Abstract
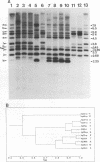
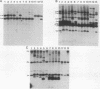
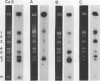
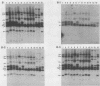
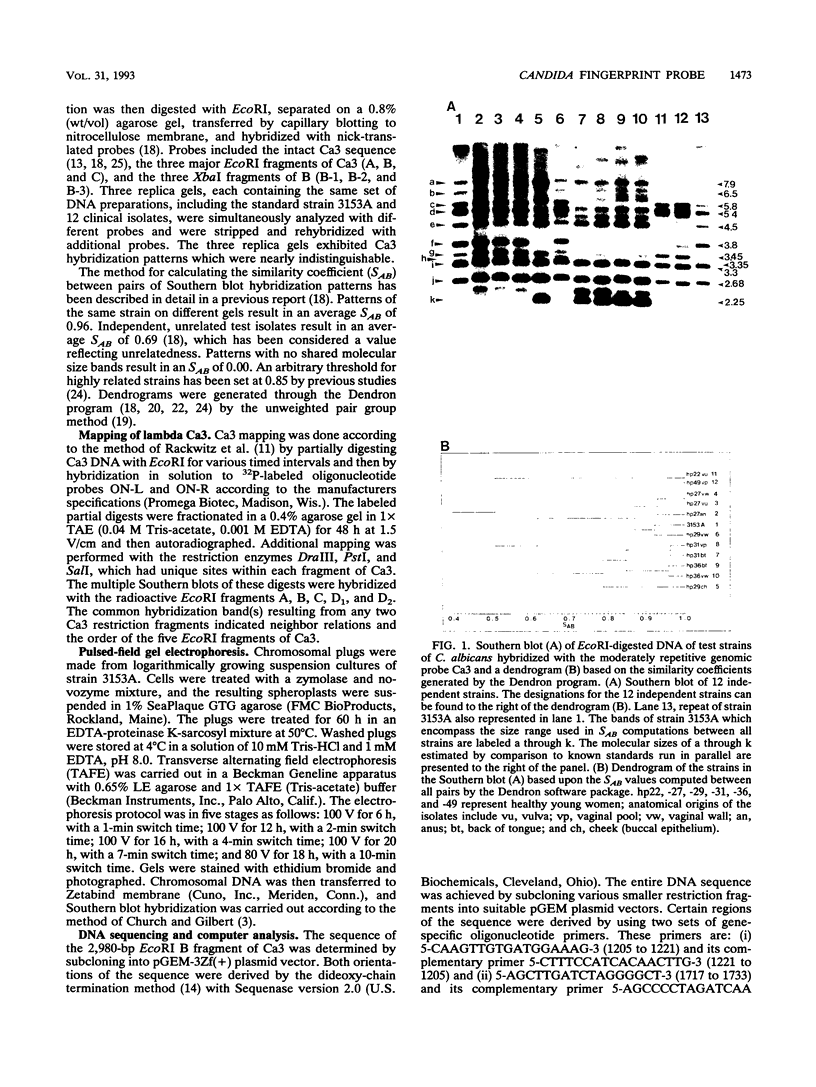
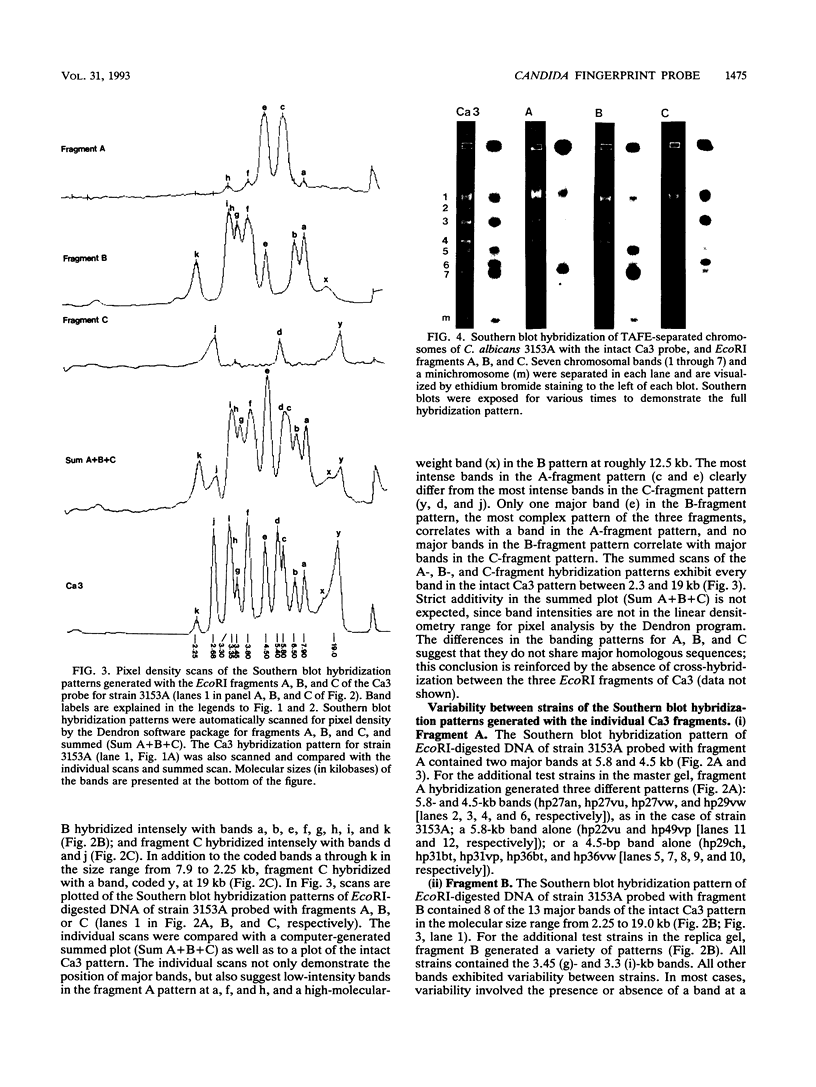
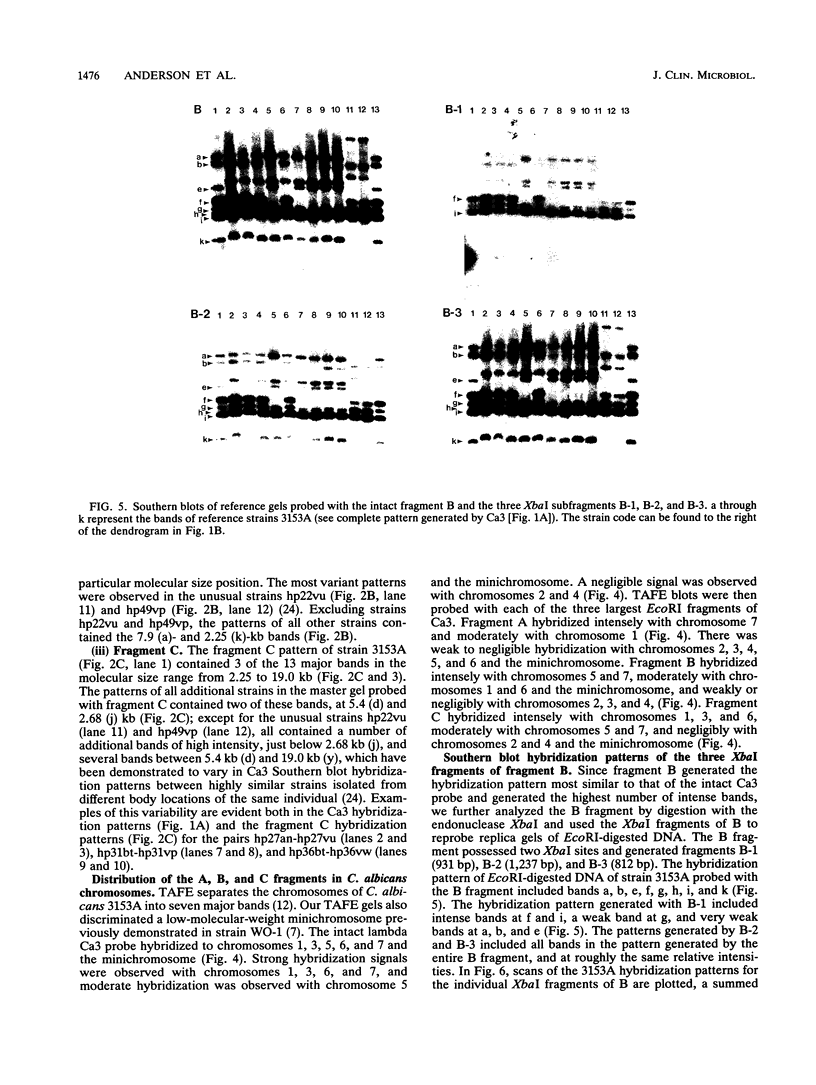
The moderately repetitive Ca3 fragment of Candida albicans has been used as an effective DNA fingerprinting probe in epidemiological studies. EcoRI digestion of Ca3 DNA results in seven fragments of 4.2 kb (A), 2.98 kb (B), 2.85 kb (C), 0.77 kb (D1), 0.77 kb (D2), 0.38 kb (E), and 0.30 kb (F). Five of these EcoRI fragments have been mapped in the 5'-3' order C B D1 A D2. The intact Ca3 probe and the three largest EcoRI fragments, A, B, and C, were individually used to probe Southern blots of EcoRI-digested DNA of a set of test strains, transverse alternating field electrophoresis-separated chromosomes of strain 3153A, and Northern (RNA) blots of test strain 3153A. Fragments, A, B, and C each generate a different Southern blot hybridization pattern with EcoRI-digested whole-cell DNA; Ca3 sequences are present in at least five of seven separable chromosomes and a minichromosome of strain 3153A; fragments A, B, and C are distributed differently on chromosomes; and fragments A, B, and C do not cross-hybridize. Ca3 hybridizes to three major transcripts of 2.8, 2.3, and 1.5 kb. Fragment A hybridizes intensely to the 1.5-kb transcript, while fragments B and C both hybridize intensely to the 2.8- and 2.3-kb transcripts. The B fragment, which contains 2,980 bp and contributes to the major portion of the Ca3 pattern, was sequenced. Both direct and inverted repeat sequence motifs were identified. These results provide us with initial insights into the evolution of the Ca3 pattern and the nature of the probe.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedell G. W., Soll D. R. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun. 1979 Oct;26(1):348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. L., Buckley H. R., Campbell C. C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia. 1975 Jul;13(2):148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Hicks J. B. Dosage of the smallest chromosome affects both the yeast-hyphal transition and the white-opaque transition of Candida albicans WO-1. J Bacteriol. 1991 Dec;173(23):7436–7442. doi: 10.1128/jb.173.23.7436-7442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaki S. H., Hicks J. B., Greenspan D., Polacheck I., MacPhail L. A., White T. C., Agabian N., Greenspan J. S. The identification and tracking of Candida albicans isolates from oral lesions in HIV-seropositive individuals. J Acquir Immune Defic Syndr. 1992 Oct;5(10):1039–1046. [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackwitz H. R., Zehetner G., Frischauf A. M., Lehrach H. Rapid restriction mapping of DNA cloned in lambda phage vectors. Gene. 1984 Oct;30(1-3):195–200. doi: 10.1016/0378-1119(84)90120-3. [DOI] [PubMed] [Google Scholar]
- Sadhu C., Hoekstra D., McEachern M. J., Reed S. I., Hicks J. B. A G-protein alpha subunit from asexual Candida albicans functions in the mating signal transduction pathway of Saccharomyces cerevisiae and is regulated by the a1-alpha 2 repressor. Mol Cell Biol. 1992 May;12(5):1977–1985. doi: 10.1128/mcb.12.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Stevens D. A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Stevens D. A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987 Apr;25(4):675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J., Odds F. C., Wiselka M. J., Nicholson K. G., Soll D. R. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J Clin Microbiol. 1992 Apr;30(4):935–941. doi: 10.1128/jcm.30.4.935-941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J., Voss E., Soll D. R. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990 Jun;28(6):1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Galask R., Isley S., Rao T. V., Stone D., Hicks J., Schmid J., Mac K., Hanna C. Switching of Candida albicans during successive episodes of recurrent vaginitis. J Clin Microbiol. 1989 Apr;27(4):681–690. doi: 10.1128/jcm.27.4.681-690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Galask R., Schmid J., Hanna C., Mac K., Morrow B. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J Clin Microbiol. 1991 Aug;29(8):1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Langtimm C. J., McDowell J., Hicks J., Galask R. High-frequency switching in Candida strains isolated from vaginitis patients. J Clin Microbiol. 1987 Sep;25(9):1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R. Molecular biology of Candida pathogenesis. Mol Cell Biol Hum Dis Ser. 1992;1:131–172. doi: 10.1007/978-94-011-2384-6_5. [DOI] [PubMed] [Google Scholar]
- Soll D. R., Staebell M., Langtimm C., Pfaller M., Hicks J., Rao T. V. Multiple Candida strains in the course of a single systemic infection. J Clin Microbiol. 1988 Aug;26(8):1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]