Abstract
TopBP1, acting in concert with DNA containing bulky base lesions, stimulates ATR kinase activity under physiologically relevant reaction conditions. Here, we analyze the roles of the three components in ATR activation: DNA, base damage and TopBP1. We show that base adducts caused by a potent carcinogen, benzo[a]pyrene diol epoxide (BPDE), constitute a strong signal for TopBP1-dependent ATR kinase activity on Chk1 and p53. We find that the C-terminus of TopBP1 binds preferentially to damaged DNA and is sufficient to mediate damaged DNA-dependent ATR activation in a manner similar to full-length TopBP1. Significantly, we find that stimulation of ATR by BPDE-damaged DNA exhibits strong dependence on the length of DNA, with essentially no stimulation with fragments of 0.2 kb and reaching maximum stimulation with 2 kb fragments. Moreover, TopBP1 shows preferential binding to longer DNA fragments and, in contrast to previous biochemical studies, TopBP1 binding is completely independent of DNA ends. We find that TopBP1 binds to circular and linear DNAs with comparable affinities and that these DNA forms elicit the same level of TopBP1-dependent ATR activation. Taken together, these findings suggest a cooperative activation mechanism for the ATR checkpoint kinase by TopBP1 and damaged DNA.
INTRODUCTION
DNA damage checkpoints are signal transduction pathways that delay or arrest cell cycle progression in response to DNA damage or inhibition of replication. Checkpoints aid in maintaining genomic integrity and cell survival in unicellular organisms and are known or presumed to prevent genomic instability, cancer and death in multicellular organisms. ATM and ATR are members of the phosphoinositide 3-kinase-related protein kinase (PIKK) family of protein kinases that function in the early stages of checkpoint signaling pathways. In general, the checkpoint response to double-strand breaks is initiated by ATM, whereas the checkpoint response to base adducts and inhibition of replication is induced by ATR (1–3). It is commonly accepted that single-stranded DNA resulting from uncoupling of the replicative helicase and DNA polymerase because of base lesions or dNTP depletion during the S-phase (4), from processing of double-strand breaks (5,6), or from damage removal in the form of ∼30-nt long oligomers in the G1 and G2 phases (7–9) constitutes the signal for the ATR-mediated DNA damage checkpoint response. While there is strong evidence that single-stranded DNA coated with replication protein A (RPA) is a signal for ATR activation (10,11), there are also in vivo and in vitro data indicating that the base lesion itself acts as a signal for the ATR-mediated DNA damage checkpoint response (12–14).
The development of partially reconstituted checkpoint systems with purified proteins has been instrumental in obtaining mechanistic details about these important signal transduction pathways (15–18). Using a minimal in vitro system, Dunphy and coworkers (15) made the important discovery that the topoisomerase IIβ binding protein 1 (TopBP1) is an essential co-activator of ATR. Subsequently, we demonstrated that under more physiologically relevant reaction conditions the TopBP1-dependent ATR kinase activity on the Chk1 signal transduction kinase was strongly stimulated by DNA, in particular DNA containing bulky base adducts induced by the model carcinogen N-acetoxy-2-acetylaminofluorene (N-Aco-AAF) (14). In the current study, we investigate whether other bulky base lesions act as checkpoint signals similarly to AAF-guanine adducts, and identify that the C-terminus of TopBP1 is sufficient for damaged DNA- and TopBP1-dependent stimulation of ATR kinase activity on Chk1 as well as p53 substrates. Finally, we present evidence that cooperative binding of TopBP1 to DNA may be essential for its function as the ATR co-activator. Collectively, our data support the view that TopBP1 is capable of recognizing unprocessed bulky DNA lesions, recruiting ATR to the damage site, and activating ATR by a cooperative mechanism.
MATERIALS AND METHODS
Antibodies and purification of checkpoint proteins
Chk1 phospho-S345 (#2348) and p53 phospho-S15 antibodies (#9248) were purchased from Cell Signaling Technology (Danvers, MA, USA), and Chk1 (sc-8408) and p53 (sc-6243) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Native ATR, which contains ATRIP but is free of other checkpoint proteins and other PIKK family members, was purified from HeLa cell-free extracts as previously described (14). GST-TopBP1-His, GST-TopBP1 fragments, GST-p53 and His-Chk1 kinase dead (Chk1-kd) were all purified by standard procedures as previously described (14).
Preparation of DNA substrates
For benzo[a]pyrene diol epoxide (BPDE)-damaged DNA, pUC19 plasmid (50 μg/ml) was treated with 50 μM BPDE (NCI Chemical Carcinogen Reference Standard Repository, Midwest Research Institute, Kansas, MO, USA) in 10 mM Tris–HCl, pH 7.5 and 1 mM EDTA at 37°C for 16 h in the dark. The reaction was followed by ether extraction and ethanol precipitation to remove the nonreacted excess BPDE. This treatment produces ∼20 adducts/plasmid as previously reported (19). To generate various sizes of DNA fragments, PCR was performed with pUC19 plasmid as a template.
DNA-binding assays
Assays were carried out as described previously (14). Briefly, DNA substrates were 5′-end labeled with [γ-32P]ATP and then mock-treated or treated with BPDE as described above. For circular DNA, labeled DNA was ligated and purified by agarose gel electrophoresis. Purified proteins (3 pmol) on glutathione beads were incubated at 37°C for 10 min with the DNA (0.6 fmol or 1 ng) in buffer B (10 mM Tris–Cl, pH 7.7, 1 mM EDTA, 0.5% NP 40) containing 0-300 mM NaCl. For DNA-binding assays in kinase reaction buffer, comparable amounts of TopBP1 and DNA to those used in kinase assays were used. After the incubation, the beads were washed three times with buffer B, and bound DNA was eluted by incubation with 0.1 μg/μl proteinase K at 37°C for 15 min. The DNA was resolved in a 0.8% agarose gel, dried, visualized by autoradiography and quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA) and ImageQuant 5.2 software. The averages from independent experiments were graphed, and the error bars indicate the standard deviation of the mean.
Kinase assays
The procedure was essentially as previously described (14). Briefly, kinase assay reactions contained 14 mM HEPES, pH 7.9, 3 mM MgCl2, 1 mM ATP, 0.5 mM DTT, 5% glycerol, 1% polyethylene glycol (6000), 35 mM KCl, 50 mM NaCl and 1 μM microcystin in 10 μl final volume. For low-ionic strength conditions, the concentration of NaCl was reduced to 10 mM. Purified ATR (0.4 nM) was preincubated in the reaction buffer for 15 min at 30°C with the indicated amounts of recombinant full-length TopBP1 or TopBP1 fragments and with various DNA substrates where indicated. After the preincubation, 10 nM Chk1-kd, or p53 where indicated, was added into the reaction, incubated for 20 min at 30°C, terminated by the addition of SDS–PAGE loading buffer, and separated by SDS–PAGE. Chk1 or p53 phosphorylation was detected by immunobloting using the phospho-S345 or phospho-S15 antibody, respectively. Levels of phosphorylation were quantified using ImageQuant 5.2 software after scanning immunoblots. The highest level of Chk1 or p53 phosphorylation in each experiment was set equal to 100, and the levels of phosphorylated protein in the other lanes were determined relative to this value. The averages from 2–3 independent experiments were graphed, and the error bars indicate the standard deviation of the mean.
RESULTS
DNA-binding properties of TopBP1
We previously found that full-length TopBP1 bound with higher affinity to DNA-containing AAF-guanine adducts than to undamaged DNA (14). Since it was reported that TopBP1 contains multiple DNA-binding domains (20), we decided to determine which region of TopBP1 is responsible for the damaged DNA-binding activity. To this end, we generated several bacterial constructs to express full-length TopBP1 or fragments of the protein encompassing varying domains from the N- and C-termini (Figure 1A). TopBP1 contains eight BRCA1 carboxyl-terminal (BRCT) motifs that are known to mediate protein–protein interactions (21) and a region between the sixth and seventh BRCT domains that is sufficient for binding to and activation of ATR (15,22). Fragment A includes the two N-terminal BRCT domains; fragment B carries the ATR-activating domain but lacks a full BRCT motif; fragment C contains the ATR-activating domain as well as the two C-terminal BRCT motifs; fragment D has the N-terminal six BRCT motifs but lacks the ATR-interacting domain. All of these constructs were purified by GST affinity chromatography and were of high purity with the exception of the full-length protein and the large D fragment, both of which contain some degradation or premature termination products (Figure 1B).
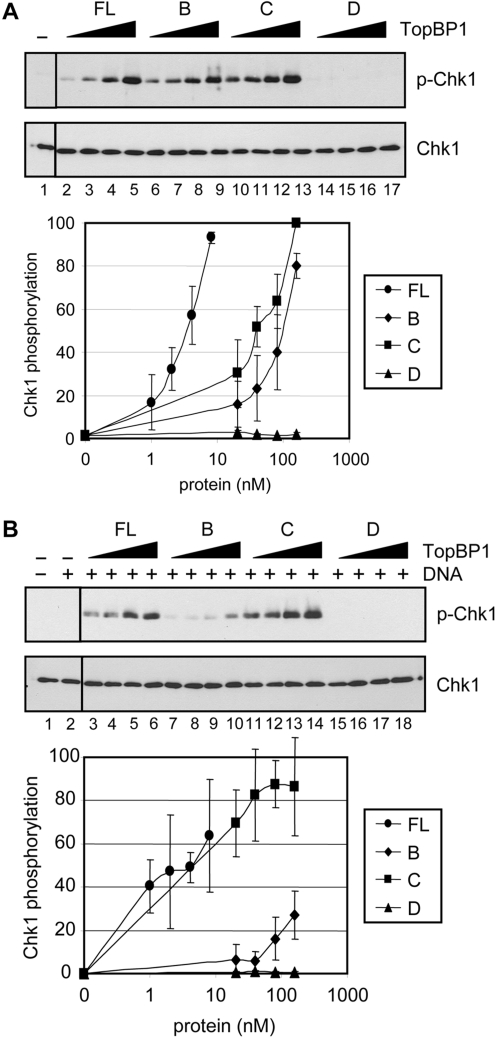
Figure 1.
TopBP1 fragments used in this study and their DNA binding properties. (A) Schematic of human TopBP1 and its fragments that were purified for structure–function experiments. The amino acid positions are indicated, and the eight boxes indicate the BRCT motifs. The ATR activating domain between BRCT domains 6 and 7 is indicated. (B) The GST-fusion proteins visualized by SDS–PAGE followed by Coomassie blue staining. (C) Preferential binding of TopBP1 and TopBP1 fragments to BPDE-damaged DNA. (Top panel) Unmodified (UM) or BPDE treated pUC19 plasmid DNAs (1 ng) which had been labeled with [γ-32P]ATP were incubated with 3 pmol of full-length TopBP1 (FL) bound beads or beads carrying 3 pmol of the TopBP1-A, -B, -C or -D fragments in buffer containing 200 mM NaCl. The bound DNA was eluted by proteinase K, analyzed by agarose gel electrophoresis, and visualized by autoradiography. The input lanes contain fifty percent of DNA added to the reaction. The results from two experiments were quantified and plotted.
We performed DNA pull-down assays to investigate the effect of DNA damage on binding by full-length TopBP1 and the fragments. As seen in Figure 1C, full-length TopBP1 (lanes 3 and 4), the C-terminal fragment carrying the ATR-activating domain and the last two BRCT motifs (fragment C) (lanes 9 and 10), and the N-terminal fragment of the protein (fragment D) containing the first six BRCT motifs (lanes 11 and 12) bind to damaged DNA preferentially, with the full-length protein exhibiting the highest affinity. In contrast, the N-terminal fragment carrying the first two BRCT motifs (fragment A), which was previously shown to have DNA-binding activity (20), fails to exhibit measurable binding to either undamaged or damaged DNA under our experimental conditions (lanes 5 and 6). Fragment B, containing the activation domain but no BRCT motifs, also fails to bind undamaged or BPDE-damaged DNA (lanes 7 and 8), which is in agreement with our previous results with N-Aco-AAF-damaged DNA (14). Thus, we conclude that the two DNA-binding domains located in the N- and C-terminal halves of TopBP1 contribute to the preferential binding of this protein to DNA containing bulky base lesions such as BPDE–guanine adducts. We then proceeded to analyze the effect of BPDE damage on ATR activation mediated by full-length TopBP1 and the TopBP1 fragments.
Stimulation of the ATR kinase by TopBP1 fragments
It is known that in buffers of low ionic strength, both full-length TopBP1 and the ATR-activating domain of TopBP1 can activate the ATR kinase in the absence of DNA (14,15). We directly compared the ATR-stimulatory activities of the TopBP1 fragments under low ionic strength reaction conditions, and the results are shown in Figure 2A. We find that full-length TopBP1 is the most efficient in stimulating the ATR kinase (lanes 2–5), and that fragments B and C (lanes 6–9 and lanes 10–13, respectively) which carry the ATR-activating domain are also capable of stimulating ATR, albeit at <10% of the efficiency of the full-length protein. The D fragment, which does not contain the ATR-activating domain, fails to stimulate ATR (lanes 14–17).
Figure 2.
Stimulation of the ATR kinase by TopBP1 and its fragments. (A) TopBP1 fragments containing the ATR-activating domain stimulate ATR in the absence of DNA. ATR (0.4 nM) was incubated with Chk1 (10 nM) in the presence of full-length TopBP1 (1–8 nM) or the indicated TopBP1 fragments (20–160nM) under low ionic strength conditions (45 mM total salt concentration). ATR kinase activity was determined by immunoblotting for phospho-Chk1 (S345) and Chk1 as indicated. The graph shows quantitative analysis of the data from three independent experiments conducted under identical conditions. (B) DNA stimulates the kinase activity of ATR in the presence of full-length TopBP1 or the C-terminal fragment. Kinase assays were performed as described in Figure 2A, except with 5 ng of BPDE-damaged DNA under high ionic strength conditions (85 mM total salt concentration). The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed.
We previously reported that under conditions of high ionic strength, TopBP1 failed to stimulate ATR, and that some stimulation was observed only when DNA was included in the reaction mixture and that the strongest stimulation was seen when DNA was damaged by N-Aco-AAF (14). To gain further insight into the role of damaged DNA binding by TopBP1 in ATR activation, we tested the fragments of TopBP1 for their abilities to stimulate ATR in the presence of BPDE-damaged DNA. As shown in Figure 2B, in addition to full-length TopBP1 (lanes 3–6), the C fragment (lanes 11–14), which has DNA-binding activity as well as the activation domain, also efficiently stimulates ATR in the presence of BPDE-damaged DNA. DNA slightly stimulates the ATR kinase in the presence of high concentrations of the B fragment (lanes 7–10), which carries the activation domain but lacks DNA-binding activity. DNA has no effect on the ATR kinase in the presence of the D fragment (lanes 15–18), which binds DNA but lacks the activation domain. These data support our previous model that the formation of a damaged DNA–TopBP1–ATR ternary complex is essential for ATR stimulation by damaged DNA in vitro (14).
Since the results in Figure 2B demonstrate that the C-terminal one-third of TopBP1 is sufficient for mediating damaged DNA-dependent stimulation of the ATR kinase, we wished to determine whether the observed stimulation by the C fragment of TopBP1 is indeed damaged-DNA specific, as was previously reported with full-length TopBP1 (14). For this purpose, we tested increasing amounts of either unmodified or BPDE-damaged DNA in ATR kinase reactions containing full-length TopBP1 or the C fragment (Figure 3). In the presence of full-length TopBP1, BPDE-damaged DNA stimulates the ATR kinase ∼5-fold more than unmodified DNA (lanes 7–10 versus lanes 3–6), which is consistent with what we previously reported for DNA damaged with N-Aco-AAF (14). Importantly, in the presence of the C fragment of TopBP1, BPDE-damaged DNA stimulates the ATR kinase ∼9-fold more than unmodified DNA (lanes 17–20 versus lanes 13–16). Thus, we conclude that like full-length TopBP1, the C fragment preferentially binds to DNA containing bulky base lesions and specifically stimulates ATR kinase activity. Therefore, the C-terminal one-third of TopBP1 is sufficient for damaged DNA-dependent ATR activation.
Figure 3.
TopBP1 C-terminal fragment stimulates the ATR kinase in a manner dependent on the presence of damaged DNA. Kinase assays were carried out with ATR (0.4 nM), Chk1 (10 nM) and unmodified (UM) or BPDE-damaged DNA (5–40 ng) in the presence of full-length TopBP1 (0.5 nM) or TopBP1 C-fragment (10 nM) under high ionic strength conditions. The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed.
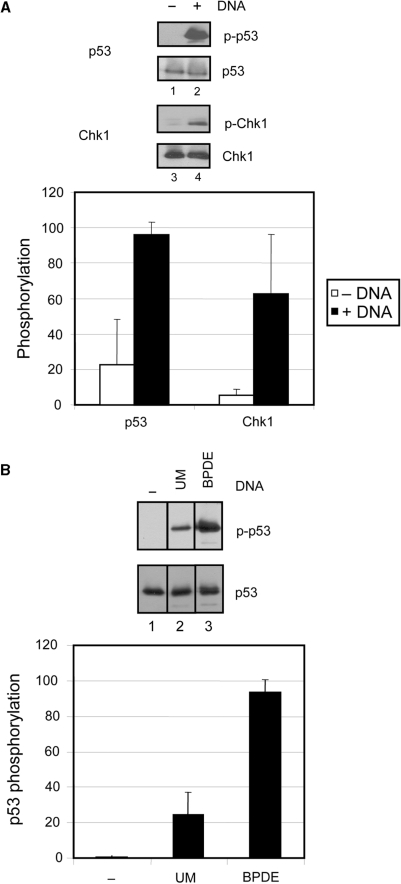
We next examined whether the TopBP1- and DNA-dependent activation of ATR is specific to the Chk1 substrate or whether it is a general mechanism applicable to other known ATR substrates as well. Therefore, we tested another key checkpoint protein and ATR substrate, p53, in our assay, and the results are shown in Figure 4A. We observe ∼5-fold more phosphorylation of serine 15 of p53 in the presence of DNA (lane 2) than in the absence (lane 1), which is comparable to the level of DNA-dependent stimulation of Chk1 phosphorylation (lanes 3 and 4). As with the Chk1 substrate, the DNA-dependent p53 phosphorylation by ATR occurs in the presence of either full-length or the C-fragment of TopBP1 (Figure 4A and B). More importantly, BPDE-damaged DNA specifically stimulates TopBP1-dependent ATR kinase activity ∼4-fold more than undamaged DNA (Figure 4B, lanes 2 versus 3). Therefore, the TopBP1- and damaged DNA-dependent stimulation of ATR is not specific to the Chk1 substrate.
Figure 4.
Damaged DNA-dependent stimulation of ATR is independent of the substrate. (A) Addition of DNA stimulates the ATR kinase activity toward another key downstream target, p53 in the presence of TopBP1. Kinase assays were carried out with ATR and TopBP1-C fragment in the absence or presence of 5 ng of unmodified circular DNA in reactions containing 5 nM Chk1 (lane 1 and 2) or 5 nM p53 (lane 3 and 4) as described in Figure 3. The average levels of Chk1 and p53 phosphorylation from three independent experiments were quantified and graphed. (B) Phosphorylation of p53 is strongly stimulated by BPDE-damaged DNA. Kinase assays were performed with ATR, p53 and 5ng of unmodified (UM) (lane 2) or BPDE-damaged DNA (BPDE) (lane 3) in the presence of full-length TopBP1 as described in Figure 3. The average levels of p53 phosphorylation from two independent experiments were quantified and graphed.
Cooperativity in ATR activation
It was recently shown that the MRN (Mre11-Rad50-Nbs1)-mediated cooperative binding of ATM to DNA greatly stimulated the ATM kinase activity (23). To determine whether ATR exhibited cooperativity similarly to ATM, BPDE-damaged DNAs ranging in size from 0.2 to 2.6 kb were tested at identical DNA mass (and therefore adduct) concentrations for activating ATR in the presence of TopBP1. The results are shown in Figure 5A. Fragments of 0.2 kb had no effect on TopBP1-dependent stimulation of the ATR kinase (lanes 18–20), whereas, at longer DNA sizes, the stimulation increased essentially uniformly, approaching maximum in the 2–2.6 kb range (lanes 3–11). It is important to note that the number of DNA ends in the reaction does not contribute to the DNA length-dependent stimulation of ATR, as there is no difference in the ATR kinase stimulation induced by linear and circular DNAs (Figure 5A, lanes 3–8 and Figure 6B), and the length effect is also observed with equal molar quantities of DNA (data not shown). We have also observed similar DNA length-dependent stimulation of ATR in the presence of TopBP1-C fragment and with undamaged DNA (data not shown). Thus, it appears that, as in the case of ATM, cooperative binding of ATR to DNA is required for activation of the kinase activity of this PIKK member.
Figure 5.
Efficient ATR activation and DNA binding by TopBP1 depend on the length of DNA. (A) TopBP1-dependent ATR stimulation increases with DNA length. Kinase assays were performed as described in Figure 3, except with 1.25, 2.5, and 5 ng of DNAs of different lengths ranging from 0.2 to 2.6 kb. The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed. (B) TopBP1 preferentially binds to longer DNA. DNA-binding assays were carried out as described in Figure 1C, except with equal molar or mass amounts of 0.2 or 2.6 kb DNA fragments in buffer containing 300 mM NaCl. The results from two experiments were quantified and plotted. (C) TopBP1 preferentially binds longer DNAs under kinase reaction conditions. DNA-binding assays were performed with 0.15 pmol of TopBP1 and 3.5 fmol of 0.2 or 2.6 kb DNA fragments under conditions used for kinase reactions. The results from two experiments were quantified and graphed.
Figure 6.
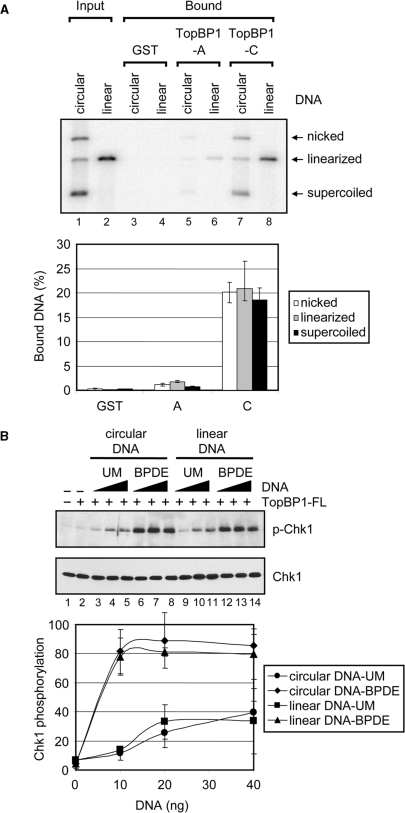
DNA binding and ATR stimulatory activities of TopBP1 are independent of DNA ends. (A) TopBP1 has no preferential binding to DNA ends. Covalently closed circular and linear DNA that had been labeled with [γ-32P]ATP were incubated with GST alone, the TopBP1-A or TopBP1-C fragment immobilized on glutathione beads. After extensive washing, the bound DNA was eluted and analyzed on an agarose gel containing ethidium bromide. The gel was dried and visualized by autoradiography. Note that the ‘covalently closed DNA’ sample contains covalently closed, nicked and linear DNAs and hence gives a fair representation of the relative affinities of these three forms to the TopBP1 fragments. The input lanes contain fifty percent of DNA added to the reaction. The results from five experiments were quantified and plotted. (B) DNA double-strand breaks has no effect on TopBP1-dependent ATR activation. Kinase assays were carried out with ATR, Chk1 and TopBP1 in the presence of unmodified (UM) or BPDE-damaged circular or linear DNA as described in Figure 3. The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed.
To determine whether TopBP1 exhibits cooperative DNA-binding properties, we performed DNA pull-down assays with different size fragments of DNA. The results are shown in Figure 5B and C. We observe preferential binding of TopBP1 to longer BPDE-modified DNA fragments under conditions of either equal DNA mass or molar quantities of the two DNA fragments (Figure 5B, lane 4). Under these experimental conditions, we observe negligible binding to unmodified DNAs of either length (lane 3). Because these DNA-binding experiments were carried out under conditions different from those of the kinase assay, we then repeated the DNA-binding experiment in kinase reaction buffer, and obtained similar results (Figure 5C). There was significantly more binding of TopBP1 to the longer DNA fragments whether the DNAs were unmodified (lane 3) or BPDE-modified (lane 4). Therefore, we conclude that TopBP1 preferentially binds to longer DNAs which results in cooperative recruitment and activation of ATR.
DNA binding and ATR stimulatory activities of TopBP1 are independent of DNA ends
It was previously reported that TopBP1 binds preferentially to DNA duplex ends and nicks in vitro, indicating that TopBP1 may have an important role in recognition of DNA breaks (20). However, we observed no preferential binding to DNA ends under our experimental conditions (Figure 5B). Since our results seem to be contradictory to the previous report, we wished to directly address whether TopBP1 has different binding affinities for circular and linear DNAs. Therefore, we performed DNA pull-down assays according to the previously described procedures with the A and C fragments which are equivalent to the TopBP1 fragments used in the previous report (20). The results are shown in Figure 6A. The C-terminal fragment (lanes 7 and 8) binds about 10-fold more efficiently to DNA than the N-terminal fragment (lanes 5 and 6); and importantly, there are no significant differences between the affinities of either fragment to linear, nicked or circular DNA, leading us to conclude that TopBP1 has no preference for DNA ends. It is unclear why our results differ from the previous report showing similar DNA-binding affinities for the two equivalent TopBP1 fragments as well as an absolute dependence on DNA ends for binding (20). We note that we have also conducted these experiments using electromobility shift assays which also confirm results (data not shown).
Since we did not observe a noticeable difference in the binding affinity of TopBP1 to circular or linear DNA in the DNA pull-down assays, we used the kinase assay to probe for a difference in the ability of these two forms of DNA to stimulate TopBP1-dependent ATR kinase activity. Figure 6B shows that circular (lanes 3–8) and linear (lanes 9–14) DNAs, which were either mock- or BPDE-treated, were nearly identical in their ability to stimulate ATR kinase activity in the presence of TopBP1. The presence of unmodified DNA (lanes 3–5 and lanes 9–11) in the kinase reaction resulted in >10-fold stimulation of ATR activity relative to no DNA (lane 2), and the addition of BPDE-modified DNA (lanes 6–8 and lanes 12–14) resulted in ∼8-fold further stimulation over unmodified DNA, independent of whether the DNAs were linear or circular. We conclude from these results that in contrast to the MRN-mediated cooperative activation of ATM which depends on free DNA ends, DNA termini are not required for TopBP1 DNA binding or DNA-dependent cooperative activation of ATR.
DISCUSSION
In this study, we demonstrate that TopBP1 has two DNA binding sites and binds DNA with no preference for DNA termini. Moreover, we find that both sites aid in preferential binding of TopBP1 to DNA damaged by BPDE. In addition, we demonstrate that while under conditions of low ionic strength any TopBP1 fragment that carries the ATR-binding domain (15,22) is sufficient for ATR activation, only the C-terminal fragment that contains the ATR-activating domain together with a DNA-binding domain can stimulate the ATR kinase in a manner dependent on the presence of BPDE-damaged DNA under physiologically relevant ionic strength. Interestingly, whereas the N-terminal half of TopBP1 is conserved through evolution from yeast to humans and plays an essential role in replication initiation, the C-terminal half of TopBP1, containing the ATR-activation domain, is only conserved in metazoans and is essential for checkpoint activation (24,25). In fact, the C-terminal half of TopBP1 is sufficient for Chk1 phosphorylation induced by oligonucleotides in Xenopus egg extracts (24). The budding yeast TopBP1 homolog, Dpb11, while lacking sequence homology to the ATR-activation domain, is still able to activate the Mec1ATR kinase in an in vitro system (26,27). However, the yeast Dpb11TopBP1 also lacks the C-terminal region that we have identified to be important in the human protein for mediating DNA-dependent ATR activation, and does not support DNA-dependent Mec1ATR activation (26). While results from our reconstituted checkpoint system indicate that the C-terminus of TopBP1 is sufficient for direct binding to damaged DNA and activation of ATR kinase activity, there are recent reports that the N-terminus of TopBP1 is required for its recruitment and resulting activation of ATR via an interaction with Rad9 in mammalian cell lines (28) and Xenopus egg extracts (29). Work is underway to develop an in vitro system that depends on Rad9 and the other factors identified genetically for optimal activation of the ATR-mediated checkpoint response.
An unexpected finding of this study has been the DNA length dependence of TopBP1 stimulatory activity. Although a similar length dependence was reported for ATM autophosphorylation, in that case there was an absolute requirement for DNA ends for cooperative binding and autophosphorylation of ATM (23). In contrast, in the case of ATR there is no requirement for DNA termini. It must also be noted that the length effect on ATM activation was ascribed to chromatinization of the DNA added to the egg extract, the minimum requirement for DNA of 0.2 kb coinciding with the DNA length required for efficient formation of nucleosomes, which are presumed to be required for recruitment of ATM to DNA flanking double-strand breaks and subsequent activation (23). Clearly, this is not the case for ATR in our system in which purified proteins are used in the checkpoint reconstitution. However, studies on ATM activation using purified proteins also demonstrated a very similar DNA length dependence for ATM activation (18). In that report, fragments of 0.384 kb had a minimal effect on MRN-dependent stimulation of ATM kinase, and the stimulation increased essentially uniformly with longer DNA sizes, with the maximum at 2.3 kb. Therefore, it appears that both ATM and ATR share similar DNA length-dependence of MRN and TopBP1 stimulatory activity, respectively. It should also be noted that in the case of ATM, by increasing the DNA concentration of a 0.2 kb fragment, the stimulatory approached that achieved by a 2 kb fragment (23), whereas in the case of ATR, increasing the amount of the 0.2 kb fragment did not affect the outcome. This is in line with the argument that the length-dependent cooperative effects of DNA with double-strand breaks, in the case of ATM, and DNA with base damage, in the case of ATR, result from different mechanisms.
We use the word cooperativity as an operational term, not in the strict mechanistic sense, because at present, we do not have a mechanistic model for the DNA-induced cooperativity of the ATR kinase. Although ATM and ATR have distinctly different modes of damage sensing and activation, our in vitro findings regarding the cooperative activation of the ATR kinase are in line with the recent report that the lac operator/lac repressor-mediated binding of budding yeast Ddc2ATRIP (and therefore Mec1ATR) to DNA exhibits a similar type of cooperativity for Mec1ATR kinase activation in vivo (30). It was found that at least 40 repressor/operator complexes with the repressor-fused Ddc2ATRIP were required for significant Mec1ATR activation as measured by Rad53Chk1/2 phosphorylation. There is also evidence for cooperative activation of ATR in vitro in Xenopus egg extracts where Chk1 phosphorylation is dependent on the size of the single stranded DNA gap (11).
While in wild-type human and yeast cells, in addition to ATR (Mec1), the 9–1–1 complex and TopBP1 (Dpb11) are required to act coordinately to initiate the checkpoint response after DNA damage, the requirement for the 9–1–1 complex, TopBP1 or DNA damage can be circumvented under special reaction conditions in vitro (14–16,26,27), by overexpressing the TopBP1 ATR activating domain in vivo (15), or by artificially tethering these checkpoint proteins to the DNA in vivo (28,30). Hence, we believe that our in vitro system in which damaged DNA-bound TopBP1 recruits ATR and activates its kinase function is a reasonable approximation to ATR activation in vivo, and provides a useful platform for mechanistic studies of the ATR-mediated DNA damage checkpoint.
FUNDING
National Institutes of Health (GM32833). Funding for open access charge: National Institutes of Health (GM32833).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ö. Serçin for providing GST-p53 protein and M. Kemp for critical reading and useful comments.
REFERENCES
- 1.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 6.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Falconi MM. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl Acad. Sci. USA. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, et al. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 9.Stiff T, Cerosaletti K, Concannon P, O’Driscoll M, Jeggo PA. Replication independent ATR signalling leads to G2/M arrest requiring Nbs1, 53BP1 and MDC1. Hum. Mol. Genet. 2008;17:3247–3253. doi: 10.1093/hmg/ddn220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 11.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unsal-Kacmaz K, Makhov AM, Griffith JD, Sancar A. Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:6673–6678. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol. Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc. Natl Acad. Sci. USA. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 16.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 18.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 19.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 20.Yamane K, Tsuruo T. Conserved BRCT regions of TopBP1 and of the tumor suppressor BRCA1 bind strand breaks and termini of DNA. Oncogene. 1999;18:5194–5203. doi: 10.1038/sj.onc.1202922. [DOI] [PubMed] [Google Scholar]
- 21.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat. Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- 25.Yan S, Lindsay HD, Michael WM. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 2006;173:181–186. doi: 10.1083/jcb.200601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc. Natl Acad. Sci. USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 30.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol. Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]