mutant, TNA44, conjugation and screening were carried out as described above. The tnaA gene deletion in the mutant TNA44 was confirmed by PCR using the primers TN-S1 and TN-X2 (Table 2).
2.4. Invasion Assay
Human BMECs were routinely cultured in RPMI 1640 medium (Mediatech, Herndon, Va, USA) containing 10% heat inactivated fetal bovine serum, 10% Nu-serum, 2 mM glutamine, 1 mM sodium pyruvate, essential amino acids, vitamins, penicillin G (50 μg/mL), and streptomycin (100 μg/mL) at 37°C in 5% CO2 [7]. Invasion assays were performed as previously described [6, 7, 18]. The number of intracellular bacteria was determined on blood agar plates after the extracellular bacteria were killed by incubation of the monolayers with experimental medium containing gentamicin (100 μg/mL) for 1 hour. Results were expressed as percent invasion (100 × (number of intracellular bacteria recovered)/(number of bacteria inoculated)).
2.5. Protein Extraction and 2DE
Protein extraction and 2DE were carried out as described [25] with minor modifications. Briefly, E. coli strain E44 and the ibeR deletion mutant BR2 were cultured in BHI medium overnight without agitation. The bacterial cells were harvested at the stationary-phase (OD = 2.5−3.0) by centrifugation at 6000 × g for 10 minutes. Denaturing protein extraction (phenol extraction procedure) was performed according to Saravanan and Rose [26]. The lyophilized pellets were dissolved in rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 0.2% pH 3–10 Biolytes, 1% DTT; 1 mg dry pellets for 200 μL buffer) and shaken on vortex for 1 hour at room temperature. The first and second dimensions of the PAGE were performed at least in triplicate (to reduce the likelihood of differences based solely upon gel-to-gel variability) according to the standard protocols developed and defined by Bio-Rad. Solubilized total E. coli protein samples (200 μL each) were loaded on 11 cm immobilized pH gradient (IPG) strips (pH 4–7). Rehydration/loading was done passively (no voltage) for 1 hour, followed immediately by 14 hours of active rehydration (50 V) at 20°C. Isoelectric focusing of the IPG strips was performed at 20°C using a 50 μA current limit per strip to prevent damage to the strip and the instrument. Electrophoresis was carried out as follows: step 1, 250 V for 20 minutes; step 2, a rapid ramp to 8000 V; step 3, focusing at 8000 V for 55 000 Vhr; step 4, hold at 400 V. Due to latent ionic components in the sample the actual running voltage was only approximately 6500 V. After the first dimensional run was completed, the IPG strips were equilibrated in buffer I (6 M urea, 0.375 M Tris-HCl pH 8.8, 2% SDS, 20% glycerol, and 2%DTT) for 15 minutes and then in buffer II (6 M urea, 0.375 M Tris-HCl pH 8.8, 2% SDS, 20% glycerol, and 2%DTT) for additional 15 minutes. The second dimension SDS-PAGE was performed with 15% resolving gels and 5% stacking gels (160 × 180 × 0.5 mm). The gels were stained with 0.1% coomassie brilliant blue (CBB) R-250. The 2DE gels were scanned at a 200 bpi resolution with Typhoon scanner (Amersham Biosciences, NJ, USA), and analyzed with ImageMaster 2D Platinum version 6.0 (GE Healthcare BIO-Science, NJ, USA). Only those protein spots having differences in density of 1.5-fold or greater between the groups were chosen. Moreover, all protein spots selected for analysis were shown to have significant difference in protein density (mean ± SEM, P < .01) by software of SPSS 10.0.
2.6. In-Gel Protein Digestion
Protein bands were excised from preparative coomassie blue-stained gels and washed several times with destaining solutions (25 mM NH4HCO3 for 15 minutes and then with 50% acetonitrile containing 25 mM NH4HCO3 for 15 minutes). Gel pieces were then dehydrated with 100% acetonitrile, dried, and then incubated with a reducing solution (25 mM NH4HCO3 containing 10 mM dithiothreitol) for 1 hour at 56°C and subsequently with an alkylating solution (25 mM NH4HCO3 containing 55 mM iodoacetamide) for 45 minutes at room temperature. After reduction and alkylation, gels were washed several times with the destaining solutions and finally with pure water for 15 minutes before being treated again with 100% acetonitrile. Depending on the protein content, 2-3 μL of 0.1 μg/μL modified trypsin (Promega, Wiss, USA, sequencing grade) in 25 mM NH4HCO3 was added over the gel spots and incubated for 30 minutes. About 7–10 μL of 25 mM NH4HCO3 was then added to cover the gel spots and incubated at 37°C overnight. The in-gel digestion products were extracted with formic acid/acetonitrile solutions followed by evaporation. Samples were desalted using mZipTip C18 pipette tips (Millipore, Mass, USA) before MS/MS analysis.
2.7. Protein Identification by LC-MS/MS
The sample was resuspended in 10 μL of 0.1% formic acid, injected via an autosampler (Surveyor, ThermoFinnigan, Calif, USA) and subjected to reverse phase liquid chromatography using ThermoFinnigan Surveyor MS-Pump in conjunction with a BioBasic 18 100 × 0.18 mm reverse-phase capillary column (ThermoFinnigan, Calif, USA). Mass analysis was done using a ThermoFinnigan LCQ Deca XP Plus ion trap mass spectrometer equipped with a nanospray ion source (ThermoFinnigan, Calif, USA) employing a 4 cm metal emitter (Proxeon, Odense, Denmark). Spray voltage of the mass spectrometer was set to 2.9 kV and capillary temperature was set at 190°C. The column equilibrated for 5 minutes at 1.5 μL/min with 95% Solution A and 5% Solution B (A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile) followed by a linear gradient was initiated 5 minutes after sample injection ramping to 65% Solution A over 45 minutes. Solution A was increased to 80% over the subsequent 5 minutes and held at 80% for 5 minutes, after which the column was reequilibrated back to 5% Solution A (aqueous). Mass spectra were acquired in the m/z 400–1800 range. A data-dependent acquisition mode was used where each of the top five ions for a given scan was subjected to MS/MS analysis. The protein identification was conducted with the MS/MS search software Mascot 1.9 with confirmatory or complementary analyses using TurboSequest as implemented in the Bioworks 3.2. E. coli genome sequences at the National Center for Biotechnology Information (NCBI) were used as the primary search databases and searches were complemented with the NCBI nonredundant protein database.
2.8. Indole Assay
For determination of indole production, we followed the method of indole conversion into indigo as described previously [12, 27], with minor modifications. All strains E44, BR2, TNA44, MG1655, and RH90 were transformed with the pStyABB plasmid, which constitutively expresses the StyAB protein converting indole to indigo. The bacteria were incubated in M9 medium containing 0.4% glucose and 100 μg/mL ampicillin overnight with shaking. Then the bacteria were collected by centrifugation and resuspended to OD600 = 0.2 in BHI medium supplied with 100 μg/mL ampicillin, and incubated at 37°C without shaking. To determine the indigo formation at different time points, batch-grown cells were harvested every 2 hours by centrifugation. Then the bacteria were lysed in DMSO for 30 minutes. The samples were read at 600 nm to determine the indigo concentration by comparison to a standard curve.
2.9. Determination of Resistance to Environmental Stress
Bacteria were grown in BHI broth at 37°C overnight without shaking, and collected by centrifugation. The number of cells was measured on the basis of their OD at 600 nm. Bacteria were suspended and diluted to 107 cells/mL in PBS for the following assays. For heat shock, 100 μL of bacteria was heated at 54°C for 3 minutes. For alkali endurance, the bacterial suspension was mixed with equal volume of Tris buffer (1 M, pH = 10.0) and 8 volumes of water (final concentration, 100 mM, pH 10.0) and incubated at 37°C for 30 minutes. For acid endurance, 1/10 volume of the bacterial suspension was mixed with LB containing acetic acid (final concentration, 90 mM, pH 2.8) and incubated at 37°C for 20 minutes. For high osmolarity challenge, bacteria were mixed with an equal volume of 4.8 M NaCl (final concentration, 2.4 M) and incubated at 37°C for 1 hour. For oxidative stress, bacteria were harvested and resuspended in an equal volume of PBS containing 10 μM H2O2 incubated at 37°C for 30 minutes. After exposure to these stresses, bacteria were diluted in 0.9% saline and plated in duplicate on LB agar plates. The surviving rate with stress was calculated from the ratio of the bacterial number under stress condition to the bacteria number under nonstress condition. The surviving rate without stress was calculated from the bacteria number grown on plates.
3. Results and Discussions
3.1. The ibeR Regulatory Gene is Required for Invasion of Human BMEC by Meningitic E. coli K1 Strain E44
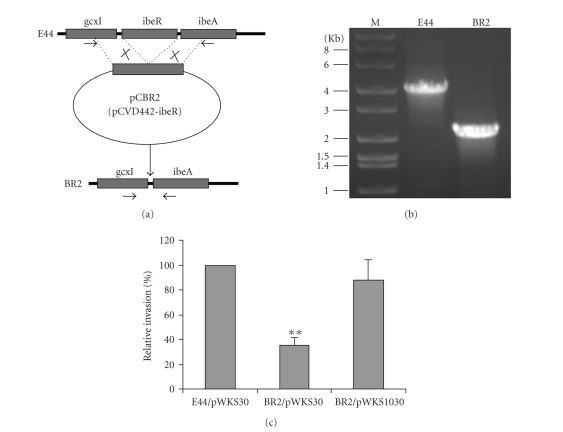
The gene ibeR in E. coli K1 E44 is predicted as the only regulatory protein present in the ibeRAT operon in GimA by bioinformatics approaches [16]. To determine the role of ibeR gene in the growth-phase-dependent invasion of BMEC by meningitic E. coli K1, an isogenic in-frame deletion mutant of ibeR was made by chromosomal gene replacement with the recombinant suicide plasmid pCBR2 carrying a 2.2 kb DNA fragment with ibeR internal deletion (Figure 1(a)). The 2.2 kb DNA fragment was generated by ligation of two PCR amplicons (1.2 and 1.0 kbs) flanking the 1.8 kb ibeR coding region. The ibeR deletion mutant was obtained by mating E44 with SM10 λpir carrying pCBR2. The mutant colony morphology on LB agar plates and growth rate in LB broth were the same as the parent strain E44. The deletion of ibeR was confirmed by colony PCR and DNA sequencing (Figure 1(b)). In order to examine the virulence phenotype of the ibeR deletion mutant, a comparative study of the invasiveness of E44 (parent strain), BR2, and the complemented BR2 was carried out. As shown in Figure 1(c), the relative invasion rate of BR2 was significantly reduced as compared to that of E44 and the plasmid pWKS1030 carrying the ibeR gene was able to complement the noninvasive phenotype of BR2, suggesting that the ibeR gene contributes to the E. coli E44 invasion process.
Figure 1.
Generation and characterization of ibeR deletion mutant from the meningitic strain E44. (a) Generation of the ibeR deletion mutant (BR2). Two DNA fragments flanking ibeR were amplified and ligated into the suicide vector pCVD442 to construct pCBR2. The ibeR deletion mutants were generated through gene allele exchanges and suicide vector loss. The arrowheads indicate primer locations. (b) Verification of the ibeR deletion by PCR. The parent strain E44 and the ibeR deletion mutant BR2 were verified though colony PCR with primers listed in Table 2. (c) Invasion of HBMEC with the E. coli parent E44 carrying pWKS30, the ibeR mutant BR2 with pWKS30, and the complemented BR2 carrying the ibeR locus in pWKS30. E44/pWKS, BR2/Pwks, and BR2/pWKS1030 were incubated with HBMEC monolayers and the standard invasion assay was carried out as described in Section 2. The results are expressed as relative invasion%. Columns marked with ∗∗ are significantly different (P < .01).
3.2. 2D Proteomic Analyses of IbeR-Regulated SP Protein Expression in Meningitic Strain E44
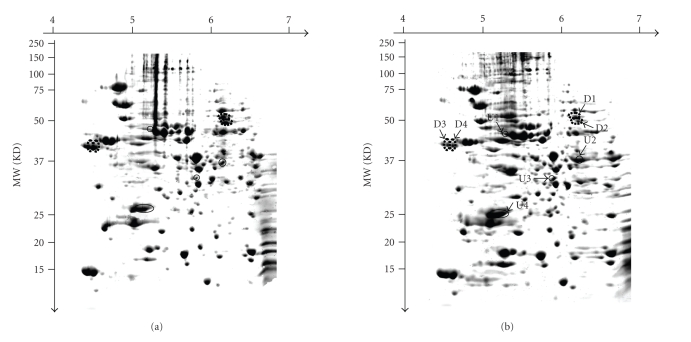
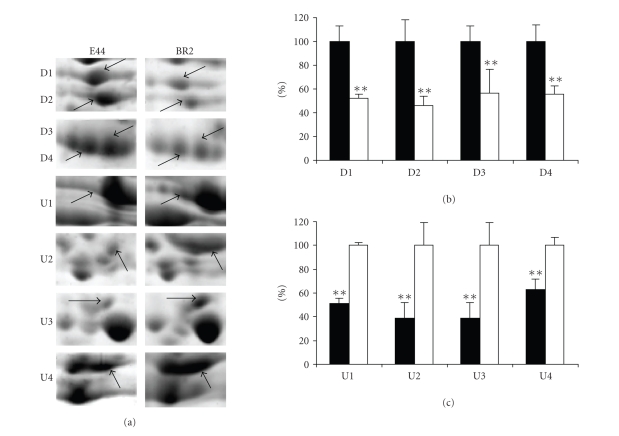
To determine the role of ibeR in regulating SP gene expressionof meningitic E. coli K1, the wild type E. coli E44 and its ibeR deletion mutant BR2 were cultured in BHI broth overnight. The total proteins of each strain were extracted from the cells as described in Section 2. The whole cell extracts were analyzed on the 2D protein gels. Approximately 800 spots were detected on a gel image. The experiment was performed three times with two sets of independently grown cultures. Only spots showing the same pattern in three independent runs were retained and quantified using the software ImageMaster 2D Platinum version 6.0. Figures 2(a) and 2(b) showed the protein patterns of E44 and BR2, respectively. All the upregulated spots and downregulated spots satisfying the criteria as mentioned above were marked on both the 2D maps. They were excised and identified by LC-MS/MS (Table 3). Eight protein spots were found to be differentially expressed in BR2 as compared to its parent strain E44. Among them, 4 protein spots were significantly upregulated in BR2 including elongation factor EF-Tu (TufB, spot U1), glyceraldehyde-3-phosphate dehydrogenase A (GapA, spot U2), outer membrane protein 3a (OmpA, spot U3), and alkyl hydroperoxide reductase (AhpC, spot U4), while 4 protein spots were of decreased abundance in BR2, including dihydrolipoamide dehydrogenase (LpdA, spot D1), tryptophanase (TnaA, spot D2), and two isoforms of outer membrane protein C (OmpC, spot D3, and D4). Figure 3(a) showed the enlargements of each changed protein marked with black arrows and spot numbers. The relative ratios of each downregulated protein and upregulated protein were shown in Figures 3(b) and 3(c).
Figure 2.
2DE maps of E. coli strains with stationary-phase cultures in BHI medium. (a) E44 (wild type strain). (b) BR2 (the ibeR deletion mutant). The upregulated proteins were circled with a solid line and marked with U1–U4; the downregulated proteins were circled with a broken line and marked D1–D4.
Table 3.
Identification of differentially displayed proteins in 2D maps.
| Spot number | Protein ID | Access number | Mass(KD)/PI (theriol) | Mass(KD)/PI (analytic) | Peptides matched | Sequence coverage | Function category |
|---|---|---|---|---|---|---|---|
| downregulated | |||||||
| D1 | dihydrolipoamide dehydrogenase (LpdA) | gi | 15799800 | 50.9/5.79 | 51.0/6.05 | 249 | 47% | Central metabolism (CM): E3 component of pyruvate dehydrogenase complex |
| D2 | tryptophanase (TnaA) | gi | 15804305 | 53.8/5.88 | 49.0/6.10 | 92 | 34% | Response to environmental modifications (REMs), and CM: initiation of indole signaling |
| D3 | outer membrane protein C (OmpC) | gi | 15802768 | 40.5/4.55 | 40.5/4.55 | 53 | 50% | REM: osmotically regulated porin |
| D4 | outer membrane protein C (OmpC) | gi | 15802768 | 40.5/4.55 | 40.5/4.60 | 7 | 20% | REM: osmotically regulated porin |
| upregulated | |||||||
| U1 | elongation factor EF-Tu (TufB) | gi | 15803852 | 43.4/5.3 | 43.5/5.30 | 55 | 45% | REM: binding and transport of aminoacyl-tRNA |
| U2 | glyceraldehyde-3-phosphate (G-3P) dehydrogenase A(GapA) | gi | 15802193 | 35.7/6.61 | 36.0/6.20 | 94 | 32% | CM: oxidation and phosphorylation of G-3P to 1,3-bisphosphoglycerate |
| U3 | outer membrane protein 3a (OmpA) | gi | 15800816 | 37.3/5.99 | 30.0/5.80 | 35 | 48% | REM: maintaining cell envelope integrity |
| U4 | alkyl hydroperoxide reductase(AhpC) | gi | 15800320 | 20.9/5.03 | 22.0/5.15 | 57 | 49% | REM: a primary scavenger of endogenous H2O2 at a low (10−5 M) concentration |
Figure 3.
Comparative analysis of the protein spots showing significant changes. (a) Enlargement of differentially expressed protein spots (indicated with arrows) from Figure 2. (b) The relative level of down-regulated proteins. (c) The relative level of up-regulated proteins. The protein spot intensities of E44 were showed as black columns, and the protein spot intensities of BR2 were showed as white columns. Columns marked with ∗∗ are significantly different (P < 0.01).
We classified the proteins into two main categories on the basis of their roles in the SP growth of E. coli cells: (a) response to environmental modifications (including TnaA, TufB, OmpC, OmpA, and AhpC) and (b) central metabolism (including LpdA and GapA). Since ibeR was hypothesized as an SP-regulator contributing to the growth regulation and virulence of E44, its role in the invasion process and resistance to stress conditions should be further characterized. TnaA, a tryptophanase, degrades tryptophan, resulting in the formation of indole, which has been proposed to act as an extracellular signal in stationary phase cells of E. coli [28, 29]. Production of indole, via the enzymatic activity of TnaA, is also induced during biofilm formation [30]. TnaA, which was controlled by RpoS in other E. coli strains [27], is one of the most important transcriptional regulators for the gene expressions in SP cells [31]. TufB (EF-Tu) is responsible for binding and transporting the appropriate codon-specified aminoacyl-tRNA to the aminoacyl (A) site of the ribosome [32, 33]. In addition to its function in translation elongation, elongation factor Tu is implicated in protein folding and protection from stress like a chaperone molecule [34].
OmpC, as well as OmpF, is a porin protein present on the outer membrane of E. coli, responding to the osmotic challenge. OmpR, as a regulator, activates transcription of ompF and ompC [35], and changes the ratio of these two, so that the total level of porin proteins remains approximately constant [36, 37]. OmpF, which produces slightly wider pores (1.2 nm) than does OmpC, predominates at low osmotic strength, whereas OmpC (1.1 nm) predominates at high osmotic strength [38]. In E. coli, the expression of OmpC is repressed at low osmolarity and induced at high osmolarity. It has been proposed that the smaller pores formed by OmpC could reduce the diffusion of larger hydrophobic and negatively charged molecules when bacteria encounter high osmolarity conditions as in the host compartments. Presumably, this protein is very important for the stress resistance of E. coli in the stationary phase. In this study, the downregulation of OmpC resulting from the ibeR deletion might decrease resistance to high osmolarity. In addition, it had been reported that OmpC is involved in invasion of epithelial cells by Crohn's disease-associated E. coli strain LF82 and Shigella flexneri [39, 40], suggesting that OmpC might be involved in E44 invasion of the host tissue barriers. OmpA is a major protein in the outer membrane of both pathogenic and nonpathogenic E. coli [41]. As shown in our previous study, the ompA-deletion mutant of E44 was significantly more sensitive than that of its parent strain to SDS, cholate, acidic environment, and high osmolarity [41]. OmpA is downregulated upon entry into SP by sigmaE, which plays a central role in maintaining cell envelope integrity both under stress conditions and during normal growth [42, 43]. We demonstrated here that OmpA was upregulated in the ibeR mutant BR2 (Figure 3), suggesting that OmpA expression is suppressed upon entry into SP by IbeR in a manner similar to sigmaE.
Alkyl hydroperoxidase (AhpC) functions as a primary scavenger of endogenous H2O2 at a low (10−5 M) concentration [44]. All of ahpC, katG, and katE genes are known to participate in the antioxidant defense mechanism against H2O2-induced stress in E. coli. It has been reported that SP-inducible RpoS regulates katE gene expression and OxyR regulates ahpC and katG genes [45, 46]. Our previous study has demonstrated that E. coli K1 RS218 had a nonsense mutation in its rpoS gene, resulting in a negligible katE activity, but no obvious difference in katG [5]. In this study, the increase in ahpC expression indicated that the ibeR deletion led to an increased oxidative stress in SP compared with the wild type strain, suggesting that ibeR is involved in the resistance to oxidative stress upon entry into SP.
Lipoamide dehydrogenase (LpdA), which is the same as dihydrolipoamide dehydrogenase (DLDH), makes up the E3 component of pyruvate dehydrogenase complex, 2-oxo glutarate dehydrogenase, and branched-chain 2-oxo acid dehydrogenase complexes. DLDH has been identified as virulence factors contributing to the pathogenesis of bacterial infections caused by Mycobacterium tuberculosis and Streptococcus pneumoniae because it enhances their survival within the host cells [47, 48]. As shown in Figure 3, LpdA was downregulated in the ibeR deletion mutant BR2, suggesting that this enzyme might be involved in the virulence of meningitic E. coli K1 via enhancing the pathogen survival within the host. Recently, GapA in E. coli has been identified as one of a few proteins, which harbors functionally important thiol groups against oxidative stress [49]. As GapA is upregulated in BR2 (Figure 3), IbeR may be involved in the negative control of gapA in SP.
In summary, all these proteins contribute to growth-related carbon source metabolism or stress resistance. They are associated with the SP regulation. Among these proteins, TnaA is the most important one as it produces the signal molecule indole and is regulated by RpoS [27]. Since E44 carries a loss-of-function mutation in its rpoS gene [5], there should be alternative signaling pathway(s) to complement the functional deficiency of RpoS in this pathogenic E. coli strain. Our proteomic analyses showed that the TnaA expression was significantly affected by IbeR, which might be functionally equivalent to RpoS. Therefore, our focus for further studies was placed on how TnaA is regulated by IbeR.
3.3. Indole Production is Controlled by IbeR via Regulation of TnaA
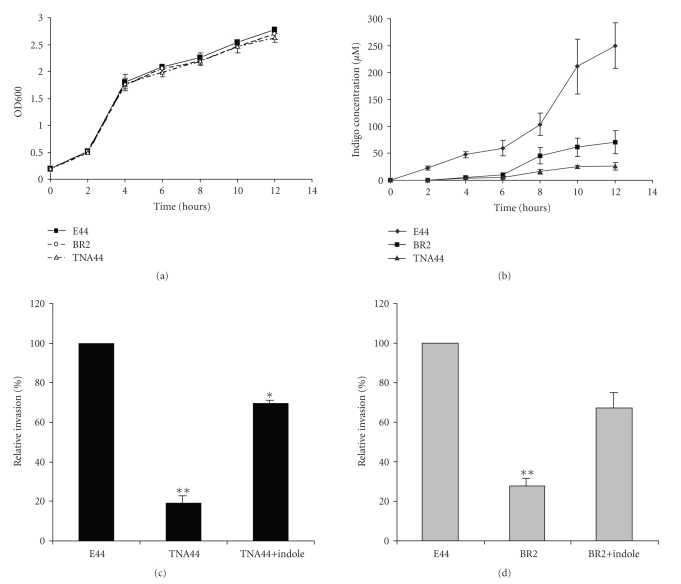
To test our hypothesis that IbeR is an RpoS-like regulator, the tnaA in-frame deletion mutant TNA44 was generated with the same gene replacement approach that was used for the ibeR deletion mutant. TNA44 was obtained by mating E44 with SM10 λpir carrying the recombinant suicide plasmid pCTNA2 which contains the truncated tnaA gene. The virulence phenotype of TNA44 was examined with invasion assays. Although overall growth rates did not differ between the mutants (BR2 and TNA44) and their parent strain E44 (Figure 4(a)), the invasive capability of TNA44 (19%) and BR2 (35%) was significantly reduced as compared to that of E44 (100%) (Figures 4(c)-4(d)). These data suggest that TnaA is an important downstream regulator that is required forIbeR-modulated E. coli K1 invasion.
Figure 4.
Role of TnaA in indole production and bacterial invasion. (a) Growth curves of E44, TNA44, and BR2, (b) indole production (IP) of E coli strains E44, BR2, and TNA44, (c) the invasion phenotypes of E44, TNA44, and indole(100 μM)-complemented mutant TNA44, and (d) the invasion phenotypes of E44, BR2, and indole(100 μM)-complemented mutant BR2. Columns marked with * are significantly different (P < .05), ∗∗ are significantly different (P < .01).
As our proteomics analysis had shown that tnaA expression was induced by IbeR in SP and the tnaA and ibeR deletion resulted in a deficiency in indole production in SP-cultures (Figure 4(b)), we further tested whether the TnaA product indole, as an SP extracellular signal molecule, played a role in the process of invasion. Indole is converted to indigo (which is not further degraded in E. coli) by several monooxygenases, thus providing an easy method for its determination [12]. A plasmid pStyABB, carrying the gene for styrene monooxygenase, was used to monitor the indole production through its conversion to indigo. The plasmid was transformed into E. coli strains E44, BR2, and TNA44, and the indole production was measured at different time points for these stains in BHI media (Figure 4(b)). The production of indole in E44 was revealed by indigo accumulation. By contrast, the indole production was almost abolished in the tnaA deletion mutant and severely reduced in the ibeR deletion mutant (Figure 4(b)). These results demonstrated that the indole production was controlled by ibeR through tnaA. To examine whether indole could compliment the noninvasive phenotype of tnaA and ibeR deletion mutants, indole was supplied in the BHI medium at 100 μM to the TNA44 and BR2 SP cultures. The result showed that indole was able to significantly enhance the relative invasion rate of TNA44 (19% to 69%) and BR2 (35% to 65%) as compared to that of E44, suggesting that indole could partly compliment the noninvasive phenotype of TNA44 (Figure 4(c)) and BR2 (Figure 4(d)). Lacour and Landini have shown that the rpoS gene in E. coli K12 strain MG1655 controls the production of indole, which acts as a signal molecule in SP cells, via regulation of TnaA, the indole-producing enzyme [27]. As TnaA is regulated by IbeR in E44, it is most likely that IbeR is a novel regulator to complement the functional deficiency of RpoS in E44.
3.4. The Role of ibeR in Stress Conditions
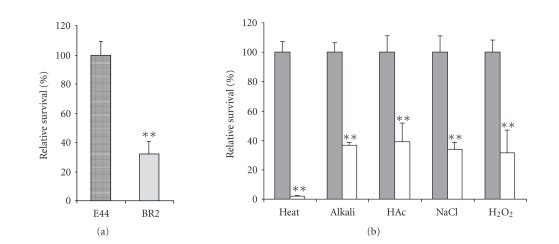
It has been reported that RpoS is able to positively and negatively control expression of a large set of genes when bacteria enter into the SP [46, 50, 51]. During such transition, bacteria undergo physiological changes that allow their SP organisms to survive better in such insults as heat, high-osmotic environment, starvation, UV radiation, H2O2, and acid than their exponential counterpart [50, 52]. The loss of RpoS resulted in the decrease of stress resistance and cell survival in the SP [5, 53]. Although our study showed that the loss of ibeR did not affect the growth rate in BHI medium, the survival rates of the ibeR deletion mutant BR2 in the SP significantly decreased as compared to that of the wild type strain E44 even without any stress treatment (Figure 5(a)). Our proteomics analysis also revealed that the most significant proteomic changes in the ibeR deletion mutant were related to bacterial response to environmental modifications. For example, AhpC, as a primary scavenger of endogenous H2O2, was upregulated in the ibeR deletion mutant, implying that the loss of ibeR resulted in the decreased survival rates of bacterial cells under an oxidative stress in the SP. OmpC, as a porin protein, was downregulated in BR2, perhaps resulting in the decreased resistance to osmolarity stress. These results suggested that ibeR plays a regulatory role in response to stress conditions in E44 that carries a nonsense mutation in rpoS. To examine the function of ibeR in response to stress environments, we performed several survival assays under different stress conditions, including heat shock (54°C for 3 minutes), alkali endurance (Tris, pH = 10 for 30 minutes), acid endurance (acetic acid, pH = 2.8 for 20 minutes), high osmolarity challenge (2.5 M NaCl for 1 hour), and oxidative stress (10 μM H2O2 for 30 minutes). In all the survival experiments, the wild type strain E44 showed higher survival rates than the ibeR deletion mutant, indicating that the ibeR gene is required for all these stress resistances (Figure 5(b)). Especially in the heat shock assay, the loss of ibeR resulted in over 95% cells death, indicating that ibeR played a vital role in temperature sensitivity in this strain. In the other stress treatments, the ibeR deletion also significantly reduced the survival rates (more than 60% cell death) of BR2 as compared to that of the wild type strain E44, suggesting that IbeR had a global regulatory role in the resistance to acid, alkali, high osmolarity and oxidative stress. In the survival assays for the E. coli control strain MG1655, the rpoS deletion mutant RH90 also decreased the survival levels in these five stress conditions, showing the similar patterns like ibeR in response to stress environments (data not shown). Combining the proteomics analysis and the stress survival studies, we conclude that IbeR is an RpoS-like regulator to control gene expression of proteins that are critical for stress-resistance and cell survival in the SP in E44, which carries a loss-of-function mutation in the rpoS gene.
Figure 5.
The relative survival rate of the BR2 mutant comparing with E44. (a) Both strains in stationary phase without any stress treatment, (b) both strains were tested with different environmental stress including heat shock (54°C for 3 minutes), alkali (Tris pH = 10.0 for 30 minutes), acid endurance (acetic acid, pH = 2.8 for 20 minutes), high osmolarity challenge (2.4 M NaCl for 1 hour), and oxidative stress (10 μM H2O2 for 30 minutes). Gray columns: E44; white columns: BR2. Columns marked with ∗∗ are significantly different (P < .01).
4. Concluding Remarks
Currently, most E. coli meningitis studies are done with SP cultures in which the pathogen invasion of human BMEC is significantly greater than the log phase cultures. In most strains of E. coli, RpoS plays a central role in regulating the SP regulatory genes for protecting cells against starvation and stress damage. RpoS, the major SP regulator, has also been shown to regulate the expression of microbial virulence genes in various bacteria including E. coli K1 (O157 : H7), Salmonella typhimurium, Shigella flexneri, Yersinia enterocolitica, Vibrio cholera, and Borrelia burgdorfer, [5, 53, 54]. Surprisingly, however, RpoS was found to be inactive in meningitic strain E44 [5]. The current proteomic studies may provide an answer to the long-standing question regarding the SP gene regulation in E44. Combining the proteomics analysis, virulence determination, and the stress survival studies of the ibeR mutant BR2, we have demonstrated for the first time that IbeR serves as an RpoS-like regulator to control gene expression that is critical for stress-resistance and cell survival in the SP of E44.
IbeR is not a structural homologue of RpoS as IbeR and RpoS do not share any significant sequence homology. RpoS (also known as σ38, σs, or KatF) is a global regulator in E. coli, which is the second principal σ subunit after the major σ70 factor [5]. In E44, however, IbeR appears not to be a master regulator on the basis of its genomic prevalence and functional spectrum. The prevalence of the GimAlocus carrying ibeR is highly dependent on the origin of the strain and on the subgroup it belongs to (A, B1, B2, and D) (4). In all the studies, where the presence of this locushas been analyzed, GimA was found to be restricted to the B2 subgroup, a subgroup that includes strains with the highest virulence in mice and the highest level of virulence determinants (4). Our proteomic studies showed that a limited number of genes were regulated by IbeR, suggesting that IbeR is a regulator with a narrow functional spectrum. In other E. coli strains, either pathogenic or probiotic strains, functional heterogeneity of RpoS in stress tolerance was widely observed [5, 53, 55]. Those studies have shown that some E. coli strains can maintain their stress tolerance capability or significantly modulate their stress resistance phenotype independent of their rpoS genotypes. Such adaptation processes compromising the RpoS-dependent stress responses may have significant impact on bacterial survival in environments, as well as in the host's stomach and intestine [53, 55]. IbeR, a regulator in the GimA regulon, may contribute to bacterial virulence adaptation process in E44 to complement the functional deficiency of RpoS.
Another significant finding of our proteomic studies is that IbeR in meningitic strain E44 is able to upregulate TnaA, which is controlled by RpoS in other E. coli strains [56]. The virulence determination of the tnaA mutant showed that TnaA and its product indole were required for E44 invasion of human BMEC. The generation of indole, via the tryptophanase activity of TnaA, was also observed during the formation of biofilm in E. coli and other bacteria [28, 56]. In addition to the initiation of indole-mediated signaling, TnaA (tryptophanase) is able to catabolize tryptophan, cysteine, and serine to pyruvate [29, 56]. The Three proteins significantly upregulated by IbeR are TnaA, LpdA, and OmpC, all of which are directly or indirectly involved in pyruvate metabolism. LpdA (dihydrolipoamide dehydrogenase) is the E3 component of pyruvate dehydrogenase complex. OmpC, an osmotically regulated porin, may facilitate nutrient uptake [56]. On the other hand, the three operons (ptnIPKC, cglDTEC, and gcxKRCI) in the GimA regulon may also directly or indirectly contribute to pyruvate metabolism by converting dihydroxyacetone, glycerol, and glycerate to pyruvate [16]. It has been shown that the capability to catabolize carbon source is an important parameter in the ability to persist and compete in stationary phase [29]. This raises the possibility that signaling by indole, which is regulated by IbeR via TnaA, may play a critical role in the pathways that prepare the pathogens for a nutrient-poor environment (e.g., CNS) when the carbon source becomes limited for energy production.
Acknowledgments
The authors would like to thank Dr. K. S. Kim for providing brain endothelial cells. This project was financially supported by Public Health Service Grants R01-AI40635 (S.H.H.) and R01-NS047599 (A.J.).
References
- 1.Huang S-H, Jong AY. Cellular mechanisms of microbial proteins contributing to invasion of the blood-brain barrier. Cellular Microbiology. 2001;3(5):277–287. doi: 10.1046/j.1462-5822.2001.00116.x. [DOI] [PubMed] [Google Scholar]
- 2.Weber JR, Tuomanen EI. Cellular damage in bacterial meningitis: an interplay of bacterial and host driven toxicity. Journal of Neuroimmunology. 2007;184(1-2):45–52. doi: 10.1016/j.jneuroim.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Huang S-H, Stins MF, Kim KS. Bacterial penetration across the blood-brain barrier during the development of neonatal meningitis. Microbes and Infection. 2000;2(10):1237–1244. doi: 10.1016/s1286-4579(00)01277-6. [DOI] [PubMed] [Google Scholar]
- 4.Huang SH, Germon P, Jong A. Focus on Meningitis Research. New York, NY, USA: Nova Science; 2004. [Google Scholar]
- 5.Wang Y, Kim KS. Effect of rpoS mutations on stress-resistance and invasion of brain microvascular endothelial cells in Escherichia coli K1. FEMS Microbiology Letters. 2000;182(2):241–247. doi: 10.1111/j.1574-6968.2000.tb08902.x. [DOI] [PubMed] [Google Scholar]
- 6.Huang S-H, Wan Z-S, Chen Y-H, Jong AY, Kim KS. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. The Journal of Infectious Diseases. 2001;183(7):1071–1078. doi: 10.1086/319290. [DOI] [PubMed] [Google Scholar]
- 7.Huang S-H, Chen Y-H, Fu Q, et al. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infection and Immunity. 1999;67(5):2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasadarao NV, Wass CA, Weiser JN, Stins MF, Huang S-H, Kim KS. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infection and Immunity. 1996;64(1):146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Huang S-H, Wass CA, Stins MF, Kim KS. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infection and Immunity. 1999;67(9):4751–4756. doi: 10.1128/iai.67.9.4751-4756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg MS, Calderwood SB, Donohue-Rolfe A, Keusch GT, Kaper JB. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade HEp-2 cells. Infection and Immunity. 1990;58(6):1565–1571. doi: 10.1128/iai.58.6.1565-1571.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infection and Immunity. 1991;59(12):4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connor KE, Dobson AD, Hartmans S. Indigo formation by microorganisms expressing styrene monooxygenase activity. Applied and Environmental Microbiology. 1997;63(11):4287–4291. doi: 10.1128/aem.63.11.4287-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng C-H, Cai M, Shin S, et al. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infection and Immunity. 2005;73(5):2923–2931. doi: 10.1128/IAI.73.5.2923-2931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman JA, Badger JL, Zhang Y, Huang S-H, Kim KS. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infection and Immunity. 2000;68(9):5062–5067. doi: 10.1128/iai.68.9.5062-5067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badger JL, Wass CA, Weissman SJ, Kim KS. Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infection and Immunity. 2000;68(9):5056–5061. doi: 10.1128/iai.68.9.5056-5061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S-H, Chen Y-H, Kong G, et al. A novel genetic island of meningitic Escherichia coli K1 containing the ibeA invasion gene (GimA): functional annotation and carbon-source-regulated invasion of human brain microvascular endothelial cells. Functional & Integrative Genomics. 2001;1(5):312–322. doi: 10.1007/s101420100039. [DOI] [PubMed] [Google Scholar]
- 17.Zou Y, He L, Huang S-H. Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochemical and Biophysical Research Communications. 2006;351(3):625–630. doi: 10.1016/j.bbrc.2006.10.091. [DOI] [PubMed] [Google Scholar]
- 18.Huang S-H, Wass C, Fu Q, Prasadarao NV, Stins M, Kim KS. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infection and Immunity. 1995;63(11):4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou Y, He L, Chi F, Jong A, Huang S-H. Involvement of Escherichia coli K1 ibeT in bacterial adhesion that is associated with the entry into human brain microvascular endothelial cells. Medical Microbiology and Immunology. 2008;197(4):337–344. doi: 10.1007/s00430-007-0065-y. [DOI] [PubMed] [Google Scholar]
- 20.Bächler C, Schneider P, Bähler P, Lustig A, Erni B. Escherichia coli dihydroxyacetone kinase controls gene expression by binding to transcription factor DhaR. The EMBO Journal. 2005;24(2):283–293. doi: 10.1038/sj.emboj.7600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silver RP, Aaronson W, Sutton A, Schneerson R. Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infection and Immunity. 1980;29(1):200–206. doi: 10.1128/iai.29.1.200-206.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westenberg DJ, Gunsalus RP, Ackrell BA, Sices H, Cecchini G. Escherichia coli fumarate reductase frdC and frdD mutants. Identification of amino acid residues involved in catalytic activity with quinones. The Journal of Biological Chemistry. 1993;268(2):815–822. [PubMed] [Google Scholar]
- 23.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Molecular Microbiology. 1991;5(1):49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Huang SH, Jong A, Summersgill JT. Handbook of Proteomic Methods. Totwa, NJ, USA: Humana Press; 2003. [Google Scholar]
- 26.Saravanan RS, Rose JKC. A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics. 2004;4(9):2522–2532. doi: 10.1002/pmic.200300789. [DOI] [PubMed] [Google Scholar]
- 27.Lacour S, Landini P. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. Journal of Bacteriology. 2004;186(21):7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Canadian Journal of Microbiology. 2003;49(7):443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Ding X, Rather PN. Indole can act as an extracellular signal in Escherichia coli. Journal of Bacteriology. 2001;183(14):4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Ni L, Somerville RL. A stationary-phase protein of Escherichia coli that affects the mode of association between the trp repressor protein and operator-bearing DNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(12):5796–5800. doi: 10.1073/pnas.90.12.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hengge-Aronis R. Back to log phase: σS as a global regulator in the osmotic control of gene expression in Escherichia coli. Molecular Microbiology. 1996;21(5):887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 32.Miller DL, Weissbach H. Molecular Mechanisms of Protein Biosynthesis. New York, NY, USA: Academic Press; 1977. [Google Scholar]
- 33.Brot N. Molecular Mechanisms of Protein Biosynthesis. New York, NY, USA: Academic Press; 1977. [Google Scholar]
- 34.Caldas TD, El Yaagoubi A, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. The Journal of Biological Chemistry. 1998;273(19):11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- 35.Norioka S, Ramakrishnan G, Ikenaka K, Inouye M. Interaction of a transcriptional activator, OmpR, with reciprocally osmoregulated genes, ompF and ompC, of Escherichia coli. The Journal of Biological Chemistry. 1986;261(36):17113–17119. [PubMed] [Google Scholar]
- 36.Lugtenberg B, Peters R, Bernheimer H, Berendsen W. Influence of cultural conditions and mutations on the composition of the outer membrane proteins of Escherichia coli. Molecular and General Genetics. 1976;147(3):251–262. doi: 10.1007/BF00582876. [DOI] [PubMed] [Google Scholar]
- 37.Alphen WV, Lugtenberg B. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. Journal of Bacteriology. 1977;131(2):623–630. doi: 10.1128/jb.131.2.623-630.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiology and Molecular Biology Reviews. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rolhion N, Carvalho FA, Darfeuille-Michaud A. OmpC and the σE regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Molecular Microbiology. 2007;63(6):1684–1700. doi: 10.1111/j.1365-2958.2007.05638.x. [DOI] [PubMed] [Google Scholar]
- 40.Bernardini ML, Sanna MG, Fontaine A, Sansonetti PJ. OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infection and Immunity. 1993;61(9):3625–3635. doi: 10.1128/iai.61.9.3625-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y. The function of OmpA in Escherichia coli. Biochemical and Biophysical Research Communications. 2002;292(2):396–401. doi: 10.1006/bbrc.2002.6657. [DOI] [PubMed] [Google Scholar]
- 42.Udekwu KI, Wagner EGH. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Research. 2007;35(4):1279–1288. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. Journal of Molecular Biology. 2006;364(1):1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. Journal of Bacteriology. 2001;183(24):7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. Journal of Bacteriology. 2001;183(15):4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schellhorn HE, Audia JP, Wei LI, Chang L. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. Journal of Bacteriology. 1998;180(23):6283–6291. doi: 10.1128/jb.180.23.6283-6291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith AW, Roche H, Trombe M-C, Briles DE, Håkansson A. Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Molecular Microbiology. 2002;44(2):431–448. doi: 10.1046/j.1365-2958.2002.02883.x. [DOI] [PubMed] [Google Scholar]
- 48.Deghmane A-E, Soulhine H, Bach H, et al. Lipoamide dehydrogenase mediates retention of coronin-1 on BCG vacuoles, leading to arrest in phagosome maturation. Journal of Cell Science. 2007;120(16):2796–2806. doi: 10.1242/jcs.006221. [DOI] [PubMed] [Google Scholar]
- 49.Brandes N, Rinck A, Leichert LI, Jakob U. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Molecular Microbiology. 2007;66(4):901–914. doi: 10.1111/j.1365-2958.2007.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loewen PC, Hengge-Aronis R. The role of the sigma factor σS (KatF) in bacterial global regulation. Annual Review of Microbiology. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 51.Farewell A, Kvint K, Nyström T. Negative regulation by RpoS: a case of sigma factor competition. Molecular Microbiology. 1998;29(4):1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 52.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72(2):165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 53.Bhagwat AA, Tan J, Sharma M, et al. Functional heterogeneity of RpoS in stress tolerance of enterohemorrhagic Escherichia coli strains. Applied and Environmental Microbiology. 2006;72(7):4978–4986. doi: 10.1128/AEM.02842-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. Evidence that RpoS (σS) in Borrelia burgdorferiv is controlled directly by RpoN (σ54/σN)∇. Journal of Bacteriology. 2007;189(5):2139–2144. doi: 10.1128/JB.01653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coldewey SM, Hartmann M, Schmidt DS, Engelking U, Ukena SN, Gunzer F. Impact of the rpoS genotype for acid resistance patterns of pathogenic and probiotic Escherichia coli. BMC Microbiology. 2007;7, article 21:1–13. doi: 10.1186/1471-2180-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lelong C, Aguiluz K, Luche S, et al. The Crl-RpoS regulon of Escherichia coli. Molecular & Cellular Proteomics. 2007;6(4):648–659. doi: 10.1074/mcp.M600191-MCP200. [DOI] [PubMed] [Google Scholar]