Abstract
Recent advances in both clone fingerprinting and draft sequencing technology have made it increasingly common for species to have a bacterial artificial clone (BAC) fingerprint map, BAC end sequences (BESs) and draft genomic sequence. The FPC (fingerprinted contigs) software package contains three modules that maximize the value of these resources. The BSS (blast some sequence) module provides a way to easily view the results of aligning draft sequence to the BESs, and integrates the results with the following two modules. The MTP (minimal tiling path) module uses sequence and fingerprints to determine a minimal tiling path of clones. The DSI (draft sequence integration) module aligns draft sequences to FPC contigs, displays them alongside the contigs and identifies potential discrepancies; the alignment can be based on either individual BES alignments to the draft, or on the locations of BESs that have been assembled into the draft. FPC also supports high-throughput fingerprint map generation as its time-intensive functions have been parallelized for Unix-based desktops or servers with multiple CPUs. Simulation results are provided for the MTP, DSI and parallelization. These features are in the FPC V9.3 software package, which is freely available.
INTRODUCTION
Although draft sequencing [i.e. whole-genome shotgun (1)] has become steadily more cost-effective in recent years, restriction fragment fingerprint maps remain an important component of many genome sequencing projects, where fingerprint maps are typically assembled using the FPC software package [fingerprinted contigs (2,3)]. One reason that FPC maps continue to be constructed is that many genomes (e.g. maize, wheat, barley) are too large and repeat-rich for shotgun sequencing; indeed, maize was recently sequenced using the clone-by-clone approach based on the FPC map (4). However, draft sequence can augment the clone-by-clone approach by providing sequence to aid in selecting the minimal tiling path (MTP), covering regions that are missed by the large clones, and detecting errors in both the map and sequence. The FPC map can be used to select a MTP for finished high-quality sequence of the whole genome or the gene-rich regions, where the gene-rich regions can be identified by having the draft sequence anchored to the FPC map. For small genomes, draft sequencing can generally provide good coverage of the genome; however, having a good FPC map can aid these projects by finding problems with the assembly of the sequence and providing a MTP of clones for difficult regions.
A second benefit of having an FPC map is that bacterial artificial chromosome (BAC) libraries remain essential for much laboratory work, and it is often beneficial to assemble the BACs into a map for more accurate anchoring; i.e. even if draft sequence exists, direct anchoring of a single BAC to the draft sequence can be ambiguous since BAC end sequences (BESs) can be repetitive, but a whole FPC contig can usually be anchored unambiguously. Pooled-BAC sequencing (5) is another promising approach to de novo sequencing of large genomes, and the FPC maps can be used to select the BAC pools.
A third benefit for large genomes is that FPC maps can be used for comparative analysis with a related sequenced genome. For example, Gregory et al. (6) aligned the mouse FPC map using BESs to the human genome. The SyMAP program (7) systematized this process, computing synteny between any FPC map and sequenced genome, and displaying the results in an interactive Java program. Green et al. (8) developed an approach to generate universal probes to create sequence ready FPC maps of orthologous regions from multiple related species.
As sequencing has improved, so has fingerprinting. In fact, high information content fingerprinting (HICF) (9–12) has itself benefited from advances in sequencing technology. It has enabled high-throughput fingerprinting on a sequencing machine, eliminating the need for manual band-calling. Also, it is now typical to have BESs generated from the same plates of clones that are fingerprinted, providing an immediate set of survey sequences which are located on the FPC map, and which can later be used to anchor and order draft sequenced contigs.
As a result of these developments it is increasingly common for a species to have both a fingerprint map and draft sequence, along with BESs to link them together; indeed, this is the case for soybean (13), sorghum (http://www.phytozome.org/soybean.php), medicago (14), poplar (15), xenopus (http://www.nih.gov/science/models/xenopus/) and is soon to be the case for several other species such as brachypodium (16) and cotton (17). It is obviously desirable to integrate these dual, complementary resources in order to maximize the quality of each.
Although it would be possible to incorporate information from the FPC map directly during the draft assembly process (18,19), the general practice is to perform separate assemblies and then compare the sequence and map afterwards in order to correct errors and assist with ordering and orientation of the draft contigs. Currently, this is carried out on a case-by-case basis using custom methods, which has evident drawbacks. First, there is not a standardized approach for evaluating the sequence in terms of the map and vice versa. Second, every laboratory needs to develop the software, resulting in duplication of effort. Third, it would clearly be advantageous to have support for this analysis within FPC, a tool which is already familiar to physical mapping researchers, and which already contains a full-featured display tailored to FPC contigs. To meet this need, we have added a comprehensive draft sequence integration (DSI) module to FPC, which can align and display draft sequences within the FPC contig display, detect alignment discrepancies indicative of mis-assemblies, assist in ordering and orienting draft contigs, and merge FPC contigs based on spanning draft sequenced contigs. The DSI functions were tested on simulations (see Results section), and were developed in part for the integration of the soybean HICF FPC map (13) with the soybean draft sequence produced by the Department of Energy Joint Genome Institute (DOE-JGI) (www.phytozome.org/soybean.php) (manuscript in preparation).
Fingerprinted maps have been used for selecting MTP clones for sequencing since 1988 (20). Now, with many genome projects generating both draft sequence and FPC maps, this dual resource can be used to select MTPs for regions of interest or to generate a finished sequence of the entire genome. In support of this approach, FPC V7 (21) included a function for selecting a MTP using BES-plus-draft sequence; however, this is not sufficient at lower draft coverages because the sparseness of BESs (e.g. ∼7.5 kb average separation for a 10× coverage and 150 kb clones) means that few draft contigs will span two BESs [e.g. with 2× coverage, 800 bp reads and perfect overlap detection, the mean contig length predicted by the Lander–Waterman model (22) is 2.6 kb]. Hence, the MTP module has been upgraded to add support for selecting a MTP using both fingerprint overlap and BES-plus-draft sequence. The algorithm models the methods used by a human technician to select a MTP interactively. The MTP module has been tested with simulations (see Results section), and applied to regions of the soybean map (Schmutz,J. et al., manuscript in preparation).
Both the MTP and DSI modules can take as input the results from comparing the draft sequence to the BESs, which can be done within FPC by using the BSS (blast some sequence) function. This function can also create in silico markers by comparing any sequences to the BESs, and then integrating them into the map. The BSS was first released in FPC V6 (21) and has since been modified to provide a simple yet very flexible interface which can handle most scenarios of sequence-to-map anchoring via BESs.
In conjunction with keeping FPC abreast with the latest sequencing approaches, it has also been important to support high-throughput map generation. Towards this end, the time-intensive functions of FPC have been parallelized. Ness et al. (23) parallelized the first part of the assembly algorithm, which compares all pairs of clones to each other, for a distributed processing environment (i.e. multiple computers networked together, often referred to as ‘compute clusters’). Recently, Unix-based desktop and server machines with multiple CPUs (or multi-core CPUs) have become affordable by most labs, making it unnecessary to resort to a compute cluster. Therefore, we have parallelized the pair-wise comparison to run easily on one machine with multiple CPUs, and also parallelized the second part of the algorithm that orders the clones. The remaining computationally intensive functions have also been parallelized, where the most important is the ends-to-ends merging algorithm, which is very important for HICF assembly as it generally requires multiple executions of the function with progressively less stringent cutoffs.
The FPC software is freely available at http://www.agcol.arizona.edu/software/fpc.
METHODS
FPC has three main functions for building contigs: (i) the Build function assembles the contigs, (ii) the DQer function detects possible chimeric contigs and breaks them apart and (iii) the Ends→Ends function detects possible contigs to merge and either lists them or automatically merges them. All three functions compare clones using the ‘Sulston’ score (20) to determine if they overlap, where the Sulston score estimates the probability that the bands in common are a coincidence. Bands are considered in common (or shared) if they have the same length within a user-specified tolerance. The user sets a ‘cutoff’ on the Sulston score that FPC uses to determine if two clones overlap. These functions will be referred to in the following; for more detailed information see ref. (2).
We first describe the BSS module, as it is used for both the DSI and MTP modules. All three modules assume that the name of a BES is the same as the clone name with an added suffix, e.g. clone a0001a01 may have BESs named a0001a01.f and a0001a01.r. A fixed-length library name prefix is also permitted.
The BSS (blast some sequence) module
The BSS function parses alignments of query sequences against BESs from clones in the FPC map using one of several search tools [BLAT (24), BLAST (25) or MegaBLAST (26)]. The primary uses of the function are: (i) anchoring of markers, ESTs or other survey sequences to the FPC map (as in silico markers or clone remarks); (ii) alignment of draft sequence against the BESs to aid in computing an MTP; (iii) aligning the sequence of a BAC against the BESs in order to confirm the ordering of the sequence contigs of the BAC; and (iv) aligning draft sequence to the BESs to be used by the DSI module.
The BSS was first described in ref. (21) and has since undergone a major revision in order to simplify its user interface while also augmenting functionality. A feature has been added for splitting the results by contig, which makes viewing the results easier. The filtering function for blast hits has been substantially upgraded, permitting iterative application of numerous filters and allowing complex filters to be constructed very easily, for example: add the alignment results to the map as in silico markers, but only those that have at least 75% of the marker aligned, align to at least two clones in a contig, and hit no more than five contigs. The upgraded BSS interface consists of two windows, where the first is a window to specify the parameters and execute the search, while the second window displays the output in a sortable table, and provides menus to filter and add results to FPC. These two simple windows and the filters cover the various possible needs of the applications enumerated above.
The DSI module
The BESs ‘anchor’ the draft sequence to the FPC map, as the BESs are either assembled with the sequence contigs or can be aligned to them, and the BESs are generated from the clones in the map. The input to the DSI module consists of ‘description’ files giving the draft sequence contig lengths, and ‘association’ files giving the locations of BESs on the draft sequence. If the sequenced contigs are ordered in supercontigs, the arrangement is specified in the description file. Note that the actual draft sequence is not necessary, as FPC only reads the description files. The association files may result from either assembling the draft with the BESs or aligning the BESs to the draft using the BSS module.
The alignment of draft sequence to the FPC map using BESs is complicated by several sources of error, including sequence error and repetitive sequence in both the BESs and draft, along with FPC contig assembly errors and even clone naming errors. In addition, paired BES evidence is not consistently available since (i) many clones do not have two BESs, (ii) shorter draft contigs may not contain two BESs from one clone and (iii) many BESs may have been masked and not included in the draft assembly. The alignment algorithm therefore should search for clusters of anchoring BESs, with detection thresholds adjustable by the user. A further requirement is that the algorithm should be able to resolve multiple alignment regions between a given draft and FPC contig; for example, a sequence may span an entire FPC contig with an alignment that appears valid on a large scale, but closer inspection may reveal that it is interrupted by an inversion or missing segment, indicating an assembly error. To meet these criteria, the alignment function uses a double sliding window algorithm which identifies both draft sequence and FPC contig regions of a window size w that have at least a minimum number m of BES anchors between them. The FPC window size is estimated based on the number of consensus bands times the average genomic distance per fingerprint band (a parameter which the user needs to estimate and provide). The window size w defaults to 250 kb, and the minimum anchor count m defaults to 5, where both are adjustable by the user.
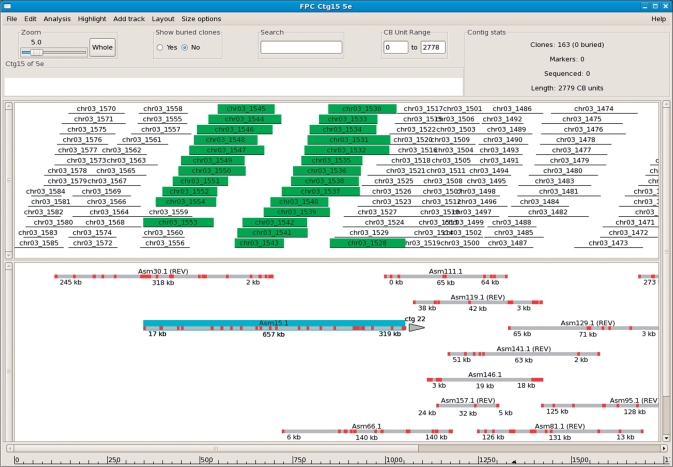
In order to view the alignment of the sequenced contigs to the FPC contigs, the track capability of the FPC contig display has been enhanced by the addition of a new ‘sequence’ track (Figure 1). There are now five types of tracks: markers, clones, remarks, framework markers and sequence. One or more tracks of any type can be created and placed anywhere on the display, and objects in a track with certain characteristics can be highlighted or hidden. In a sequence track, if a sequence contig also aligns to other FPC contigs, links are provided to the nearest aligning contigs left and right along the sequence, allowing the user to navigate easily among the contigs aligning to a given draft sequence (e.g. Asm15.1 in Figure 1 has an arrow on the right end with ‘ctg22’ above it indicating the link).
Figure 1.
DSI alignment within FPC. The top track shows the clones of FPC contig 15, while the bottom track shows the aligned draft sequence contigs. Draft contig Asm15.1 was clicked with the mouse causing it to highlight in blue, and the associated clones (those having BESs contained in Asm1.1) are also highlighted in the clone track. Alignments that have a reversed orientation are indicated by the notation ‘REV’, as seen on Asm30.1 at the left. The alignment of Asm15.1 illustrates detection of an assembly error, as described in the text.The draft contig lines are drawn to match the extent of the clones with which they share BES anchors; this makes small contigs appear larger than they actually are. Also, the alignment start/end labels reflect the locations of the first/last BES anchors, so they typically do not start exactly at 0 kb or end at the sequence endpoint.
A number of analysis functions have also been developed to assist the user in extracting full information from the sequence alignments. The most important of these is the MisAssembly function, which identifies probable assembly errors, either in the draft sequence contig or FPC contig. An assembly error is indicated when a draft sequence alignment terminates in the middle of an FPC contig, but not at the endpoint of the sequence; in other words, given correct assemblies and alignments, an alignment should not terminate until one of the contigs (draft or FPC) terminates.
It is important to realize that errors in the sequence and errors in the FPC contigs are indistinguishable on the basis of alignments alone. Once the error is identified, further analysis is needed to determine whether the problems lie in the FPC or the sequence contig. Typically, this analysis will consist of attempting to break the FPC contig apart at more stringent cutoff values and checking whether the fragmented contig aligns to the draft in a consistent way. If the inconsistency persists at stringent cutoffs, it is more likely to be a sequence problem. Additional data, e.g. genetic marker information, can also aid in determining the source of error.
Whereas the MisAssembly function determines where sequence and FPC contigs have been incorrectly joined, the data can also indicate where new joins should be made between sequence or FPC contigs. The functions Ctg-Joins and Seq-Joins scan for indications of possible FPC contig and draft sequence joins, respectively. The former is indicated by two FPC contigs aligning to adjacent (or overlapping) regions of the same sequence contig, while the latter is indicated by the opposite situation, in which the ends of two sequence contigs are adjacent along an FPC contig.
Three additional functions provide information to fill alignment gaps or locate smaller assembly errors. The Unaligned-FPC-Contigs function lists FPC contigs having no alignments under the current settings; these may correspond to gaps in the draft assembly. The Unaligned-Draft-Contigs function lists draft contigs having no alignments, which may correspond to gaps in the FPC map coverage. The Misplaced-BES function lists BESs which are located within aligning regions of the draft sequence, but whose clone is located elsewhere in the FPC map. These BESs may be misassembled into the sequence, or their clones may be incorrectly placed in FPC (or misnamed).
The last analysis component of the DSI module is an extension of the Ends→Ends function to optionally require sequence confirmation for merges (i.e. not only must clones near the ends of the contigs overlap based on fingerprints, but also the two contigs in question must align adjacently along the same draft sequence). Merges based on this strong dual evidence can then be performed quickly and automatically.
The MTP module
The FPC MTP selection module was first published in ref. (21), where it required draft sequence and BES, and used a two-step process as follows: (i) the draft sequence was first aligned to the BESs using the BSS function, and (ii) the MTP clones were selected using the overlap information provided by the pairs of BESs bridged by draft sequences. Two BESs bridged by a sequence contig, coupled with close proximity of the clones in the FPC map, is strong evidence of contiguity and produces a robust MTP. However, a complete MTP cannot generally be found in this way, at least at low draft coverage levels, since the BES coverage is sparse (e.g. approximately one every 7.5 kb for a 10× map), and few draft contigs are likely to span two BESs. The MTP function has therefore been extended to make use of fingerprint band overlap data in conjunction with the earlier BES-plus-draft overlaps, giving preference to the latter.
MTP selection based on fingerprints is not a trivial problem, as fingerprints contain considerable error and bands may randomly appear shared (i.e. have approximately the same length). Random shared bands are a major problem in agarose fingerprinting, e.g. the average number of shared bands between two random clones in the maize agarose map is 7.7 from an average 28 bands per clone, or a 28% random overlap (i.e. 28% of all bands between two random clones will match by chance). The HICF method generates 3–4 times as many bands as the standard agarose approach and also records the end base of each band, resulting in much more information per fingerprint. Moreover, the fraction of randomly shared bands is considerably lower than in agarose; e.g. in the maize HICF map, the average number of shared bands is 10.8 from an average 98 bands per clone, a random overlap of 11%. However, HICF also has more error, at least in comparison to agarose maps that have very consistent band calling (since agarose fingerprinting mainly relies on manual band-calling, the quality of fingerprints can vary greatly). In maize, for example, for which there is both an agarose map and an HICF map (4,12), duplicate fingerprints were generated and the reproducibility measured showing 89% reproducibility for agarose (unpublished data) and 75% for HICF (12). The sources of error in HICF fingerprints are not currently well characterized (9). In spite of the poorer reproducibility of HICF fingerprints, their greater information content and smaller random overlaps lead to superior contig assemblies and MTP selection (see Results section).
As a result of the error and uncertainty in the bands, the FPC clone coordinates are highly approximate and cannot be used by themselves to estimate clone overlaps, although we can assume that clones that are near each other in the map have a greater likelihood of overlapping than those that are distant. Hence, neighboring clones are evaluated using an approach that closely follows that typically used in manual MTP selection. The algorithm requires a certain minimum number of overlapping bands between a candidate MTP pair, and these bands are confirmed with a ‘spanner’ clone and two ‘flanking’ clones (Figure 2).
Figure 2.
An MTP clone pair, with confirming spanner and flankers. The tick marks represent bands and vertically aligned tick marks represent shared bands. All the bands are shown for candidate MTP clones CL and CR. Only the shared bands are shown for the spanner and flanker. Three bands are not confirmed by either the spanner or the flanker clones.
The core MTP algorithm is as described in ref. (21) with modifications to incorporate pairs of overlapping clones based on fingerprints. All candidate MTP pairs are first computed and assigned a pair overlap score, and then the algorithm selects the spanning set of pairs having smallest total overlap score. Candidate fingerprint MTP pairs are selected as follows: every pair of clones (CL, CR) within a user-specified distance from each other on an FPC contig is examined, and for each pair a spanner and two flanking clones are selected which best account for all bands in the pair. An overlap score is computed for each pair which factors in both the measured overlap (based on shared bands) and the quality of confirmation by the spanner and flankers; pairs with poor confirmation (many bands not matched) are more likely to be false-positives, i.e. not genuinely overlapping. The score uses the following variables: M is the number of matching bands between the pair; S is the number of bands in the spanner clone that do not match bands in either CL or CR; U is the number of bands in CL and CR not found in either the spanner or two flanker clones; and the variables WM, WS, WU are the corresponding weight factors (penalities). The score for clone pair (C1, C2) is then
where the weight factors WM, WS, WU have been set to 1, 10, 2, respectively, after extensive testing in simulations.
FPC parallelization
The Build (assembly) algorithm has two parts, both of which have been parallelized. First is the N × N comparison of the fingerprints of all clones to cluster them into contigs. This uses the POSIX ‘pthread’ library so that the threads share a common memory. The algorithm uses a sparse matrix where each row represents a clone and each node of the matrix represents a valid overlap. Each thread receives a preassigned set of clones to be analyzed, and all threads use a shared pool of preallocated nodes. The second part of the algorithm orders the clones within contigs, which uses a greedy algorithm since the optimal ordering problem has been shown to be NP-hard (27). Since the greedy algorithm can get into a local minimum (i.e. it cannot be guaranteed that it is the best solution, and can sometimes be a bad solution), it is executed T times with a different seed clone each time, and the best solution is used. The T executions are run in parallel, which is implemented using the Unix ‘fork’ function, where a fork creates a ‘thread’ that is a distinct Unix process with its own copy of the memory.
Since the parallelization uses standard Unix functions, it does not require the installation of special packages to compile or execute the parallel FPC. The user simply launches FPC with the appropriate flag and the desired level of parallelization, e.g. ‘fpc -p 4’, and it will execute in parallel the main assembly algorithms just described, as well as several others which also require large numbers of fingerprint comparisons. Specifically, the Build, Incremental Build Contigs, DQer, ReBuild, Ends→Ends and KeySet→FPC all execute in parallel.
The most important function in FPC besides the Build is the Ends→Ends merge-detection function, since it is used extensively in the iterative ‘step-down method’ for assembly of HICF fingerprints (12). The reason that HICF assembly requires a different strategy than agarose assembly is due to the higher level of error in HICF fingerprints. Agarose fingerprints can be assembled at a relatively low cutoff and then the DQer functions can be run to break apart false joins based on their content of ‘Q clones’ (clones that have a questionable alignment in the contig). This approach does not work well with HICF since the high rate of error in HICF fingerprints causes many correctly assembled clones to be marked as Q clones. Consequently, HICF requires an almost opposite approach of assembling at a stringent cutoff and then gradually merging contigs using the Ends→Ends function, with the intent of avoiding false joins. The Ends→Ends function is parallelized using the Unix fork function, where each thread performs the computations for a subset of the contig pairs (divided so as to equalize the number of overlap calculations between the clones at the ends of contigs) and transmits the result to the master process.
Simulations
In order to develop and test the MTP and DSI modules, simulation software that was originally developed for comparing fingerprinting methods (28) was extended to include MTP functions and simulated draft assemblies. The software takes as input a long sequence (e.g. a sequenced chromosome) and generates simulated FPC project data from it through several steps. First, a clone library of specified coverage and size range is created by randomly selecting pairs of EcoRI restriction sites. BESs are also generated for the clones, using a distribution of lengths derived from laboratory data in Oryza subspecies (29). Second, fingerprints are generated for each clone using in-silico digestion for the fingerprint method in question [in the present study, either agarose or SNaPshot HICF (11)]. Third, error is added to the fingerprints, both by adding Gaussian random values to the bands and by removing a given percentage of bands (12.5% for HICF, 6% for agarose) and replacing them with random bands. We note that, although it is not currently possible to exactly simulate the sizing of HICF fragments by automated sequencing machines (12), realistic fingerprints can still be generated, as the exact values of the fragment sizes are not important for assembly. Fourth, the fingerprints are assembled using a fixed cutoff of 1e-12 for agarose and 1e-40 for HICF. Fifth, simulated draft read sets of various coverage levels are generated from the original input sequence (the simulated reads are 800 bp segments chosen randomly from the source sequence). For MTP testing, the BSS is used to align the BESs to the draft, where the alignments are used to select the MTP. For the DSI testing, PCAP (30) is used to assemble the simulated reads plus BES, and the results are aligned to the FPC contigs. Simulations used rice (Oryza sativa) chromosome 3 (36.1 Mb), fly (Drosophila melanogaster) chromosome 3L (23.8 Mb), and human chromosome 21 (35.4 Mb), all of which were downloaded from Genbank.
RESULTS
Draft sequence integration
In order to verify and illustrate the DSI functions, simulated 7× draft reads, BESs and 10× HICF FPC assembly were generated from the O. Sativa chromosome 3 (36.1 Mb), as described in Methods section. These coverage levels were chosen as illustrative of typical sequencing and mapping projects; smaller coverages would result in smaller FPC and draft contigs, with correspondingly less integration. The exact amount of integration to be expected depends on the length of the draft contigs and the density of FPC-associated BES embedded in those contigs; the latter in turn depends upon the clone coverage, the success rate of the fingerprinting and BES sequencing, and the fraction of BES which are masked or rejected or other reasons during the draft assembly process.
The 7× plus BES assembly resulted in 233 contigs, spanning 34.1 Mb with a maximum contig size of 850 kb and L50 value 92 kb. The FPC assembly resulted in 55 contigs covering essentially the full rice chromosome, along with three singleton clones.
Using default settings, 156 (67%) of the draft contigs could be aligned to the FPC map. The total aligned sequence length (taking into account alignments that did not span the whole length of a draft contig) was 30.1 Mb, or 90% of the total draft assembly length. The average size of an aligning draft contig was 156 kb. The alignments to one of the FPC contigs are shown in Figure 1.
The default alignment settings require five BES anchors within a 250 kb region along both the sequenced contig and FPC contig. Errors in the assembly, either from the sequenced or the FPC contigs, can therefore be detected only if they give rise to a cluster of five or more misplaced BESs. Inspection of the PCAP assembly output files found four such groupings of errors, all of which were detected by the DSI MisAssembly function (which gave no false-positives); three other erroneous sequence regions involving five or more misplaced BESs were not detected because the BESs were not clustered on the FPC side (i.e. the misplaced BESs were drawn from different contig regions). One detected error is illustrated in Figure 1, where Asm15.1 partially aligns to FPC contig 15. Asm15.1 has total length 657 kb, but the alignment to contig 15 covers only the portion from 7 kb to 319 kb; the remainder of the sequence aligns to contigs 22. Since the alignment breaks in the middle of contig 15, there is an assembly error on one side or the other; in this case, it is the draft contig which has an incorrect join at this point.
The alignments provide valuable information to order and orient the draft contigs. The orderings suggested by the midpoints and by the left ends of the aligned regions were compared, with the midpoints found to provide more reliable information, giving the true ordering of all the aligned draft contigs except for three small contigs (19–42 kb) that were mis-orderd. The ordering of sequential contigs that are much smaller than a clone length is difficult to determine since the only information known is the clones which are hit.
The DSI module also estimates the orientation of each draft contig relative to the FPC contig by computing the Pearson correlation function using the coordinate pairs of the individual BES anchors along both draft and FPC contig. Since the 5′–3′ orientation of FPC clones is not known, the FPC coordinates of a BES are taken to be the midpoint of the associated clone; this ambiguity, along with error in clone ordering within a contig, diminishes the accuracy of the orientation determination. Of the 156 aligned sequences, the orientation was computed correctly for 141 (90%). Greater certainty is possible for longer draft contigs, as the 41 aligned contigs greater than 200 kb were all oriented correctly.
Minimal tiling path
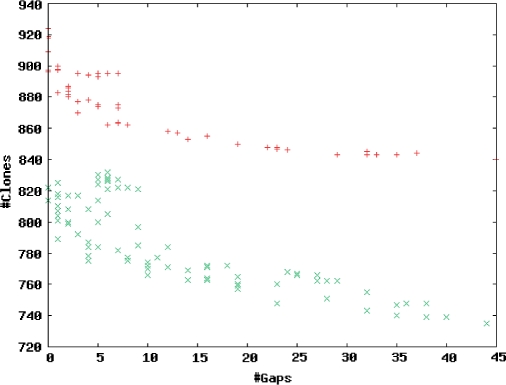
Figure 3 depicts the range of results achievable from the MTP function with different choices of the parameters MinOlap (the minimum amount that the two clones must overlap in FPC coordinates) and MinShared (the minimum number of shared bands), shown for both HICF and agarose. As can be seen, the number of gaps in the MTP has an inverse relationship to the number of clones selected. Although some parameter choices are less optimal than others (producing more clones for a given gap count), the difference is not great except for gap counts below 10, indicating that to remove all gaps is not a realistic goal. The gaps counted are only the false-positive overlapping pairs, i.e. clone pairs picked as overlapping by the MTP software, but not actually overlapping in reality; additional gaps are likely to exist between the FPC contigs.
Figure 3.
Effect of different parameters and fingerprint error levels on MTP performance for agarose and HICF. Aggregated MTP clone count and gap count are plotted from simulations of rice chr3, human chr21 and fly chr3L (total genomic sequence 95.2 Mb) for 10× clone coverage for HICF (‘×’) and agarose (‘+’). For HICF, the FPC MinOlap parameter was varied from 0 to 10 and the MinShared parameter was varied from 0 to 30, where MinOlap is the minimal amount of overlap between two clones based on FPC coordinates and MinShared is the minimal number of shared bands. For agarose, MinOlap varied from −6 to 10 and MinShared varied from 0 to 12.
The results in Figure 3 guided the choice of default parameters for the MTP function. For HICF, these correspond to the lowest data point having 10 gaps, namely MinOlap = 6 and MinShared = 14; for agarose, the lowest data point achieving 14 gaps was used, generating defaults MinOlap = 0 and MinShared = 6. These relatively conservative settings reflect the large cost of filling unexpected gaps, as compared to the steadily decreasing cost of sequencing the tiling BACs themselves. Figure 3 also shows that HICF fingerprinting leads to ∼10% more efficient MTP selection relative to agarose, i.e. 10% fewer MTP clones for the same number of gaps. When gaps between contigs are factored in, the advantage of HICF grows, since HICF assemblies also generate fewer (and larger) contigs.
Table 1 shows in more detail the results which can be anticipated from the MTP algorithm, using fingerprints only and default parameters. In simulations based on human chromosome 21, rice chromosome 3 and fly chromosome 3L (see Methods section for details), the average MTP overlap for HICF at 10× coverage varied from 29 to 36 kb, and at 20× from 23 to 28 kb. The number of gaps between MTP pairs (i.e. false-positive overlaps) was between 1% and 3% of MTP clone pairs. MTPs selected using agarose FPC maps have significantly higher overlaps on average, ∼51–53 kb for 10× and 34–37 kb for 20×, while having comparable gap percentages. As the coverage increases, the amount of overlap decreases because there is a larger pool of clone pairs to choose from; however, a small overlap increases the chance of a gap, so the number of gaps can actually increase with coverage. Table 2 shows sample results from adding draft sequence at coverage levels 1×, 2×, 4× and 7×. With increasing coverage level, a steady decrease is seen in the average overlap length, along with a steady increase in the number BSS pairs (i.e. MTP pairs whose overlap was determined through alignment to a draft sequence). The BSS pairs have very small average overlaps, especially at lower coverage levels (the average overlap of BSS pairs grows with coverage simply because the draft contig length grows, allowing clone pairs with more overlap to be found as BSS pairs). The smaller overlaps of BSS pairs, however, are somewhat offset by the increasing average overlap of the fingerprint pairs. This occurs because the gaps between BSS pairs must be filled in with fingerprint pairs, but the arbitrary placement of the gaps generally will not permit the fingerprint overlaps to be optimized to the extent possible when picking an entire MTP of fingerprint pairs.
Table 1.
MTP performance tested in simulations using only fingerprint data (see Methods section)
| Method | Speciesa | Clone coverage (×) | No. of contigs | No. of MTP clones | Average overlap (kb)b | No. of gapsc | MTP coveraged (%) |
|---|---|---|---|---|---|---|---|
| HICF | human21 | 10 | 37 | 263 | 30 | 1 | 95 |
| HICF | human21 | 20 | 14 | 265 | 28 | 5 | 98 |
| Agarose | human21 | 10 | 136 | 315 | 51 | 7 | 95 |
| Agarose | human21 | 20 | 55 | 286 | 36 | 10 | 97 |
| HICF | rice3 | 10 | 55 | 277 | 29 | 6 | 98 |
| HICF | rice3 | 20 | 11 | 262 | 23 | 6 | 99 |
| Agarose | rice3 | 10 | 159 | 327 | 52 | 3 | 97 |
| Agarose | rice3 | 20 | 68 | 292 | 34 | 8 | 98 |
| HICF | fly3L | 10 | 35 | 192 | 36 | 2 | 97 |
| HICF | fly3L | 20 | 8 | 177 | 26 | 2 | 99 |
| Agarose | fly3L | 10 | 99 | 211 | 53 | 4 | 95 |
| Agarose | fly3L | 20 | 45 | 196 | 37 | 4 | 98 |
| Gapse (%) | |||||||
| HICF | Average | 10 | - | - | 31 | 1.2 | - |
| HICF | Average | 20 | - | - | 20 | 1.8 | - |
| Agarose | Average | 10 | - | - | 52 | 1.6 | - |
| Agarose | Average | 20 | - | - | 36 | 2.8 | - |
aHuman chr21 is 35.4 Mb, rice chr3 is 36.1 Mb and fly chr3L is 23.8 Mb.
bAverage overlap of the clone pairs selected for the MTP.
cNumber of gaps between clone pairs selected for the MTP, i.e. false-positive overlaps.
dMTP coverage of the genomic sequence.
eGap percentage is the number of gaps divided by the number of MTP clone pairs.
Table 2.
MTP performance with simulated draft sequence and fingerprints
| Coverage | Draft coverage (×) | No. of MTP clones | No. of Gapsa | Average overlap (kb)b | Average FP overlap (kb)c | Average BSS overlap (kb)d | No. of FP pairs | No. of BSS pairs |
|---|---|---|---|---|---|---|---|---|
| HICF (×) | ||||||||
| 10 | 0 | 263 | 1 | 30 | 30 | 0.0 | 226 | 0 |
| 10 | 1 | 265 | 0 | 28 | 35 | 0.4 | 184 | 44 |
| 10 | 2 | 264 | 1 | 28 | 36 | 1.0 | 167 | 60 |
| 10 | 4 | 261 | 0 | 26 | 42 | 7.7 | 104 | 120 |
| 10 | 7 | 258 | 0 | 20 | 55 | 17.0 | 18 | 203 |
| 20 | 0 | 263 | 5 | 28 | 28 | 0.0 | 248 | 0 |
| 20 | 1 | 273 | 1 | 26 | 38 | 0.6 | 169 | 89 |
| 20 | 2 | 272 | 2 | 25 | 42 | 1.5 | 144 | 113 |
| 20 | 4 | 271 | 1 | 22 | 47 | 8.2 | 84 | 172 |
| 20 | 7 | 259 | 0 | 15 | 71 | 13.3 | 10 | 234 |
| Agarose (×) | ||||||||
| 10 | 0 | 315 | 7 | 51 | 50 | 0.0 | 179 | 0 |
| 10 | 1 | 320 | 7 | 47 | 58 | 0.3 | 151 | 33 |
| 10 | 2 | 319 | 3 | 44 | 62 | 1.3 | 127 | 56 |
| 10 | 4 | 317 | 0 | 41 | 71 | 6.2 | 91 | 90 |
| 10 | 7 | 318 | 0 | 38 | 104 | 13.5 | 44 | 138 |
| 20 | 0 | 286 | 10 | 36 | 36 | 0.0 | 231 | 0 |
| 20 | 1 | 279 | 5 | 28 | 41 | 0.4 | 150 | 74 |
| 20 | 2 | 282 | 1 | 26 | 48 | 2.3 | 115 | 112 |
| 20 | 4 | 274 | 0 | 24 | 61 | 6.9 | 62 | 157 |
| 20 | 7 | 270 | 1 | 21 | 113 | 11.5 | 16 | 199 |
The simulation used human chr21 sequence and draft sequence coverages from 0×–7×; see Methods section for simulation details. The last four columns show the numbers and average overlaps of the MTP clone pairs selected based on fingerprint and sequence data; see Results section for further discussion.
aNumber of gaps between clone pairs selected for the MTP, i.e. false-positive overlaps.
bAverage overlap for both FP and BSS clone pairs.
cAverage overlap between clone pairs selected from fingerprint overlaps.
dAverage overlap between clone pairs selected from sequence overlaps, i.e. identified using the BSS routine with draft sequence and BESs.
FPC parallelization
To time the speedup of the parallelization of the assembly algorithm, a dataset was created from 50 832 HICF maize fingerprints, which assembled into 190 contigs. The parameters were gel length 25 500, tolerance 7, cutoff 1e-50 and 50 tries of the clone ordering algorithm. The option to precompute the Sulston score was used, as for the HICF data it speeds up the N × N comparison of the serial assembly from 4 h 29 min to 1 h 45 min (note that this does not benefit agarose, which uses an alternative optimization that works well when there are less than 60 bands).
Timing tests for the parallelized assembly functions were carried out and the results are shown in Table 3. All tests were run on a Dell Poweredge 6650 4-processor 2.8 GHz machine with 5 GB of memory. The overall speedup on 4 processors was 3.5× for the N × N comparison and 3× for the clone ordering. A speedup of 4× on 4 processors is not possible in practice as the startup cost and serial portions of the code prevent it (this well-known limitation is referred to as ‘Amdahl's law’). Moreover, the clone ordering step does not execute in parallel for small contigs as it is not worth the overhead. Regardless, the 3× decrease in time from 3 h 38 min to 1 h 16 min is significant. As the clone ordering is twice as time-consuming as the N × N comparisons, it has proved highly advantageous to have both parts of the algorithm parallelized, resulting in an overall 3.2× speedup.
Table 3.
Results of timing experiments on the FPC assembly and Ends→Ends algorithms
| Processors |
||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Build assembly algorithm | ||||
| N × N comparison | 1 h 45 min | 0 h 57 min | 0 h 38 min | 0 h 30 min |
| Speedup | 1 | 1.8 | 2.76 | 3.5 |
| Clone ordering | 3 h 48 min | 2 h 9 min | 1 h 34 min | 1 h 16 min |
| Speedup | 1 | 1.76 | 2.4 | 3 |
| Total time | 5 h 33 min | 3 h 6 min | 2 h 12 min | 1 h 46 min |
| Speedup | 1 | 1.8 | 2.5 | 3.2 |
| Ends→Ends algorithm | ||||
| Comparison | 2 h 51 min | 1 h 26 min | 58 min | 0 h 44 min |
| Speedup | 1 | 1.9 | 2.9 | 3.9 |
Times are in hours (h) and minutes (min). The speedup is in comparison to using one processor.
To time the Ends→Ends function, the full maize database of 350 253 clones was assembled at 1e-57 resulting in 3951 contigs. The Ends→Ends was run with a 1e-25 cutoff, which merged 2298 contigs. The execution time was 2 h 51 min for 1 processor and 43 min for 4 processors, resulting in a speedup of 3.9×.
DISCUSSION
FPC has been used for over a decade for building physical maps. Its success stems in part from the fact that it has been constantly upgraded to keep current with the latest technologies and to integrate multiple types of data. FPC was first developed at the Sanger Center as a second generation program to replace Contig6 (20), which was used for building the physical map of Caenorhabditis elegans (31). The initial maps built with FPC used the Image program (32) for detecting the fingerprinted bands. FPC V4.2 had enhancements to use markers in its assembly, framework markers for ordering and anchoring contigs, and to perform incremental assembly (2). FPC V4.2 was used for building the agarose based physical map of human (33) from which the human genome was sequenced using a BAC-by-BAC approach (34). Subsequent sequencing projects of large genomes began using a hybrid of BAC-by-BAC and whole-genome shotgun, e.g. the mouse (6) and rat (35). To support this hybrid approach, FPC V6-V7 (21) contained the first versions of the BSS and MTP modules, which facilitate using sequence in combination with markers and fingerprinted clones; these modules have now been upgraded as described above. In addition, the new DSI module has been developed for the integration of draft sequenced contigs with the map. FPC has also kept current with the need for high-throughput fingerprinting with complete support for HICF (12) and multi-processor computations for time-intensive algorithms.
The DSI module, developed in conjunction with the soybean DOE-JGI draft sequencing project (www.phytozome.org/soybean.php; manuscript in preparation), comprises functions tailored to the needs of map-assisted draft sequencing. The most important of these are the sequence alignment and mis-assembly detection functions. The full-featured graphical display of the results is also essential for the more detailed analysis that is often required to determine whether a given error lies in the draft sequence or the FPC map. Both draft sequence and FPC map are subject to errors, but the information used in their respective assemblies is so different that errors are often complementary, enabling each to correct the other; for example, the soybean FPC map resolves ordering difficulties in the repeat-laden pericentromeric regions, where sequencing has proved difficult (Steven Cannon, personal communication). The soybean sequence, in turn, revealed a number of problems in the FPC map. Aside from error correction, the immediate graphical feedback on the regions of agreement is also helpful in providing confidence in the assembly.
The MTP function is a valuable adjunct to either BAC-by-BAC or pooled-BAC sequencing efforts, for part or all of a genome. Since the MTP algorithm not only emulates the confirmation process used by a skilled human (i.e. verification with spanner and flankers), but also tries every possible combination of spanners and flankers, and every possible tiling path through the contig, there is little reason to expect that manual selection (using fingerprint data alone) could achieve a better result. As sequencing decreases in cost relative to human effort, the value of utilizing automation wherever possible during production increases correspondingly. The declining cost of sequencing also influenced the more conservative default settings in FPC V9.3, which generates far fewer gaps than the previous settings, at the cost of higher clone overlaps.
Though the N × N comparison function of the assembly algorithm was parallelized for distributed machines by Ness et al. (23), we have parallelized both parts of the algorithm for now-standard multi-processor machines. As the clone layout function of the assembly algorithm takes twice as much time as the N × N comparison, it was important that it be parallelized for maximum efficiency. Additionally, the function that compares the end clones from all contigs to identify possible merges was parallelized. Not only are 4 processor machines relatively inexpensive so that a greater than 3× speedup can be obtained without needing an expensive high-performance computer, but the current FPC implementation requires no special software or assembly.
An additional small, but very useful new feature of FPC is the chimeric fingerprint detection function. FPC maps are very vulnerable to chimeric fingerprints (12,36), i.e. fingerprints compromised by well-to-well contamination within plates. Such fingerprints are very likely to cause contig assembly errors, which are quite difficult to correct after the fact; therefore, it is best to eliminate chimerics to the extent possible before contig assembly. A function has been added to FPC to detect clones from neighboring wells that have similar fingerprints [an equivalent function is found in the Genoprofiler package (36)]. The function can be found in the Search Commands list for clones, on the main FPC window.
Not only has FPC kept abreast of current technologies, it also offers a complete environment for building and studying fingerprint physical maps, so the user does not have to turn to separate programs for further editing and analysis needs. The core algorithms included in FPC are those for assembly and ordering of the clones, selecting the MTP, aligning sequence to the map and detecting assembly problems. However, since error in the data prevents fully automated resolution of all problems, FPC goes beyond core algorithms to provide a complete suite of manual editing features. In order to visualize the relations between thousands of entities in the map, FPC provides an excellent graphical display where the user has considerable flexibility in viewing the data. In addition, in order to make the map easily accessible to the community, web-based display tools have been developed (37). It is this full-featured, evolving support which has made FPC indispensable to physical mapping over its 10 year history. The FPC V9.3 software, tutorials and documentation are available at www.agcol.arizona.edu/software/fpc.
FUNDING
National Science Foundation (0501877, 0213764). Funding for open access charge: University of Arizona.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank all the FPC users who have provided us with feedback over the years.
REFERENCES
- 1.Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- 2.Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soderlund C, Longden I, Mott R. FPC: a system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- 4.Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, Hurwitz B, Yu Y, Collura K, Gill N, Sanmiguel P, Mullikin JC, Maher C, Nelson W, Wissotski M, et al. Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 2008;9:R45. doi: 10.1186/gb-2008-9-2-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory SG, Sekhon M, Schein J, Zhao S, Osoegawa K, Scott CE, Evans RS, Burridge PW, Cox TV, Fox CA, et al. A physical map of the mouse genome. Nature. 2002;418:743–750. doi: 10.1038/nature00957. [DOI] [PubMed] [Google Scholar]
- 7.Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas JW, Prasad AB, Summers TJ, Lee-Lin SQ, Maduro VV, Idol JR, Ryan JF, Thomas PJ, McDowell JC, Green ED. Parallel construction of orthologous sequence-ready clone contig maps in multiple species. Genome Res. 2002;12:1277–1285. doi: 10.1101/gr.283202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y, Johnson MD, Chen WQ, Wong D, Chen YJ, Benson SC, Lam JY, Kim YM, Shizuya H. Five-color-based high-information-content fingerprinting of bacterial artificial chromosome clones using type IIS restriction endonucleases. Genomics. 2001;74:142–154. doi: 10.1006/geno.2001.6547. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, Johnson MD, Colayco R, Chen YJ, Melnyk J, Schmitt H, Shizuya H. Contig assembly of bacterial artificial chromosome clones through multiplexed fluorescence-labeled fingerprinting. Genomics. 1999;56:237–246. doi: 10.1006/geno.1998.5734. [DOI] [PubMed] [Google Scholar]
- 11.Luo MC, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OD, Dvorak J. High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics. 2003;82:378–389. doi: 10.1016/s0888-7543(03)00128-9. [DOI] [PubMed] [Google Scholar]
- 12.Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, Kim H, Wing RA, Messing J, Soderlund C. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 2005;139:27–38. doi: 10.1104/pp.105.061978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoemaker RC, Grant D, Olson T, Warren WC, Wing R, Yu Y, Kim H, Cregan P, Joseph B, Futrell-Griggs M, et al. Microsatellite discovery from BAC end sequences and genetic mapping to anchor the soybean physical and genetic maps. Genome. 2008;51:294–302. doi: 10.1139/G08-010. [DOI] [PubMed] [Google Scholar]
- 14.Cannon SB, Crow JA, Heuer ML, Wang X, Cannon EK, Dwan C, Lamblin AF, Vasdewani J, Mudge J, Cook A, et al. Databases and information integration for the Medicago truncatula genome and transcriptome. Plant Physiol. 2005;138:38–46. doi: 10.1104/pp.104.059204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 16.Garvin DF, Gu Y-Q, Hasterok R, Hazen SP, Jenkins G, Mockler TC, Mur LAJ, Vogel JP. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 2008;48:S69–S84. [Google Scholar]
- 17.Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007;145:1303–1310. doi: 10.1104/pp.107.107672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren RL, Varabei D, Platt D, Huang X, Messina D, Yang SP, Kronstad JW, Krzywinski M, Warren WC, Wallis JW, et al. Physical map-assisted whole-genome shotgun sequence assemblies. Genome Res. 2006;16:768–775. doi: 10.1101/gr.5090606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havlak P, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Weinstock GM, Gibbs RA. The Atlas genome assembly system. Genome Res. 2004;14:721–732. doi: 10.1101/gr.2264004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulston J, Mallett F, Staden R, Durbin R, Horsnell T, Coulson A. Software for genome mapping by fingerprinting techniques. Comput. Appl. Biosci. 1988;4:125–132. doi: 10.1093/bioinformatics/4.1.125. [DOI] [PubMed] [Google Scholar]
- 21.Engler FW, Hatfield J, Nelson W, Soderlund CA. Locating sequence on FPC maps and selecting a minimal tiling path. Genome Res. 2003;13:2152–2163. doi: 10.1101/gr.1068603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 23.Ness SR, Terpstra W, Krzywinski M, Marra MA, Jones SJ. Assembly of fingerprint contigs: parallelized FPC. Bioinformatics. 2002;18:484–485. doi: 10.1093/bioinformatics/18.3.484. [DOI] [PubMed] [Google Scholar]
- 24.Kent WJ, Haussler D. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 2001;11:1541–1548. doi: 10.1101/gr.183201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 27.Alizadeh F, Karp RM, Weisser DK, Zweig G. Physical mapping of chromosomes using unique probes. J. Comput. Biol. 1995;2:159–184. doi: 10.1089/cmb.1995.2.159. [DOI] [PubMed] [Google Scholar]
- 28.Nelson WM, Dvorak J, Luo MC, Messing J, Wing RA, Soderlund C. Efficacy of clone fingerprinting methodologies. Genomics. 2007;89:160–165. doi: 10.1016/j.ygeno.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Wing RA, Ammiraju JS, Luo M, Kim H, Yu Y, Kudrna D, Goicoechea JL, Wang W, Nelson W, Rao K, et al. The oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Wang J, Aluru S, Yang SP, Hillier L. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coulson A, Sulston J, Brenner S, Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1986;83:7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulston J, Mallett F, Durbin R, Horsnell T. Image analysis of restriction enzyme fingerprint autoradiograms. Comput. Appl. Biosci. 1989;5:101–106. doi: 10.1093/bioinformatics/5.2.101. [DOI] [PubMed] [Google Scholar]
- 33.Consortium IHGM. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 34.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 35.Chen R, Sodergren E, Weinstock GM, Gibbs RA. Dynamic building of a BAC clone tiling path for the Rat Genome Sequencing Project. Genome Res. 2004;14:679–684. doi: 10.1101/gr.2171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You FM, Luo MC, Gu YQ, Lazo GR, Deal K, Dvorak J, Anderson OD. GenoProfiler: batch processing of high-throughput capillary fingerprinting data. Bioinformatics. 2007;23:240–242. doi: 10.1093/bioinformatics/btl494. [DOI] [PubMed] [Google Scholar]
- 37.Pampanwar V, Engler F, Hatfield J, Blundy S, Gupta G, Soderlund C. FPC Web tools for rice, maize, and distribution. Plant Physiol. 2005;138:116–126. doi: 10.1104/pp.104.056291. [DOI] [PMC free article] [PubMed] [Google Scholar]