Abstract
Although mechanisms of single-nucleotide residue deletion have been investigated, processes involved in the loss of longer nucleotide sequences during DNA replication are poorly understood. Previous reports have shown that in vitro replication of a 3′-TGC TGC template sequence can result in the deletion of one 3′-TGC. We have used low-energy circular dichroism (CD) and fluorescence spectroscopy to investigate the conformations and stabilities of DNA models of the replication intermediates that may be implicated in this frameshift. Pyrrolocytosine or 2-aminopurine residues, site-specifically substituted for cytosine or adenine in the vicinity of extruded base sequences, were used as spectroscopic probes to examine local DNA conformations. An equilibrium mixture of four hybridization conformations was observed when template bases looped-out as a bulge, i.e. a structure flanked on both sides by duplex DNA. In contrast, a single-loop structure with an unusual unstacked DNA conformation at its downstream edge was observed when the extruded bases were positioned at the primer–template junction, showing that misalignments can be modified by neighboring DNA secondary structure. These results must be taken into account in considering the genetic and biochemical mechanisms of frameshift mutagenesis in polymerase-driven DNA replication.
INTRODUCTION
A shift of the reading frame during template-dependent DNA synthesis can lead to the addition or deletion of one or more nucleotide residues (nts) in the newly synthesized DNA. Formation of a misaligned section of the DNA, which is the first step of frameshift mutagenesis, is thought to occur via several different processes (1–4). A mechanism by which DNA polymerase (Pol) can impose single-nucleotide deletions has recently been proposed on the basis of structural studies (5). It was shown that Pol λ stabilizes a small deformation of the primer–template (P/T) junction without disrupting base pairs (bps). The active site of the protein skips one coding base on the template and binds incoming dNTP to template position n + 1. The authors called this mechanism ‘glissando’, analogizing this DNA conformational change to a continuous sliding from one pitch to another in playing a musical scale. Frameshifts that delete several bases require separation of a number of bps prior to annealing them in a new register. This process may be more like playing an ‘arpeggio’, in which the notes of a broken chord are repeated further up or down the scale. The DNA polymerases that replicate these misalignments are highly distributive, and steric requirements suggest that bulky misalignments may take place, at least partially, while the DNA is dissociated from the polymerase (6). Alternatively the rearrangement may be imposed in part by an interaction of the DNA sequence with the polymerase. In either case an initial step in investigating these larger deletions must be to determine the equilibrium conformation(s) and conformational distributions of potential DNA replication intermediate structures that form at and near P/T junctions in the ‘absence’ of replication proteins.
Here we have studied DNA constructs implicated in a 3 nt deletion of 3′-TGC, which is known to occur in DNA replication. Although 3 nt deletions are rare events, they have been observed in vitro in replication processes driven by the distributive polymerases Escherichia coli DNA Pol IV and human Pol λ. Three of the four 3 nt deletions created by Pol IV were deletions of 3′-TGC at a 3′-TGC TGC sequence (7). Replication by DNA Pol λ of an oligonucleotide containing this sequence has also been reported to delete a 3′-TGC trinucleotide sequence in single-nucleotide addition experiments (6). Pol λ, a member of DNA polymerase family X (8), has one of the highest probabilities among known DNA polymerases of producing frameshift-induced infidelities (9).
Low-energy CD (10,11) is based on the site-specific substitution of a nucleic acid base by an analogue that absorbs above 300 nm. Local interactions with neighboring nucleic acid bases produce chirality in the probe residue that can be observed in the CD spectrum above 300 nm without interference from the CD bands of DNA and protein. Low-energy CD measurements of 2-aminopurine (2-AP) have been successfully applied to characterize site-specific conformational changes that occur in the loop of box B RNA hairpin on binding to the N protein of phage λ (12) and to map local conformations in the nucleic acid framework of phage T7 transcription complex (13,14) and of DNA polymerase I Klenow fragment (Datta K. et al., manuscript in preparation). Low-energy CD spectra have also been reported for DNA oligonucleotides containing the cytosine analogue pyrrolocytosine (PC) (11). Adjacent PC residues in B-form DNA exhibit exciton coupling with characteristic opposite-signed CD bands centered at the absorption maximum (350 nm). Earlier experiments have shown that these analogues do not greatly influence DNA structure or stability, but rather behave much like their respective canonical DNA bases, A and C [references in (10,11)]. The fluorescence signals and CD spectra of these residues provide complementary information. Here we have used these spectroscopic approaches to study DNA replication intermediates that are likely to form during frameshift mutagenesis resulting from the deletion of a 3′-TGC trinucleotide sequence.
MATERIALS AND METHODS
Oligonucleotides were synthesized and HPLC-purified by Operon (Huntsville AL; Cologne, Germany) and their concentrations were determined from the molar extinction coefficients provided by the manufacturer. The concentration units used for DNA are expressed in moles of oligonucleotide molecules per liter. Unless otherwise stated, experiments were performed at 20°C in 20 mM phosphate buffer (pH 7.5) containing 0.1 M NaCl and 0.1 mM EDTA. Duplex molecules were made by heating a mixture of the ss (single-stranded) DNA components (Tm ≤ 55°C) to 60°C, followed by cooling the reaction mixture at a rate of ≤1°C/min.
Fluorescence spectra were measured with a Jobin–Yvon Fluorolog or a Photon Technology International spectrometer. Samples containing PC probe residues were excited at 350 nm and fluorescence intensity was determined at the emission maximum near 450 nm. Samples containing 2-AP residues were excited at 315 nm and fluorescence intensity was determined 360–380 nm. The shape and position of the fluorescence spectra were the same for all constructs (Supplementary Figure S1). Experiments were replicated four to six times and error bars are standard error. CD spectra were taken between 500 and 260 nm on a Jasco model 810 CD spectrometer equipped with a thermostatted cell holder. Reported CD spectra are the average of 8–20 scans and are presented as Δε, the difference in the molar extinction coefficients of left and right circularly polarized light on a per mole PC residue basis [units of liter (mole PC)−1 cm−1]. Predicted loop and bulge structures were calculated using the Mfold web server (15,16) with initial conditions set at 20°C and 0.1 M NaCl.
RESULTS
Oligonucleotides were constructed using a labeled ‘template’ strand, T, and a ‘primer’ strand, P (Figure 1). We refer to the 3′ end of the template strand as ‘upstream’ and the 5′ end as ‘downstream’, by analogy to the expected direction of DNA replication. P strands are named by reference to the position of their 3′ terminus relative to the T strand. The numbering scheme was chosen so that positive integers correspond to the number of nts in P that are complementary to the template sequence downstream from the nucleotide residues that are extruded from the template. The gT/P-5 construct corresponds to the substrate DNA used in the Pol λ experiment (6) and the other gT/P complexes represent potential intermediates of primer extension to form a 3 nt deletion.
Figure 1.
Oligonucleotide constructs and nomenclature. (a) gT indicates the template strand used to construct duplex molecules with GC bps flanking the 3 nt loop at positions [−3,−2,−1] (c) aT indicates the template strand used to construct duplex molecules with AT bps flanking the [−3,−2,−1] loop sequence. The positions of the probe residues 2-AP (Ap) or PC (PC) are indicated in the name of the template strand in square brackets. Potential looped out sequences of the T strand are underlined. (b) and (d) are primer strands (P) that are partially complementary to the gT strands (gP) or to the aT strands (aP). Primer strands of different lengths are designated by the position of the 3′-OH terminus. The numbering scheme at the top of the Figure refers to both strands. gC and aC are fully complementary to gT and aT, respectively.
Spectroscopic characterization of a 3-nt loop
We first used constructs with a single, well-defined TTT loop (gTβ) to determine the types of spectroscopic signals that we might expect to find (Figure 2). Template strands were constructed with a PC residue at either position [−4] or [1] that flank the loop sequence. These were annealed with primer strands gP of different lengths that lacked the complementary AAA sequence (Figure 1).
Figure 2.
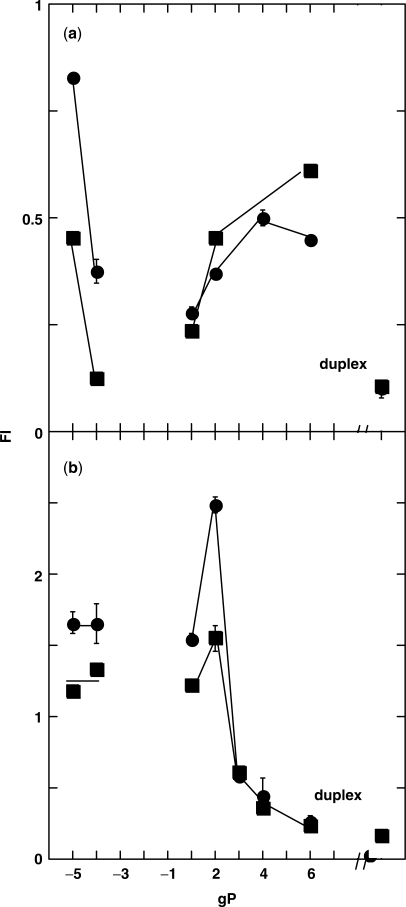
Fluorescence changes observed during loop formation. Fluorescence intensities of 1 μM duplex molecules formed with template DNA T and various length primer strands, gP (see Figure 1 for nomenclature). Fl is the fluorescence intensity of the duplex molecule relative to the signal of the ss T strand. The signal of the fully duplex molecule without extrahelical bases is shown at the right of each panel. gTβ (circles); gT (squares). Template sequences had a single PC residue at position [−4] (a) or position [1] (b).
Base-pair formation with the probe residue quenched the fluorescence of the gTβ[−4]/gP-4 complex as expected (circles, Figure 2a). The signal increased progressively for gTβ[−4]/gP2 and gTβ[−4]/gP4 as the TTT sequence forms a loop. The fluorescence intensity was the same for the gP4 and gP6 constructs and was taken as the signal of a PC residue at position [−4] of the bulge duplex.
The behavior of a PC probe at position [1], the 5′ edge of the TTT loop, was more complex (circles, Figure 2b). The fluorescence signals of duplex molecules with gP-5 and gP-4 were 1.6-fold greater than the intensity of the same signals in the single-stranded template. This behavior appears to be a consequence of the secondary structure of the gT template oligonucleotides. Mfold calculations (15,16) show stable base pairing between C1G2 and C−10G−9 (Figure 1). Formation of duplex molecules with gP-5 or gP-4 disrupts these bps and causes an apparent increase in fluorescence of the PC residue at position [1]. C−4 does not participate in base pairing in the single-stranded gTβ oligonucleotide, and the ss template strand with a probe at position [−4] did not exhibit this phenomenon (circles, Figure 2a). Hence the ‘true’ intensity of single-stranded gT oligonucleotides that have a PC residue at position [1] should be taken as the intensity observed for duplexes of the gT strand with either gP-5 or gP-4. We subsequently normalized the fluorescence signal ‘Fl’ (Figure 2) with respect to the signals of the P-5 and P-4 duplexes; this is designated as ‘rel Fl’.
Loop closure produced two kinds of changes in the fluorescence intensity of PC at position [1]. Complexes with three, four and six downstream bps, which are expected to stabilize the misaligned structure, quenched the fluorescence; the signal for gTβ[1]/gP6 was the same as that of duplex DNA. The gP2 duplex, however, exhibited 1.6-fold more intensity signal than the signals of the gP-5 and gP-4 complexes, without changing the shape of emission spectrum. We examined the effects of sequence in order to better understand this fluorescence increase.
Template oligonucleotides with several loop sequences in identical sequence context, all with a PC residue at position [1], were annealed to gP primer strands of different lengths (Figure 3). The loop sequence of gTα[1] (3′-GTC) is implicated in trinucleotide repeat disorders such as myotonic dystrophy (17). The fluorescence signal of the gT/gP2 constructs increased for all templates; the relative intensities varied with the loop sequence: GTC > TTT ≈ TCG. The fluorescence intensity generally decreased as the number of downstream bp increased and the loop closed. However, the gTα[1] complexes behaved differently (circles, Figure 3a). In this case the fluorescence signal dropped for gTα[1]/gP3, increased when gTα[1] was complexed to gP4 and then decreased again for gTα[1]/gP6.
Figure 3.
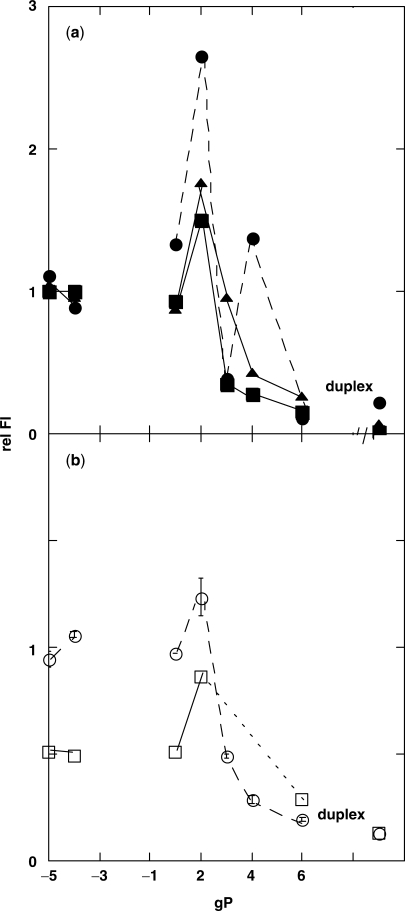
Fluorescence of duplex oligonucleotides with GC bps flanking the loop sequence. Fluorescence signals of 1 μM duplex molecules with primer strands of various lengths, gP, were normalized to the intensities of the corresponding gP-5 and gP-4 complexes (see text). (a) gTα[1] (circles), gTβ[1] (squares) and gTγ[1] (triangles). (b) gT[1] (circles) and gT[−1,1] (squares). See Figure 1 for nomenclature. In order to show the self-quenching of adjacent PC residues in ss template oligonucleotide gT[−1,1] (see text), signals of gT[−1,1] complexes are normalized relative to the intensity of gT[−1]/gP-5 and gT[−1]/gP-4.
In order to examine the influence of the neighboring bases on loop formation, we made duplex constructs using template molecules containing the same loop sequences, but with AT bps flanking the loops; a 2-AP residue was placed at position [1] of the template strand to monitor loop formation (Figure 1c). All other nts are the same as in the constructs with flanking GC bps. We did not study constructs involving the aTγ[1] template (corresponding to gTγ[1], Figure 3), because this template has a highly stable secondary structure.
Replacing the GC bps adjacent to the loop by AT bps had two effects on the fluorescence of the probe at position [1] (Figure 4). First, a fluorescence increase was not observed in the aP2 complexes with an AT flanking bp. In fact, the fluorescence signal did not significantly change for duplex molecules with primer strands aP1, aP2 and aP3, suggesting that the loop may not close in these constructs. Second, oligonucleotides with flanking AT bps appeared to require more complementary downstream bps to achieve the same degree of fluorescence quenching as oligonucleotides with GC flanking bps, indicating that the bulge may be more ‘open.’
Figure 4.
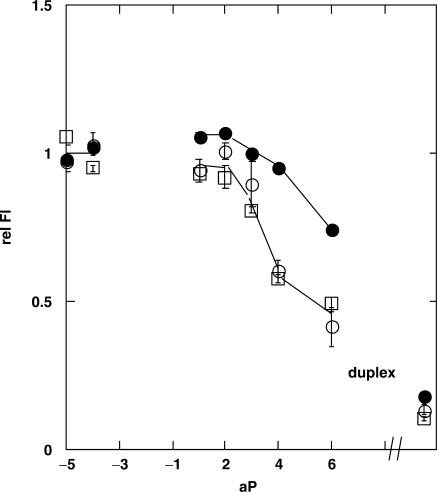
Fluorescence of duplex oligonucleotides with AT bps flanking the loop sequence. The sequences of the template strands aTα[1] (open circles) and aTβ[1] (open squares) are identical except for the loop (Figure 1). Closed circles are Fl signals from complexes of gTα[1] with the aP primers. Fluorescence intensities of 1 μM duplex molecules with various length primer strands, aP, have been normalized to the signals of the aP-5 and aP-4 complexes.
Mfold calculations (15,16) do not predict a stable looped structure for the gP2 complexes, although primer strands gP3 and longer are expected to form a stable duplex downstream from the loop. It is possible that the increased fluorescence observed in Figure 3 reflects another interaction than bp formation between the primer and the probe residue. To check this possibility we compared gTα[1]/gP constructs that can form a bp at the probe position (Figure 3), with gTα[1]/aP complexes that cannot (Figure 4, closed circles). Fluorescence of gTα[1]/aP2 did not exhibit the fluorescence increase observed with the gTα[1]/gP2 constructs. Identical results were observed for all gT template sequences (Table 1). Hence the unusual Fl signal of the gP2 complexes seems to involve specific bp formation at position [1] and possibly position [−4]. In addition the fluorescence increase that was observed for the gTα[1]/gP4 construct (Figure 3) was not observed with gTα[1]/aP4 (Figure 4), suggesting that GC base-pairing at position [1] may also contribute to this phenomenon.
Table 1.
Relative fluorescence intensities of gT[1] or aT[1] template strands complexed with aP2 or gP2 primers
| T (loop sequence) (3′ → 5′) | gT/gP2 | gT/aP2 | aT/aP2 |
|---|---|---|---|
| Tα[1] (GTC) | 2.65 ± 0.06 | 1.07 ± 0.01 | 1.01 ± 0.04 |
| T[1] (TGC) | 1.18 ± 0.11 | 0.94 ± 0.03 | nd |
| Tβ[1] (TTT) | 1.48 ± 0.06 | 0.95 ± 0.04 | 0.92 ± 0.05 |
| gTγ[1] (TCG) | 1.77 ± 0.09 | 1.05 ± 0.05 | nd |
Sequences of the oligonucleotides are in Figure 1. Values are rel Fl (see text). nd, not determined.
Loop formation at the P/T junction
Before investigating extruded bases in the 3′-TGC TGC template sequence, we measured the spectra of PC probe residues in control ss- and dsDNA constructs. Base pairing of probe residue(s) in dsDNA quenched the fluorescence signal as expected. The fluorescence signal (per mole PC residue) of the ss template oligonucleotide containing two adjacent PC residues at positions [−1,1] (gT[−1,1]) was approximately half as intense as that of an oligonucleotide with a single PC residue at position [−4] (gT[−4]) (Figure 5a and b, inserts); this difference is due in part to ‘self-quenching’ in ssDNA that has been previously observed in oligonucleotides with adjacent PC [Figure 3 and reference (11)] or 2-AP residues (D. Jose, et al., manuscript in preparation).
Figure 5.
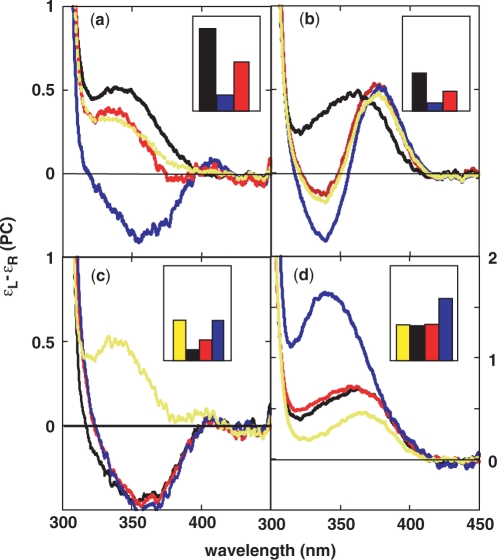
Spectroscopic characterization of bulge and P/T loop DNA constructs. (a,b) ss template DNA (black); duplex DNA with oligonucleotide gC as the complementary strand (blue); bulge DNA with complementary strand gP6 (red). The yellow lines are the best fits of the bulge CD spectra (see text). (c,d) duplex DNA with complementary oligonucleotide gP-5 (yellow), gP-4 (black), gP1 (red) or gP2 (blue). Oligonucleotide concentrations were 13 μM. Template strands have either a PC residue at position [−4], gT[−4] (a,c) or a PC dimer at positions [−1,1], gT[–1,1] (b,d). Oligonucleotide sequences and numbering schemes are shown in Figure 1. CD spectra and fluorescence intensities (inserts) are plotted per mole PC residue. Fl intensity is in arbitrary units that are the same in all Inserts.
PC has an an absorbance maximum at 350 nm which provides the opportunity to use low-energy CD spectroscopy to determine the conformation of the DNA around this probe residue (11). The low-energy CD spectra of ss template oligonucleotides, gT[−4] and gT[−1,1], both contained a single peak (Figure 5). Although the two spectra had comparable intensities [∼0.5 l (mole PC)−1(cm)−1], the gT[−4] signal had a maximum at 340 nm, while the CD peak of gT[−1,1] was located at 365 nm. This red shift of the CD maximum may reflect the presence of some exciton coupling due to partial base stacking of the PC residues in gT[−1,1], which also manifests itself in the self-quenching of the Fl in this template mentioned above. The low-energy CD spectrum of duplex DNA with a single PC residue had a negative band at 350 nm. The spectrum of dsDNA with PC residues at positions [−1,1] showed strong exciton coupling (with a symmetric low-energy peak and high-energy trough centered at the absorption maximum) characteristic of a PC dimer stacked in a right-handed B-form DNA conformation (11).
Annealing short primer strands gP with a single 5′-ACG sequence to a template strand gT containing a 3′-TGC TGC repeat can produce duplex molecules with several possible looped-out sequences (Figure 6). We used our Fl and CD methodology to probe the contributions of these various hybridization forms to the equilibrium conformation of the gP2 complex.
Figure 6.
Hybridization schemes of the gT/gP2 construct. Upper strand is gP2; lower strand represents either gT[−4] or gT[−1,1], depending on the position(s) of the PC residue (X) The different base pairing schemes are named at the left by the positions of the looped-out bases in the template strand. {0} is a possible alignment in which the two 3′ terminal bases of the primer are not base-paired with the template strand.
Fluorescence changes of duplex constructs gT/gP were similar to the changes observed for constructs Tβ/gP containing a TTT loop (Figure 2). In both cases, the fluorescence of a PC residue at position [−4] was quenched by forming a partial duplex with primer strand gP-4, and then increased as the template was annealed to progressively longer primer strands. The signal likewise increased in both gP2 complexes with the PC probe at position [1], and then decreased as the length of the primer strand increased. Quantitative differences in the signal intensities of the gTβ and gT templates may reflect the presence of additional equilibrium conformational states that are accessible within the repeat sequence (see below). The fluorescence of PC residues at positions [−1,1] also increased in the gT[−1,1]/gP2 complex and decreased in the bulge form gT[−1,1]/gP6 (Figure 3b). Hence the gTβ/gP2 and gT/gP2 constructs likely have similar conformations: in particular, it is primarily the downstream (and not the upstream) 3′-TGC sequence that forms an extra-helical loop. In agreement with this conclusion, oligonucleotide gT[−4] formed duplex molecules with gP-4, gP1 and gP2 whose CD spectra suggest that the residue at position [−4] is in a ds environment (Figure 5a and c).
Relatively minor changes were observed in the CD spectra of gT[−1,1] when annealed with gP-5, gP-4 and gP1 (Figure 5d). The fluorescence signals of these duplexes were similar to the spectrum of ss template DNA, indicating that the probe residues at position [−1,1] are located in a ss environment. In contrast, the CD spectrum of the complex of this template with the gP2 oligomer increased dramatically and the CD maximum shifted to 340 nm. In addition the fluorescence intensity of the gP2 duplex was higher than the intensity obtained with ssDNA, gT[−1,1]. These observations suggest that probe residues in the gT[−1,1]/gP2 complex are in a more open conformation than in ss gT[−1,1] (11).
Taken together these results show that the gT/gP2 duplex assumes a conformation in which the bases at position [−4] are double-stranded and the bases at positions [−1,1] are highly unstacked. This conformation, which we call a P/T loop, corresponds to the hydridized form {−3,−2,−1} (Figure 6). This structure is not predicted by Mfold calculations (15,16).
The bulge conformational equilibrium
Several observations suggest that gT/gP6 is a mixture of conformations. The CD spectrum of gT[−4]/gP6 qualitatively resembles the signal obtained with ssDNA (Figure 5a) and the CD spectrum of gT[−1,1]/gP6 is similar to that of a PC dimer in dsDNA (Figure 5b). However in both cases there are significant differences between these spectra and those observed with ss or dsDNA. In addition the fluorescence intensity of the duplex gT[−4]/gP6 is 25% larger than that of the corresponding TTT construct gTβ/gP6 (Figure 2a). These observations suggest that a bulge formed in a gT template containing the 5′-TGC TGC repeat sequence may be a mixture of hybridization schemes analogous to those in Figure 6, conferring on average a more ‘open’ conformation on the probe residue. We have attempted to quantify the contributions of the individual bp arrangements.
To a first approximation, the PC residue at position [−4] is expected to be in a duplex environment in constructs {−3,−2,−1} and {0}, and single-stranded in the other constructs. (Numbers in this and the next paragraph refer to the unpaired template bases for the gP6 complexes that correspond to the gP2 constructs in Figure 6). We fit the spectra observed for the bulge labeled at position [−4] with signals from the PC probe located in ss and dsDNA sequences (Figure 5a). Fluorescence measurements indicate that the bulge is 71% ‘single-stranded’. This is consistent with 78% ss, estimated from the best fit of the CD signal (yellow trace, Figure 5a).
The two adjacent PC residues at positions [−1,1] should be in a duplex environment in constructs {−4,−3,−2}, {−5,−4,−3} and {−6,−5,−4} (Figure 6). We initially assumed that the {−3,−2,−1} construct should have the signal shown for ssDNA in Figure 5b. However, CD spectra reconstructed from combinations of ss and ds spectra have a different shape than the bulge spectrum (data not shown). This result indicates that the unpaired scheme {0} (where the bases at positions [−1,1] should have a CD signal like ssDNA) does not contribute significantly to the conformational equilibrium. Base-pairing of the probe in the {−3,−2,−1} construct resembles that of the P/T loop, and a better fit was obtained using the fluorescence and CD signals of the gT[−1,1]/gP2 complex. Fitting the fluorescence spectrum of template labeled at [−1,1] with a linear combination of the duplex and P/T loop basis signals indicated that the bulge was 78% ds; these basis spectra also gave an excellent fit to the CD spectrum of bulge DNA with 87% dsDNA (yellow trace Figure 5b). The higher value of the duplex contribution calculated from the CD data may reflect our choice of the P/T loop for the basis spectrum, since the conformation of the gT[−1,1]/gP2 complex may not be completely homogeneous. Nevertheless, these four independent fits are remarkably consistent. Taken together they indicate that the four possible hybridization schemes with 3-nt loops (Figure 6) are approximately equally populated in the gT/gP6 bulge construct.
DISCUSSION
We report a novel DNA conformation that results from the asymmetry of duplex DNA at the P/T junction. This was readily seen in control experiments using a template strand containing an extra TTT sequence (Figure 2b). The fluorescence signal of a PC residue placed at position [1] on the downstream edge of the TTT is quenched in gTβ[1]/gP6, a bulge construct with dsDNA on both sides of the TTT loop. In contrast, the fluorescence intensity increased for the complex gTβ[1]/gP2, which has only two potential downstream GC bps (Figure 1), suggesting that the gP2 construct is in an unusual conformation. We call this structure a ‘P/T loop’, by analogy to the ‘stem-loop’ terminology commonly used for RNA secondary structure discussions. A similar fluorescence signal is observed for gP2 complexes with other extruded sequences, including duplexes formed with the 3′-TGC TGC template, indicating that this unusual conformation can be achieved for a variety of 3-nt loop constructs. Sequence-specific interactions of the flanking nucleotide residues, however, are required (Figures 3 and 4; Table 1).
The P/T loop construct with the 3′-TGC TGC template (gT/gP2) was further characterized by CD spectroscopy of PC residues placed at various positions within the template strand. The CD spectrum of PC residues located at positions [−1,1] exhibited an increased intensity and a blue shift, a signal that does not correspond to the spectra of these probe residues in ss or dsDNA (Figure 5). However similar CD spectra have been observed for adjacent PC residues located in the loop sequence of a DNA stem loop construct (N.P.J., unpublished results). They also resemble the CD spectrum of adjacent 2-AP residues in ssDNA bound to the exonuclease site of Klenow fragment of DNA polymerase I (K. Datta et al., manuscript in preparation) where these residues assume an extended ssDNA conformation (18). In all cases these CD changes were accompanied by an increase in the intensity of the Fl signal without changing the shape or position of the Fl spectrum. These results argue that the bases at positions [−1,1] of the P/T loop are in an ‘open’ conformation that is less stacked than in ssDNA. Unstacking the PC bases would also be expected to decrease the self-quenching observed between adjacent PC residues, thereby explaining the increased Fl intensity of gT[−1,1]/gP2 compared to ss gT[−1,1] (Figure 5). This open conformation may also decrease collisional quenching of the probe at position [1] by adjacent nucleotides, and cause the Fl increase observed in the gP2 complexes of other template strands (Figures 2 and 3).
The fluorescence signals of duplex constructs formed with primer strand gT[−4]/gP-4, gT[−4]/gP1 and gT[−4]/gP2 became progressively more intense; this behavior is expected for a probe residue at the 3′ edge of a loop in the T strand as it closes, judging from the analogous signal seen for formation of a well-defined TTT loop (Figure 2a). On the other hand the CD spectra of these complexes were identical and resembled the spectrum of PC in dsDNA (Figure 5c). Base stacking has a relatively small affect on fluorescence quenching of PC, which is more sensitive to H-bonding (19,20). In contrast, base stacking is a primary cause of nucleic acid CD (21). NMR studies of bulged DNA constructs (22–24) have shown that the extruded bases stack with one another and with the bases of the flanking dsDNA, thereby maintaining the right-handed helical path of the strand. The duplex compensates by bending away from the extruded bases thereby modifying the hydrodynamic properties of bulged DNA (25). A similar ‘bend’ may occur in the P/T loop. A ‘bend’ would modify the H-bonding of the PC residue with G[−4] of the primer, thereby decrease quenching; the extruded bases of gT[−4]/gP1 and gT[−4]/gP2 constructs also appear to maintain the base stacking of the PC probe residue at position [−4] of the template DNA judging by the unchanged CD spectra.
The absence of exciton coupling in the CD spectrum of gT[−1,1]/gP2 (Figure 5d) indicates that this construct forms a P/T loop corresponding to hybridization scheme {−3,−2,−1} (Figure 6), rather than the {−6,−5,−4} bulge, with 9 bp upstream, 5 bp downstream and identical extruded bases (3′-TGC) as in the {−3,−2,−1} arrangement. We found this result to be rather surprising, given that hydrogen bond interactions within a bp are usually considered rather weak compared to the larger free energies contributed by stacking between adjacent bases (26). The P/T loop {−3,−2,−1}, whose downstream duplex is so short that it has no interior bases, does not look very sturdy compared to the {−6,−5,−4} construct that carries a downstream duplex sequence representing half a turn of B-form DNA.
Although the physical mechanisms that stabilize duplex DNA are not fully understood, bases appear to stack in a way that minimizes the surface area of the cavity created in the solvent (26,27); i.e. the energy required to create a solvent cavity for stacked bases is less than the energy required to accommodate individual unstacked bases. Likewise the formation of two cavities will result in a larger total surface area than the formation of a single cavity (with the same total number of bps) because they have more ‘ends’.
Consistent with this analysis, all of the hybridization schemes for the bulge DNA construct gT/P6 (equivalent to the upper four structures in Figure 6) have the same number of total bp and ‘ends’ and are equally represented in solution (see Results section, Figure 5). Minimization of the solvation energy could also be the driving force that selects the P/T loop conformation {−3,−2,−1} among the possible hybrid structures of the gT/gP2 complex (Figure 6), if the conformation of the GC bps at positions [1,2] has a lower cost of hydration than stacked bps. A requirement for two cavities introduces additional ‘ends’ that may further destabilize the other constructs relative to {−3,−2,−1}.
If the P/T loop is stabilized by template-directed extension of the primer it becomes a bulge, which differs from the P/T loop by the presence of a significant duplex DNA on both sides of the extruded bases. A bulge can also form directly by strand slippage within a long homopolymer sequence at positions further away from the P/T junction (1). Bulged DNA has been extensively studied (22–24). Thermodynamic stability of a large bulge is primarily determined by the secondary structure of the extrahelical bases. For example, folded structures in the looped-out sequence, often involving over 100 nts, are responsible for large frameshifts associated with trinucleotide repeat disorders such as myotonic dystrophy and Huntington's disease (17). In contrast there is likely to be little or no secondary structure within short extruded loops studied here and the loop has a smaller effect on stability than does the flanking sequence.
Primer–template loop and bulge constructs are both potential substrates for the DNA replication machinery and the relative stabilities of these various misaligned P/T junctions contribute to total free energy considerations that determine the probability of a frameshift at a particular DNA sequence. It is well known that the stability of looped-out DNA constructs is influenced by the sequence that flanks the extruded bases (15,16). Our results show that it may also be important to distinguish a P/T loop from a bulge. Different bases can be preferentially extruded in these two environments, raising the possibility that the position of misalignment with respect to the P/T junction could influence the sequence-specificity of frameshift mutagenesis.
FUNDING
This work was supported by Centre Nationale de la Recherce Scientifique; a Projets Exploratoire/Prémier Soutien (PEPS) grant (to N.P.J.) and by National Institutes of Health grant (GM-15792 to P.H.v.H.). P.H.v.H. is an American Cancer Society Research Professor of Chemistry.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Emmanuelle Delagoutte, John Schellman and members of our laboratories for many helpful discussions.
REFERENCES
- 1.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J. Biol. Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 3.Bebenek K, Kunkel TA. Frameshift errors initiated by nucleotide misincorporation. Proc. Natl Acad. Sci. USA. 1990;87:4946–4950. doi: 10.1073/pnas.87.13.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J. Biol. Chem. 2004;279:45360–45368. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase lambda. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 6.Blanca G, Villani G, Shevelev I, Ramadan K, Spadari S, Hubscher U, Maga G. Human DNA polymerases lambda and beta show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry. 2004;43:11605–11615. doi: 10.1021/bi049050x. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi S, Valentine MR, Pham P, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 10.Johnson NP, Baase WA, Von Hippel PH. Low-energy circular dichroism of 2-aminopurine dinucleotide as a probe of local conformation of DNA and RNA. Proc. Natl Acad. Sci. USA. 2004;101:3426–3431. doi: 10.1073/pnas.0400591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson NP, Baase WA, Von Hippel PH. Investigating local conformations of double-stranded DNA by low-energy circular dichroism of pyrrolo-cytosine. Proc. Natl Acad. Sci. USA. 2005;102:7169–7173. doi: 10.1073/pnas.0502359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson NP, Baase WA, Von Hippel PH. Low-energy CD of RNA hairpin unveils a loop conformation required for lambda N antitermination activity. J. Biol. Chem. 2005;280:32177–32183. doi: 10.1074/jbc.M504619200. [DOI] [PubMed] [Google Scholar]
- 13.Datta K, Johnson NP, von Hippel PH. Mapping the conformation of the nucleic acid framework of the T7 RNA polymerase elongation complex in solution using low-energy CD and fluorescence spectroscopy. J. Mol. Biol. 2006;360:800–813. doi: 10.1016/j.jmb.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 14.Datta K, Von Hippel PH. Direct spectroscopic study of reconstituted transcription complexes reveals that intrinsic termination is driven primarily by thermodynamic destabilization of the nucleic acid framework. J. Biol. Chem. 2008;283:3537–3549. doi: 10.1074/jbc.M707998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.SantaLucia JJ. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 18.Beese LS, Derbyshure V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 19.Hardman SJ, Thompson KC. Influence of base stacking and hydrogen bonding on the fluorescence of 2-aminopurine and pyrrolocytosine in nucleic acids. Biochemistry. 2006;45:9145–9155. doi: 10.1021/bi060479t. [DOI] [PubMed] [Google Scholar]
- 20.Hardman JO, Botchway SW, Thompson KC. Evidence of nonbase stacking effects for the environment-sensitive fluorescent base pyrrolocytosine – comparison with 2-aminopurine. Photochem. Photobiol. 2008;84:1473–1479. doi: 10.1111/j.1751-1097.2008.00368.x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson WCJ, Tinoco IJ. Circular dichroism of polynucleotides: A general method applied to dimers. Biopolymers. 1969;8:715–731. [Google Scholar]
- 22.Hare D, Shapiro L, Patel DJ. Extra helical adenosine stacks into right-handed DNA: solution conformat ion of the d(C-G-C-A-G-A-G-C-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear overhauser effect spectra. Biochemistry. 1986;25:7456–7464. doi: 10.1021/bi00371a030. [DOI] [PubMed] [Google Scholar]
- 23.Rosen MA, Live D, Patel DJ. Comparative NMR study of A(n)-bulge loops in DNA duplexes: Intrahelical stacking of A, A-A, and A-A-A bulge loops. Biochemistry. 1992a;31:4004–4014. doi: 10.1021/bi00131a016. [DOI] [PubMed] [Google Scholar]
- 24.Rosen MA, Shapiro L, Patel DJ. Solution structure of a trinucleotide A-T-A bulge loop within a DNA duplex. Biochemistry. 1992b;31:4015–4026. doi: 10.1021/bi00131a017. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh CH, Griffith JD. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: Possible relevance to the folding of single-stranded nucleic acids. Proc. Natl Acad. Sci. USA. 1989;86:4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solie TN, Schellman JA. The interaction of nucleosides in aqueous solution. J. Mol. Biol. 1968;14:61–77. doi: 10.1016/0022-2836(68)90281-7. [DOI] [PubMed] [Google Scholar]
- 27.Cantor CR, Schimmel PR. Biophysical Chemistry. San Francisco: W.H. Freeman and Co.; 1980. pp. 328–337. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.