Abstract
Potent PCR inhibitors in blood and soil samples can cause false negative results from PCR-based clinical and forensic tests. We show that the effect of these inhibitors is primarily upon Taq DNA polymerase, since mutational alteration of the polymerase can overcome the inhibition to the extent that no DNA purification is now required. An N-terminal deletion (Klentaq1) is some 10–100-fold inhibition resistant to whole blood compared to full-length, wild-type (w.t.) Taq, which is strongly inhibited by 0.1–1% blood. Further mutations at codon 708, both in Klentaq 1 and Taq, confer enhanced resistance to various inhibitors of PCR reactions, including whole blood, plasma, hemoglobin, lactoferrin, serum IgG, soil extracts and humic acid, as well as high concentrations of intercalating dyes. Blood PCR inhibitors can predominantly reduce the DNA extension speed of the w.t. Taq polymerase as compared to the mutant enzymes. Single-copy human genomic targets are readily amplified from whole blood or crude soil extract, without pretreatment to purify the template DNA, and the allowed increase in dye concentration overcomes fluorescence background and quenching in real-time PCR of blood.
INTRODUCTION
PCR-based tests of blood and soil samples are widely used for diagnostics and forensic analyses. Of particular importance are the diagnostic PCR tests for genetic diseases, microbial and viral infections, blood typing or blood banking, as well as environmental tests and forensic human DNA identification (1–4). The effect of the main PCR inhibitors in blood and soil, hemoglobin and humic acid, is primarily associated with inactivation or inhibition of Taq DNA polymerase. Therefore, various procedures and DNA extraction kits are being used to purify DNA prior to PCR. These extra steps are time consuming, may not completely remove inhibitors or may lead to losses of target DNA.
Widely used DNA polymerases like Taq DNA polymerase and AmpliTaq Gold, a hot-start version of Taq,can be completely inhibited in the presence of less than 0.2% whole human blood (4,5). Some non-Taq DNA polymerases, however, such as rTth, Tfl, HotTub and Pwo, can tolerate higher concentrations of blood (5). Various agents have been reported to reduce the inhibitory effect of blood on Taq. It was found that an addition of betaine, bovine serum albumin, the single-stranded DNA-binding protein of the T4 32 gene, or a cocktail of protease inhibitors can partially reduce the blood inhibition and can allow Taq to work in up to 2% blood, although this effect could be sample-specific (4–8).
The inhibitory effect of blood on PCR is not yet well understood, and it has been proposed to be associated primarily with inactivation of the DNA polymerase and/or capturing or degradation of the target DNA and primers. Several major inhibitors of PCR in human blood have been characterized, such as hemoglobin, immunoglobulin G and lactoferrin (7–9,11). Protease activity in blood also could contribute to the reduced efficiency of PCR (5,9–12).
Sensitive and precise PCR detection of microorganisms in soil is necessary for agricultural purposes, infectious disease control and bioterrorism-related pathogen tests (13–16). Direct extraction of total DNA from soil samples results in a co-extraction of humic acid, known as the most potent soil inhibitor to PCR (15,16). Humic substances represent a mixture of partially characterized polyphenols that are produced during the decomposition of organic matter. Taq DNA polymerase is typically inhibited in the presence of less than 1 ng of humic acid in a PCR reaction. Other inhibitory components in soil samples include fulvic acid, polysaccarides and metal ions (16–18). Another soil-born, high-molecular weight PCR inhibitor was also identified. It forms a complex with proteins and may inhibit PCR by an interaction with Taq DNA polymerase (19). In some cases addition of bovine serum albumin to PCR can slightly reduce the inhibitory effect (20–22). A general problem with the soil samples is inconsistent data due to high variation in the concentrations of the inhibitors, depending on the soil source. This fact significantly complicates the development of standard DNA purification protocols for processing the samples before PCR.
Various procedures of DNA extraction have been developed to reduce the inhibitory effect of blood or soil components on PCR (1,18,23–32). These pre-treatment steps are generally time-consuming, labor-intensive and can be sample-specific. Moreover, some PCR inhibitors may still be present even after using DNA extraction kits. For example, about 14% of the human hepatitis B virus tests employing blood DNA purification kits could be false-negative (33).
As an alternative to the various DNA purification steps used with blood and soil samples prior to PCR, we screened for mutants of Taq polymerase that can overcome the PCR inhibition distinctive for such samples. We describe and functionally characterize such mutant enzymes and demonstrate that they can eliminate pre-PCR treatment steps. In recently reported studies Taq polymerase was also mutagenized for achieving other qualities, such as reverse transcriptase activity and ability to amplify from damaged templates (34–37).
MATERIALS AND METHODS
Site-directed mutagenesis
Part of the Klentaq1 gene was amplified with a C-terminal primer, RevTaqH, and a second internal primer spanning the amino acid change of interest. This amplified product was purified and used as a primer in a second amplification step in combination with an N-terminal primer, KT1. The N- and C-terminal primers contained NcoI and HindIII restriction sites, respectively. This second step restored the entire Klentaq gene with the desired mutation in it. The final PCR product was then treated with the restriction nuclease Dpn I, in order to eliminate the w.t. target and background plasmid template (38). The purified PCR product was cloned into the NcoI/HindIII sites of the expression vector pWB254 (39).
An analogous procedure was performed for site-directed mutagenesis of Taq, using Taq specific end primers harboring the EcoRI and BamHI restriction sites. The restriction nuclease Dpn I was used again for eliminating the w.t. background in cloning. The amplified Taq gene was cloned into the same sites of the pUC18 vector (40). All full-length Taq clones were propagated and expressed in a Taq-tolerant Escherichia coli strain, designated as R. coli that was a gift from Clontech.
Enzyme purification
The Klentaq and Taq mutant enzymes were over-expressed in our host bacterial strains X7029 and R. coli, respectively, after IPTG induction for 12–14 h. The Klentaq mutant enzymes were purified by BioRex 70 and Heparin-agarose chromatography as previously described, except that the ammonium sulfate precipitation was eliminated (39). A slightly modified protocol was utilized for purification of the Taq mutant enzymes, where 100 mM AmSO4 was used in the BioRex 70 binding step.
All enzymes were purified to at least 95% homogeneity, checked by SDS–PAGE.
Blood samples and soil extracts
Whole human blood and plasma were purchased form Innovative Research and were EDTA or heparin treated and obtained from the same donor. Whole blood was kindly provided from Mid-America Transplant Services. Blood and plasma samples were stored frozen at −75°C in 0.5 ml aliquots until use. After thawing on ice, they were mixed through gentle inversion before adding directly to PCR reactions as the last ingredient. Aliquots were refrozen and used up to five times with no noticeable effect on the enzyme or PCR performance.
For preparing soil extracts, fresh soil from different sources was suspended in 1× KLA PCR buffer in the ratio 1:10 (w/v), vortexed and heated for 30 min at 75°C, followed by centrifugation at 12 000 × g for 20 min. The supernatant was used directly in PCR tests as ‘crude soil extract’ and stored frozen. These type of extracts typically contained detectable DNA from soil microorganisms and nematodes, as well as enough humic acid to exhibit a strong inhibitory effect on PCR with added template.
PCR inhibitors
Human hemoglobin, lactoferrin and IgG serum fraction were purchased from Sigma. Hemin chloride and humic acid were purchased from Calbiochem and Fluka.
PCR amplification
The PCR reaction buffer for all the Klentaq mutant enzymes was KLA35: 50 mM Tris, pH 9.2, 16 mM AmSO4, 3.5 mM MgCl2 and 0.1% Tween-20. The Taq mutant enzymes were assayed in a modified buffer, KLA25, using pH 8.2 and 2.5 mM MgCl2. The reactions shown in Figures 1a, b, d, 2 and 6–8 were supplemented with 1.3 M betaine (Sigma). PCR reactions contained 200 nM each primer, and 200 μM each dNTP. Other DNA polymerases tested in comparison to the mutant enzymes were: Plain Taq (New England Biolabs), plain Taq and FastStart Taq (Roche), JumpStart Taq (Sigma), Fail-Safe Taq (Epicentre), AmpliTaq Gold and rTth (Applied Biosystems), Tth, Tli, and Tfl (Promega). Each enzyme was tested at 2 U per 50 μl reaction with its optimal buffer, following manufacturer's recommendations. Conventional and real-time PCR was performed in Robocycler-40 (Stratagene) and Opticon 2 (BioRad), respectively.
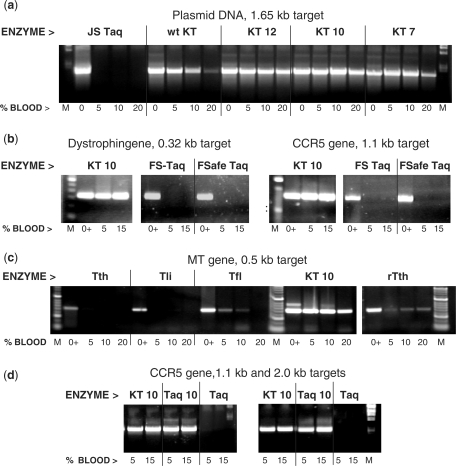
Figure 1.
The mutants of KlenTaq (KT) and Taq can perform efficient PCR in the presence of high concentrations of whole blood. The blood-tolerant Taq mutant enzymes KT 7, KT 10, KT 12 or Taq 10 were tested in PCR in the presence of 0–20% whole human blood and compared to the w.t. KlenTaq (KT) and various commercial DNA Taq enzymes: Jump Start Taq (JS Taq), Fast Start Taq (FS Taq), Fail-Safe Taq (FSafe Taq) or plain Taq (Taq), as well as Tfl, Tli, Tth and rTth DNA polymerases Positive controls (lanes 0 and 0+) contain no blood but 5 ng human DNA for the endogenous targets. Lanes M, DNA standards ladder. (a) A 1.65-kb target was amplified from 1 ng pWB254 plasmid DNA. (b)–(d) endogenous targets of the dystrophin gene (0.32 kb), HIV CCR5 receptor gene (1.1 kb or 2.0 kb) and the methyl transferase gene (0.5 kb) were amplified straight from blood. The amplified products were analyzed in a 2% agarose gel stained with ethidium bromide.
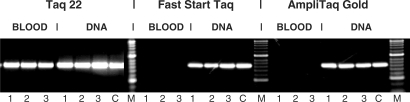
Figure 2.
The Taq mutants do not require DNA purification from blood samples prior to PCR. A 630-bp target from the human CCR5 gene was amplified from either whole blood or DNA purified from the same blood batch prior to PCR using a blood DNA extraction kit. The blood DNA was eluted in a volume equal to the initial blood volume, and equivalent volumes of either blood or DNA (2, 4 or 8 μl in 50 μl reaction, lanes 1–3, respectively) were added to the PCR. Control reactions (C) contained 4 ng human genomic DNA. The target was amplified in 35 cycles with 2 U of the BR mutant Taq 22, Fast Start Taq (Roche) or Ampli Taq Gold (Applied Biosystems), using the specific buffer for each enzyme. The amplified products were analyzed in a 2% agarose gel stained with ethidium bromide. Lanes M, DNA standards ladder.
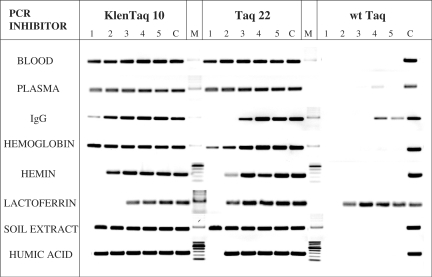
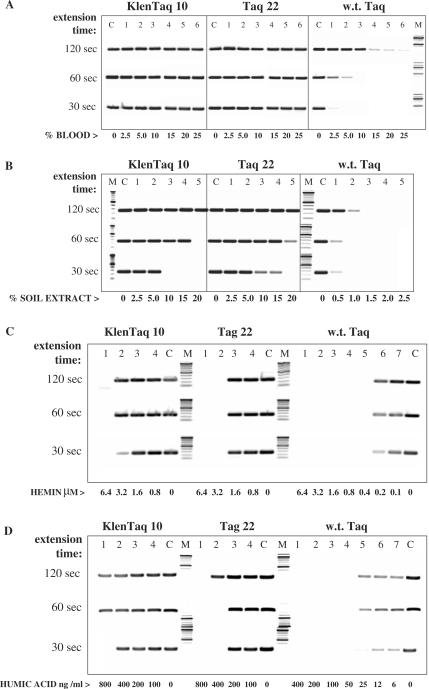
Figure 6.
Effect of blood and soil inhibitory components on DNA amplification. The 250-bp λ-DNA target was amplified with KlenTaq 10, Taq 22 or w.t. Taq in the presence of blood and soil-derived fractions or compounds, five 2-fold dilutions each (lanes 1–5), starting with the following highest concentrations: 25% whole human blood; 25% human plasma (same donor); 50 μg/ml IgG fraction; 2.5 mg/ml hemoglobin; 6.4 μM hemin; 60 μM lactoferrin; 15% crude soil extract; 400 ng/ml humic acid. Control reactions (lanes C) contained no inhibitor. Lanes M, DNA standards ladder. The amplified products were analyzed in a 2% agarose gel stained with ethidium bromide.
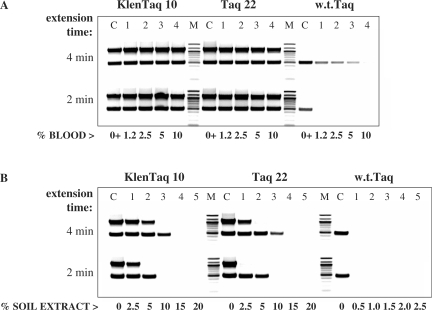
Figure 7.
Effect of the extension time on DNA amplification from blood (endogenous targets) or crude soil extract. Two targets of the human CCR5 and DNMT genes, 1.1 kb and 0.5 kb, respectively, were amplified in duplex PCR directly from 1.25% to 10% human blood with KlenTaq 10, Taq 22, or w.t. Taq (lanes 1–4, A). Control reactions (lanes C) contained 10 ng DNA and no blood. In (B), the same targets were amplified with the three enzymes from 10 ng human DNA, in the presence of 0.5–20% crude soil extract (lanes 1–5) (Note the shift in the range of the soil extract used with the w.t. enzyme). Control reactions (lanes C) contained no soil extract. Two identical samples of each reaction were amplified with 4 min or 2 min extension time in a no ramp-time PCR cycle. Lanes M, DNA standards ladder. The amplified products were analyzed in a 2% agarose gel stained with ethidium bromide.
Figure 8.
Effect of the extension time on DNA amplification in the presence of blood (exogenous targets) or soil extract. The 250-bp λ-DNA target was amplified with KlenTaq 10, Taq 22, or w.t. Taq in the presence of 2.5–25% whole blood (lanes 1–6, A), 0.5–20% crude soil extract (lanes 1–5, B), 0.8–6.4 μM hemin (lanes 1–4, C), or 6–800 ng/ml humic acid (lanes 1–7, D). (Note the shift in the range of the soil extract and humic acid used with the w.t. enzyme.) Control reactions (lanes C) contained no inhibitor. Three identical samples of each reaction were amplified with 120 s, 60 s, or 30 s. extension time in a no ramp-time PCR cycle. Lanes M, DNA standards ladder. The amplified products were analyzed in a 2% agarose gel stained with ethidium bromide.
The amount of blood or soil extract varied considering the sample type and inhibitory concentration. Whole blood (1–10%) was utilized in real-time PCR detection with varying amounts of SYBR Green I in the range of 1–64×. SYBR green I was purchased from Invitrogen at manufacturer's concentration defined as 10 000× (10 mg/ml). A positive control reaction with pure genomic DNA and final concentration of 0.5–4× SYBR was used. Post PCR for conventional PCR procedure involved 20–30 s spin to bring down precipitated blood and the supernatant was loaded in an agarose gel. No post PCR steps were required for real-time reactions, however, some reactions were run on a gel to compare and relate to real-time results.
Cycling conditions and primers: pWB254 plasmid, 1.65-kb target: 2 min/95°C, followed by 30 s/95°C, 40 s/64°C, 4 min/70°C, 35 cycles. F-primer: GAGCCATGGTCCTCCTCCACGAGTTCGGCCTTCTGG; R-primer: CGGTCCGAAAGCTTCTATCACTCCTTGGCGG. Human dystrophin gene, 0.32-kb target: 2 min/95°C, followed by 40 s/95°C, 40 s/60°C, 2 min/70°C, 40 cycles; F-primer: GGCTGTGATAGAGGCTTGTCTATA; R-primer: CTGGCCTGCACATCAGAAAAGACT. Human CCR5 0.63, 1.1 and 2-kb targets: 3 min/94°C, followed by 40 s/94°C, 60 s/60°C, and 3, 5 or 8 min/70°C, 35 cycles. F-primer (common): TGGAACAAGATGGATTATCAAGTGTCAAGTCCA; R-primers: 0.63 kb: GCAGCGGCAGGACCAGCCCCAAGATGACTATCT; 1.1 kb: AGGCTGTGTATGAAAACTAAGCCATGTGCACAA; 2 kb: AGAAGAGCTGAGACATCCGTTCCCCTACAAGAA. Human methyl transferase gene, 0.5-kb target: 2 min/95°C, followed by 30 s/95°C, 40 s/53°C, 2 min/70°C, 35 cycles. F-primer: CGAGCTACCACGCAGACATCAACC; R-primer: GGGGCACCTTCTCCAACTCATACT. Lambda DNA 0.25 kb target: 2 min/94°C, followed by 40 s/94°C, 40 s/66°C, 40 s/70°C, 35 cycles. In the experiments shown in Figure 6 the DNA extension time was 45 s, and in Figure 8 this step was 30, 60 or 120 s, as indicated. F-primer: GGGCGGCGACCTCGCGGGTTTTCGC; R-primer: CTGAATGGTACGGATACTCGCACCG. Bacillus rRNA 0.6 kb target: 3 min/94°C, followed by 40 s/94°C, 40 s/60°C, 1 min/70°C, 35 cycles. F-primer: AGGGTCATTGGAAACTGGG; R-primer: CGTGTTGTAGCCCAGGTCATA.
DNA polymerase assay
The effect of blood and soil PCR inhibitor on the catalytic activity of the Taq mutants and the w.t. enzyme was assessed by a nucleotide incorporation assay as described earlier (39), with minor modifications. The enzymes, 2 U each, were incubated for 10 min at 70°C. with 12 μg activated (partially DNAse I digested) calf thymus DNA in 50 μl 1× PCR buffer, in the presence of 50 μM each dNTPs, 1 uCi (α-32P) dATP and various concentrations of each inhibitor (as indicated in Table 1).
Table 1.
Effect of blood and soil inhibitory components on DNA polymerase activity
| Inhibitor | KlenTaq 10 |
Taq 22 |
wt Taq |
|||||||||||||||
| Blood (%) | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 |
| % Activity | 100 | 112 | 109 | 91 | 55 | 17 | 100 | 110 | 104 | 85 | 47 | 12 | 100 | 95 | 80 | 66 | 38 | 9.0 |
| Plasma (%) | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 |
| % Activity | 100 | 130 | 121 | 114 | 93 | 24 | 100 | 133 | 121 | 106 | 102 | 39 | 100 | 139 | 126 | 112 | 77 | 22 |
| Serum IgG (μg) | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 | 0 | 3.1 | 6.2 | 12.5 | 25 | 50 |
| % Activity | 100 | 135 | 131 | 99 | 96 | 53 | 100 | 136 | 114 | 89 | 70 | 59 | 100 | 124 | 83 | 80 | 74 | 53 |
| Hemoglobin (μg) | 0 | 39 | 78 | 156 | 312 | 625 | 0 | 39 | 78 | 156 | 312 | 625 | 0 | 39 | 78 | 156 | 312 | 625 |
| % Activity | 100 | 128 | 125 | 119 | 98 | 25 | 100 | 114 | 108 | 98 | 65 | 16 | 100 | 169 | 148 | 134 | 65 | 17 |
| Hemin (μM) | 0 | 16 | 32 | 64 | 128 | 256 | 0 | 16 | 32 | 64 | 128 | 256 | 0 | 16 | 32 | 64 | 128 | 256 |
| % Activity | 100 | 60 | 19 | 7.7 | 1.8 | 0.5 | 100 | 79 | 39 | 7.0 | 0.6 | 0.3 | 100 | 62 | 31 | 9.2 | 0.9 | 0.5 |
| Lactoferrin (mM) | 0 | 0.33 | 0.66 | 1.3 | 2.6 | 5.2 | 0 | 0.33 | 0.66 | 1.3 | 2.6 | 5.2 | 0 | 0.33 | 0.66 | 1.3 | 2.6 | 5.2 |
| Activity | 100 | 185 | 169 | 163 | 149 | 92 | 100 | 202 | 146 | 137 | 133 | 123 | 100 | 210 | 165 | 161 | 158 | 151 |
| Soil extract (%) | 0 | 1.5 | 3.1 | 6.2 | 12.5 | 25 | 0 | 1.5 | 3.1 | 6.2 | 12.5 | 25 | 0 | 1.5 | 3.1 | 6.2 | 12.5 | 25 |
| % Activity | 100 | 106 | 75 | 60 | 29 | 5.2 | 100 | 105 | 69 | 41 | 13 | 3.2 | 100 | 86 | 40 | 17 | 6.0 | 1.5 |
| Humic acid (μg/ml) | 0 | 0.1 | 0.2 | 0.4 | 0.8 | 1.6 | 0 | 0.1 | 0.2 | 0.4 | 0.8 | 1.6 | 0 | 0.1 | 0.2 | 0.4 | 0.8 | 1.6 |
| % Activity | 100 | 166 | 122 | 114 | 68 | 11 | 100 | 148 | 107 | 75 | 25 | 2.6 | 100 | 76 | 37 | 11 | 2.7 | 1.1 |
The same fractions and compounds used in Figure 6 were tested for their effect on the catalytic activity of the mutant and w.t. Taq enzymes in a conventional nucleotide incorporation assay. Each enzyme was incubated with activated (partially digested) calf thymus DNA and 32P-labeled dATP for 10 min at 70°C in the presence of five 2-fold increasing concentrations of each inhibitor. The enzyme activity, represented by the rate of nucleotide incorporation into DNA, is shown in bold as percent relative to the controls without inhibitor (first reactions for each enzyme and inhibitor, referred to as 100%). The activity values are average of two duplicate samples. The amount of DNA-incorporated isotope was analyzed on filter dots by Quantity One software (Molecular Imager FX, BioRad).
The reactions were stopped by a quick transfer on ice, and 5 μl aliquots of the samples were dotted on 3 mm filter paper pre-soaked with 20 mM EDTA. The filters were washed 3 × 20 min with 5% trichloroacetic acid/1% sodium pyrophosphate, dried, and exposed to a phosphoimager screen. The dot intensity, reflecting the incorporation rate of the labeled nucleotide into DNA was analyzed by Quantity One software (Molecular Imager FX, BioRad).
RESULTS
Screening for blood-resistant (BR) Klentaq1 and Taq mutant clones
We attempted to obtain mutants of both Taq and Klentaq1 DNA polymerase with high tolerance to PCR inhibitors in blood. Klentaq1, a truncated version of Taq with a N-terminal deletion of 278 amino acids, is an improved version of the w.t. enzyme, with higher fidelity and thermostability (41).
In our initial studies of PCR inhibition by blood, we found that the known Klentaq1 enzyme could amplify single-copy genomic DNA in the presence of 5–10% whole blood. This was a rather unexpected result, as the full-length Taq enzyme is inhibited by 0.1–1% blood (4,5). To our knowledge, no correlation between the N-terminal deletion of Taq and the BR feature of the enzyme has been reported. This finding encouraged us to try to obtain mutated versions of Kentaq 1 with even higher resistance to blood inhibition. We started with testing 40 arbitrary but functional Klentaq1 clones derived from a library of the Klentaq1 gene mutagenized at codons 626, 706, 707 and 708 for PCR performance in the presence of 2.5%, 5% or 10% whole human blood. Previously we had observed mutations at these positions that confer cold-sensitivity (Cs) to the enzyme, adding a ‘hot-start’ feature to its PCR performance (39). In addition, our earlier functional analysis of various Klentaq Cs-mutants indicated that codons 706–708 are of importance for the overall performance of the enzyme. After several screening rounds we identified a clone, KT 7, which reproducibly outperformed the w.t. Klentaq1, as well as the previously characterized Cs-mutants of Klentaq 1. The sequence analysis of KT7 revealed an amino acid change at position 708, E708W, in addition to the E626K and I707L Cs-mutations. To test if this change is solely responsible for the BR phenotype, we re-introduced the same mutation in the parental double Cs-mutant of Klentaq1. When challenged in PCR with increasing blood concentrations, both the original KT 7 and the newly made triple mutant showed identical performance. On the other hand, the basic BR of the double Cs-mutant was the same as the w.t. Klentaq1, indicating the importance of codon 708 for the selected phenotype.
Our library did not contain some of the possible codon changes, so we introduced the remaining ones individually until we had tested all 19 possible amino-acid variants at codon 708, starting with the double mutant E626K, I707L. The functional test of the resultant clones revealed two mutant alternatives which exhibited even higher tolerance to blood and remained functional in at least 20% whole blood, KT 10 (E708K) and KT 12 (E708L)
Figure 1A shows the performance of KT7, KT 10 and KT 12 in amplifying an exogenous DNA target in the presence of 5–20% whole blood. The three mutant enzymes outperformed the w.t. Klentaq1. Similar results were obtained with amplification of endogenous human targets, illustrated with KT 10 in Figure 1B. In control reactions commercial hot-start or plain Taq enzymes failed to perform in the presence of blood. We also tested some non-Taq thermostable DNA polymerases, Tth, rTth, Tfl and Tli, for which a high blood tolerance has been reported (5). In PCR of a human blood gene target two of these enzymes, rTth and Tfl, showed a partial BR but lower PCR efficiency in the 5–20% blood range, as compared to Klentaq 10, while Tth and Tli were not functional with blood (Figure 1C).
To further verify the functionality of the changes at residue 708, we also introduced the mutations of the KT 10 and KT 12 clones into a full-length Taq double Cs-mutant containing the E626K, I707L changes, which by itself shows no tolerance to blood. The resultant enzymes, Taq 10 and Taq 12, remained functional in at least 10–15% blood. This performance demonstrates a tolerance to blood concentrations exceeding about 100-fold that of the w.t. Taq. Figure 1D illustrates the Taq 10 performance matching that of KT 10 in amplifying blood genes from whole blood where w.t. Taq completely fails. These results demonstrate that the mutations rendering the Klentaq1 enzyme BR are functional in the context of the full-length Taq as well. In order to functionally test the entire spectrum of changes at Taq codon 708, we performed saturating mutagenesis at that codon in the full-length double Cs-mutant enzyme as well. Interestingly, we found that the best amino-acid substitutions for the BR phenotype of Taq are different than those optimal for Klentaq1. PCR tests of blood samples with various targets showed the best results with three new Taq modifications E708N, E708I and E708Q (named Taq 3, Taq 4 and Taq 22). Figure 2 illustrates the performance of Taq 22 in a PCR test where equivalent amounts of either whole blood or DNA purified from the same blood were used to amplify a target from the human CCR5 HIV receptor gene. The amplification with Taq 22 was efficient in both cases, while w.t. Taq preparations were only functional with purified DNA template. Identical results were obtained with Taq 3, Taq 4 and Klentaq 10 mutants as well (data not shown). These data suggest that the novel mutant enzymes can eliminate the need of using DNA purification steps from blood prior to PCR.
Tolerance of the BR Taq mutants to SYBR Green in PCR
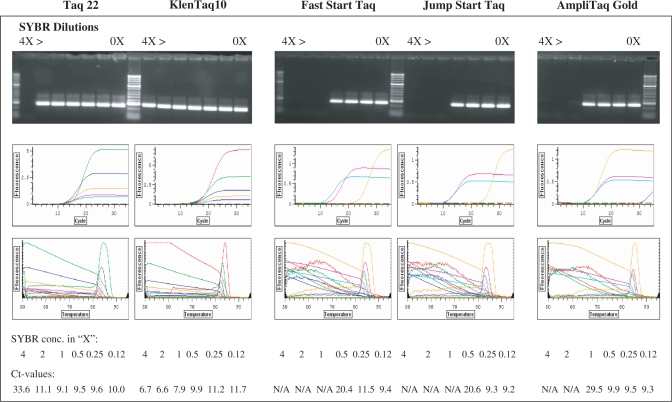
Our next step was to test the possibility of using the BR mutant enzymes in real-time PCR with crude blood samples. One obstacle we came across was that a standard qPCR protocol utilizing SYBR Green I does not work efficiently due to a pronounced quenching effect of the blood on the fluorescent dye. We tried to overcome this problem by increasing the SYBR Green concentration and in titration experiments unexpectedly found that the BR enzyme can tolerate significantly higher concentrations of the fluorescent dye than can the w.t. enzyme. Studies have shown that SYBR Green I and other fluorescent dyes used recently in qPCR exhibit some inhibitory effect on Taq enzyme (42–45). This imposes certain limitations on the fluorescence level and the sensitivity of the assay. Commercial Taq enzymes typically can tolerate not more than 0.5–1× SYBR Green in PCR. As shown in Figure 3, Fast Start and Jump Start Taqs can be inhibited even by 0.5× SYBR, showing Ct values of about 20 (yellow curves) vs. Ct of 9–11 with 0.25× and 0.125× SYBR (pink and green curves, respectively). In contrast, the mutant enzymes remained functional in several times higher concentration of the dye, up to 2× and 4× for the Taq 22 and Klentaq 10, in which range no significant shift in the Ct values was observed. Analogous results were obtained when these mutant enzymes were tested with other fluorescent dyes, such as EvaGreen, Syto9, Pico, Toto, Yoyo, LC Green and ethidium bromide (data not shown), which are alternatives to SYBR Green in qPCR (42–45).
Figure 3.
The mutant Taq and KlenTaq enzymes can tolerate high SYBR Green I concentrations in PCR. A 250-bp target was amplified from 0.5 ng lambda DNA in real time PCR with KlenTaq 10 and Taq 22 mutant enzymes as well as with three w.t. Taq enzymes: Fast Start Taq, Jump Start Taq and AmpliTaq Gold (2 U each enzyme) in the presence of various concentrations of SYBR Green. Serial dilutions of the fluorescent dye, 4×, 2×, 1×, 0.5×, 0.25× and 0.125× were tested along with no dye controls (rightmost lanes). The amplified products were analyzed both in ethidium bromide-stained agarose gel (top panels) and by real-time fluorescence incorporation (background subtracted fluorescence values of amplification and melting curves, middle and bottom panels).
Real-time PCR of crude blood samples with the BR Taq mutants
Attempting to overcome the quenching effect of blood on SYBR Green fluorescence, mentioned above, we performed series qPCR tests with blood and SYBR Green titration. We found that relatively very high input concentrations of SYBR Green are required in order to obtain an efficient qPCR detection of blood samples. We determined that the concentrations of the dye required are roughly proportional to the blood concentrations in the sample. For example, up to 64× SYBR Green was necessary to detect amplification in up to 10% blood. This finding allowed us to develop high SYBR Green concentration real-time PCR protocols for using Taq 22 and Klentaq 10 in DNA amplification from blood samples.
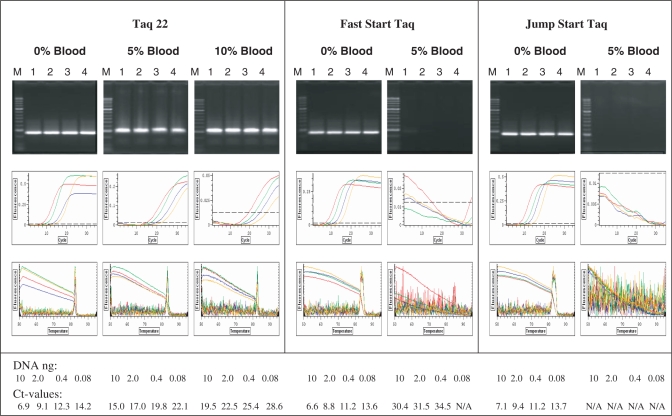
Figure 4 illustrates the application of such protocol with Taq 22 enzyme, where a DNA target was amplified in the presence of 5% and 10% blood with 32× SYBR Green. The target detection in blood was only possible with the Taq 22 mutant, while no amplification and high fluorescence background was observed with w.t. Taq enzymes. In general, the amplification efficiency in the presence of blood was reduced, as judged by a shift in the Ct values, which effect was significantly more pronounced with the w.t. Taqs, especially with the Jump Start Taq, where no Ct values were available.
Figure 4.
Real-time PCR of whole blood containing samples. A 250-bp target was amplified from Lambda DNA, using 2 U of the BR Taq 22 mutant, Fast Start Taq or Jump Start Taq enzymes. Four 5-fold dilutions of DNA, starting with 10 ng (lanes 1–4), were used in reactions containing no blood or 5% and 10% human blood. Lanes M, DNA standard ladder. PCR was performed in a real time cycler using SYBR Green I as a fluorescent dye at concentration 32×. The amplified products were analyzed in 2% agarose gel stained with ethidium bromide (top row), along with the fluorescence detection of the amplification and the melting curves (middle and bottom rows, respectively.) The red, green, blue and yellow curves correspond to background subtracted fluorescence values obtained with the four DNA dilutions in decreasing order.
Thus, a seemingly paradoxical combination of two PCR inhibitors—blood and SYBR Green, turned out to work for the Taq mutants in overcoming the PCR inhibition. To our knowledge, this is the first report on successful real-time PCR detection of genes directly in crude samples containing whole blood at 5% and higher.
High tolerance of the Taq mutants to soil-born inhibitors of PCR
PCR tests with soil samples can also be very problematic due to various potent PCR inhibitors present in the soil, such as humic acid, fulvic acid, polysaccharides and polyphenols. We attempted to find mutants of Taq polymerase that provide improved tolerance of such inhibitors. Initial screening of mutant clones from our mutagenized library of Klentaq1 gene yielded several clones that were functional in the presence of at least 10 ng humic acid per 50 ul reaction, while the w.t. Taq is inactivated by 1 ng of the inhibitor. We also screened the mini-libraries of clones representing the saturating mutagenesis at codon 708 of Taq and Klentaq1 that were used to select the best BR mutants. Interestingly, the two mutants with amino-acid changes responsible for high BR, Klentaq 10 and Taq 22, were most capable of overcoming the humic acid inhibition as well.
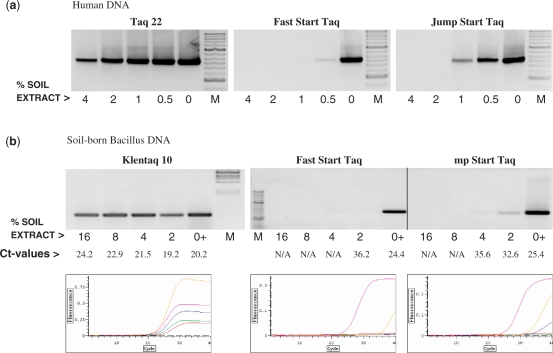
Since the humic acid is not the only soil component that inhibits Taq, we next challenged these enzymes by including crude soil extracts in PCR. As shown in Figure 5, the two mutant enzymes were able to amplify human or bacterial gene targets from soil extract concentrations that are strongly inhibiting the w.t. Taqs. The latter were only efficient after DNA was purified from the soil sample. The real-time detection showed a significant shift in the Ct values of w.t. Taq in the lowest soil concentration as compared to the controls, and no Ct values were available with the higher soil concentrations. On the other hand, the Ct values with Klentaq 10 remained similar in the whole range of soil concentrations tested (Figure 5B). Unlike the blood case, real-time detection of the products amplified with the mutant enzymes using SYBR Green I was possible with normal (1×) concentration of the dye without changing the protocol.
Figure 5.
Direct PCR detection of exogenous and soil-born target genes in crude soil samples by the Taq mutants. (a) A 630-bp target of the human CCR5 gene was amplified with the Taq 22 mutant or the w.t. enzymes Fast Start Taq and Jump Start Taq from 5 ng human DNA in the presence of serial dilutions of a crude soil extract. Control reactions contained no soil extract. (b) A 600-bp target of Bacillus rRNA gene was amplified with the KlenTaq 10 mutant, Fast Start Taq or Jump Start Taq, straight from a crude soil extract. The extract was present at the range of 2–16% in the reaction, as indicated. Control reactions (lanes 0+) contained 10 ng DNA purified from the same soil extract. The amplified products were analyzed in a 2% agarose gel stained with ethidium bromide along with DNA standards ladders (M) or by real-time fluorescence detection of SYBR Green I incorporation (b, bottom row). The pink, yellow, blue, green and red curves reflect the amplification (background subtracted fluorescence values) in the controls and in the presence of the four increasing concentrations of soil extract, respectively.
Resistance of the Taq mutants to major inhibitory components of blood and soil
We next addressed the issue of whether the resistance of Klentaq 10 and Taq 22 to inhibitors in blood and soil in PCR is due to a particular tolerance to some of the known inhibitory fractions or components found in blood and soil. In order to minimize possible template related factors in assessing the tolerance of the enzymes to such inhibitors, we picked a relatively short and easy, 250 bp Lambda DNA target. In addition, all reactions contained betaine, a known PCR facilitator and enhancer (4,6,46).
The mutant enzymes and w.t. Taq were tested in PCR in the presence of serial dilutions of whole human blood and plasma (obtained from the same donor), as well as serum IgG fraction, hemoglobin, hemin (a hemoglobin prosthetic group-containing breakdown product) and lactoferrin, for which a PCR inhibitory effect has been reported (9,11,12). The enzymes were also challenged with a crude soil extract and humic acid, the major PCR inhibitor found in soil. The results showed significantly higher resistance of the mutant enzymes as compared to the w.t. Taq to all inhibitors tested except for the lactoferrin (Figure 6). The mutant enzymes remained functional in all tested concentrations of blood and plasma (1.5–25%), hemoglobin (0.15–2.5 mg/ml), and soil extract (1–15%), and tolerated the IgG fraction at 25 μg/ml (Klentaq 10) and 12.5 μg/ml (Taq 22), hemin at 3.2 μM, lactoferrin up to15 μM (Klentaq 10) and 30 μM (Taq 22), and humic acid at 400 ng/ml (Klentaq 10) or 200 ng/ml (Taq 22). In general, Klentaq 10 proved to some extent more resistant to most of those inhibitors relative to Taq 22, consistent with its more robust performance typically observed with unfractionated blood and soil samples. On the other hand, the w.t. enzyme showed only a partial resistance to the IgG fraction (up to 6.25 μg/ml), but remained fully active in the presence of 30 μM lactoferrin, matching the tolerance of the mutants to this inhibitor.
The impressive multi-resistance of the mutant enzymes may well explain their functionality in crude blood and soil samples. These data also suggest that at least the inhibition by lactoferrin alone could be a limiting factor in their performance.
Effect of blood and soil inhibitory components on DNA polymerase activity
Another approach in analyzing Taq enzyme inhibition was to assess the effect of the blood and soil inhibitory components on the catalytic activity of the enzyme. To do so, we performed a nucleotide-incorporation assay, similar to the one used conventionally for determining DNA polymerase activity. In this assay the enzyme substrate is an activated, partially DNAse I digested DNA, where the enzyme acts mainly in filling in nicks in DNA and the DNA extension is limited to a few nucleotides. The enzyme activity is manifested by the rate of radiolabeled nucleotide incorporation into DNA. The same blood and soil-derived inhibitors, tested for their effect on PCR in Figure 6, were included in this assay. The results, shown in Table 1, revealed a general inhibition, not selective for the w.t. Taq, by human plasma, IgG fraction, hemoglobin and hemin. Remarkably, instead of inhibition, a total stimulation was observed with lactoferrin. At lower concentrations, the plasma, serum IgG and hemoglobin also had a stimulatory effect on all enzymes, probably due to a general enhancement of their activity by the increased protein concentration. On the other hand, whole blood and soil extract showed certain predominant inhibitory effect on the w.t. Taq. Relatively most selective inhibition was observed with humic acid, where the w.t. enzyme was most inhibited (bottom row of Table 1). Interestingly, humic acid present at the lowest concentrations tested (100–400 ng/ml, tolerated in PCR by Klentaq 10 and Taq 22) (Figure 6) also had certain stimulatory effect only on the activity of the mutant enzymes. The reason for this effect is unclear and remains to be analyzed. Thus, the main functional advantage for the mutants revealed by this assay was their higher tolerance to humic acid, and to some extent to whole blood and soil extract. These results imply that a more selective effect of hemoglobin, hemin and serum IgG on the w.t. enzyme should be exerted in the context of PCR.
Effect of the DNA extension time on PCR efficiency in the presence of blood and soil inhibitors
As some of the known PCR inhibitors found in blood and soil did not show a predominant effect on w.t. Taq activity in the non-PCR assay described above, we looked for another possible way some of these inhibitors may affect DNA amplification. One can assume that in crude, especially blood-containing PCR samples, the DNA template is less available, as it is likely to be bound at least by proteins. This in turn could affect the speed or processivity of the enzyme. Or, alternatively, the enzyme speed could be directly reduced by some inhibitors. Therefore, we examined whether a longer extension time could compensate for lowed speed. DNA amplification of identical samples, containing varying concentrations of a PCR inhibitor, was performed in a no ramp-time PCR cycle, using differing extension times. In the experiment shown in Figure 7, two human targets were amplified in duplex PCR directly from blood (Figure 7A), and purified human DNA was used to amplify the same targets in the presence of a crude soil extract (Figure 7B). We found that doubling the extension time from 2 min to 4 min, facilitated the performance of w.t. Taq, which was able to amplify to some extent the shorter target in 5% blood (the enzyme was not functional with the longer target). In the soil samples series, the w.t. enzyme was only functional in the control reactions containing no soil and a longer extension time did not help, while the mutant enzymes shifted their performance to a higher soil concentration when the extension time was increased. We also observed some reverse correlation between the target size and enzyme performance at a given extension time and concentration of soil inhibitors. In control reactions lacking inhibitors both targets were efficiently amplified regardless of the extension time, which shows that the target ‘difficulty’ was not an issue.
We tried to analyze this finding in more details by using the ‘easy’ 250-bp Lambda DNA target alone in similar assay using three different extension times of 30, 60 and 120 s. As shown in Figure 8A, with this target we observed a significantly more pronounced effect of the extension time on the w.t. Taq performance, ranging from no product in 2.5% blood to quite efficient amplification in 10% blood at 30 s and 120 s extension time, respectively. In contrast, the performance of the mutant enzymes in the entire blood concentration range tested (up to 25%) was not compromised even with the shortest extension time.
In soil extract (Figure 8B), however, the amplification efficiency of both the mutants and the w.t. enzyme did depend on the extension time. Using the same assay, we also tested the effect of the two major blood and soil PCR inhibitors, hemin and humic acid (Figure 8C and D). We found that none of the enzymes benefit significantly from a longer extension time in the presence of hemin, implying that this inhibitor may not interfere with the enzyme speed. On the other hand, the results with humic acids were similar to those with soil extract, where again with all enzymes a longer extension time was necessary for performance at higher inhibitor concentration.
The results of these tests shown in Figures 7 and 8 support the idea that the enzyme speed and/or processivity can be reduced in the presence of blood and soil, and demonstrate that the humic acid is responsible for this effect in soil samples. In the case of PCR in blood, longer extension time was required for an improved, yet limited performance of w.t. Taq, more pronounced with short and easy targets. Unlike the w.t. enzyme, the mutants remained functional at short extension times, consistent with a less or not affected DNA elongation speed.
DISCUSSION
In our attempt to design a better enzyme for clinical and forensic PCR applications, we obtained mutant forms of Taq and Klentaq1 DNA polymerases that remain fully functional in the presence of PCR inhibitors in blood and soil. The acquired mutant enzymes can endure concentrations of whole blood and crude soil extracts highly exceeding those tolerated by the native Taq enzyme. In addition, these mutants remain active in higher concentrations of SYBR Green and other fluorescent dyes. Evaluation of the general PCR performance of the mutant enzymes with purified DNA template showed that their sensitivity, specificity, and fidelity were not compromised relative to the w.t. enzyme (data not shown).
We observed that a large fraction of DNA targets tested benefit from adding betaine to PCR performed with the mutant enzymes in the presence of blood or soil. In the case of blood this effect was more pronounced with endogenous targets and in higher concentration of blood (usually >5%), especially for the Taq 22 mutant. Betaine is a known PCR enhancer, which lowers the melting temperature of DNA by interfering with the hydrogen bonds of the G:C pairs (46). It can also facilitate PCR in the presence of some inhibitors (3,4,6). Betaine alone, however, was not sufficient for overcoming the inhibition of w.t. Taq by high concentrations of blood or soil, as well as individual PCR inhibitors derived from blood and soil (Figures 6–8).
The mechanism of action of the PCR inhibitors present in blood and soil is not yet understood. The overall inhibition of PCR could be a collective result of inactivation and/or degradation of the enzyme and blocking or degradation of the target DNA and primers. Humic acid, fulvic acid, polysaccharides and heavy metals are among the known PCR inhibitors in soil (16,17). The humic acid is a poorly characterized mixture of polyphenolic compounds (15–17). Among the few identified major PCR inhibitors in blood cells are hemoglobin, its derivative hemin and lactoferrin, both containing iron, as well as an IgG serum fraction (9–12).
Besides inhibition, another complication of working with crude blood samples stems from the strong quenching effect of blood on the dye fluorescence in real-time PCR, most likely caused by the heme. It is also likely that the blood may cause some modification, degradation or adsorption of the dye. The mutant enzymes were able to overcome the fluorescence quenching problem, which can be explained by their tolerance to higher dye concentrations, used in our modified protocol to compensate for the low fluorescence.
We attempted to determine what functional feature(s) of the novel Taq mutants provides the advantage of their better PCR performance in the presence of those inhibitors. The fact that the specific target and mechanism of action of the PCR inhibitors is still obscure significantly complicates this issue.
Based on the differential PCR inhibition test of the major known PCR inhibitors found in blood and soil (Figure 6), the functionality of the mutants in whole blood or plasma can be explained at least by their high tolerance to hemoglobin/hemin and the serum IgG fraction, exceeding that of the w.t. enzyme about 30 times and 4 times, respectively. We exclude lactoferrin, as the mutants could not tolerate this inhibitor any better than the w.t. Taq. The resistance of the mutant enzymes to PCR inhibition in soil samples can be attributed to their high tolerance to humic acid (800 ng/ml vs. 24 ng/ml for the w.t. enzyme). On the other hand, in the DNA polymerase assay with the above inhibitors, shown in Table 1, only the humic acid, and to some extent soil extract and whole blood, had a predominant effect on the catalytic activity of the w.t. Taq. The relatively high DNA concentration used in this assay (12 μg/50 ul) might potentially diminish the overall inhibition, as DNA could outcompete some inhibitors. The complete lack of inhibition by lactoferrin could be related to such effect of DNA, although it may also indicate that this inhibitor exerts its effect only in PCR conditions. Similarly, the lack of selective inhibition of the w.t. enzyme by IgG, hemoglobin and hemin in this assay, otherwise observed in DNA amplification, suggests that they should interfere with some PCR-specific steps. Consistent with this view, we observed that a longer extension time in the PCR cycle can partially compensate for the inhibition of the w.t. enzyme both in blood and soil, as well as that of the mutant enzymes in soil. A likely explanation of this observation is that inhibitors in such samples may affect the enzyme speed and/or processivity. We found that the humic acid in soil is implicated in this effect. As mentioned above, humic acid also affects the enzyme activity in a non-PCR assay in which the enzyme speed may not be critical. The reported metal chelating potential and DNA-binding activity of the humic acid (47,48) may be involved in the inhibition.
The blood component(s) potentially interfering with the enzyme speed remains to be found. The data shown in Figure 8C suggest that the major blood inhibitor, hemin, is not involved in this inhibition aspect. Additional tests showed that this is true for hemoglobin, lactoferrin and the IgG serum fraction (data not shown). Since all four blood inhibitors were tested individually, our data do not rule out that a combination of them, if not any additional factor, may affect the enzyme speed, which would explain the results with whole blood samples shown in Figures 7 and 8A.
The Klentaq 10 mutant typically performed relatively better than Taq 22 in PCR reactions containing inhibitors. This most likely stems from the fact that the parental Klentaq 1 enzyme is more robust than the native Taq, and, as mentioned earlier, it is more resistant than w.t. Taq to blood and soil PCR inhibitors, as well as to the SYBR Green inhibition. When challenged with soil inhibitors, Klentaq 10 tolerated more humic acid, while Taq 22 outperformed Klentaq 10 in soil extract, when shorter extension times and easier-to-amplify targets were used (Figure 8B and D). A likely reason for this observation could be that the Klentaq enzyme is generally slower than the full-length Taq, which is true for their mutant versions as well (M.K., unpublished observation). This demonstrates that a higher speed of the enzyme is not sufficient for overcoming PCR inhibition. Consistent with this concept, the benefit for the w.t. Taq from a longer extension time in soil extract and humic acid was none, or still limited by the inhibitor concentration, and it was more clear in blood only with relatively shorter and ‘easy’ exogenous targets (Figures 7 and 8).
Besides the speed of the enzyme, another potential target of PCR inhibition could be the enzyme interaction with the DNA template. It has been proposed that hemin inhibits the cytoplasmic erythroid DNA polymerase by binding the enzyme and blocking the enzyme:DNA complex formation (49). We performed serial tests in a filter binding assay analogous to the one used in that report, and found that hemin and humic acid strongly affect the interaction of Taq polymerase with DNA. In this non-PCR assay the effect of the two inhibitors on the mutants and w.t. enzyme was comparable. Interestingly, we reproducibly observed a stronger DNA affinity with the mutant enzymes (data not included). Further studies could determine if this feature of the mutants is relevant to their performance, in particular to their ability to compete with template bound inhibitors.
In summary, in the scope of the assays used here we propose that both blood and soil can exhibit at least two aspects of PCR inhibition: A, affecting the catalytic activity of DNA polymerase, and B, reducing the enzyme speed/processivity, either directly or by blocking the DNA template. In the case of soil PCR, the advantage of the mutants is apparently confined more in the first aspect, based on their resistance to humic acid inhibition, while in blood PCR the mutants would outperform the w.t. Taq in the two aspects, as in addition to their combined resistance to hemoglobin/hemin and serum IgG, they seem to be capable of more efficiently traversing inhibitor-blocked DNA templates.
Single amino-acid changes at Glu708 were implicated in both desired phenotypes of blood and soil resistance of the mutants. Functional analysis revealed that the same amino-acid substitutions in that position were responsible both for the blood and soil resistance features. Interestingly, not all of the selected functional changes were equally efficient in the context of Klentaq1 and Taq DNA polymerase. Lys and Leu substitutions were efficient in both enzymes, while Asn, Gln and Ile were only functional in Taq and Trp was only functional in Klentaq1. The two phenotypes were not tightly coupled. The Klentaq 7 and 12 mutants (Glu708Trp and Glu708Leu) exhibited a significant resistance to blood but not to soil inhibitors, while another Klentaq1 mutant (Glu708Arg) was predominantly soil resistant (data not shown).
The changes of the acidic and hydrophilic w.t. Glu 708 were relatively more drastic in the cases of Lys, Leu and Ile mutants by reversing the charge or polarity. However, more subtle substitutions with the two acidic amides Gln and Asn were highly efficient in producing the desired phenotype in Taq as well. It still remains to be determined how the amino-acid substitutions at codon 708 correlate with the SYBR Green resistance aspect of the mutants, and whether the enzyme speed is affected by the dye.
Our results strongly indicate that Glu708 is critical for the catalytic function. In accordance with this, we found that the substitutions Pro and Cys, potentially causing either kink in the chain or formation of disulfide bonds, inactivated both the Klentaq1 and Taq enzyme.
The position of Glu 708, depicted in Figure 9, falls in an alpha-helix region on the surface of the enzyme, known as ‘P’ domain (residues 704–717). This domain is situated about 40 residues apart from the ‘finger’ domain which binds the incoming dNTPs and interacts with the single-stranded DNA template (50,51). The residue locates at the hinge region, which is believed to allow movement of the finger domains at microsecond time-scale as the polymerase evaluates the correctness of base pairs before incorporation. During incorporation, the fingers move even to collapse the enzyme active site for reaction (51,52). The coincidence of resistance phenotype and speed phenotype may indicate that some PCR inhibitors interact at the surface of the enzyme in such a way that they slow down the movement of the fingers, which effect may be reduced with the 708 mutations.
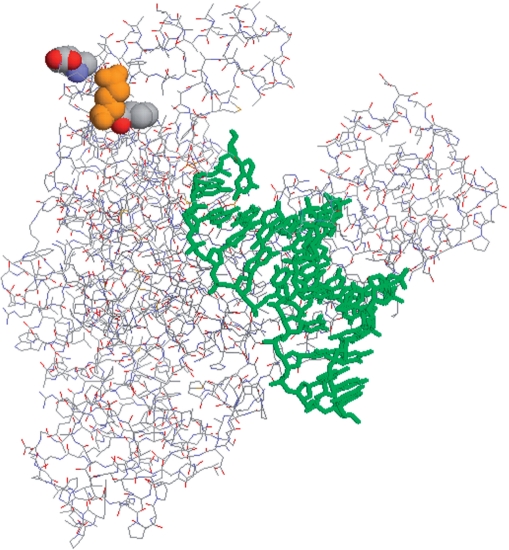
Figure 9.
Location of the mutagenized codons in Klentaq. Crystal structure from pdb file 2KTQ (51) emphasizes the location of the residues mutated in KlenTaq 10. Using full-length Taq numbering, w.t. residues are rendered in spacefill for the mutated positions 626 707 (Cs-mutations), and 708 (orange, blood and soil-resistance mutation) using the program Rasmol 2.7.1 (53). Upper right is the thumb domain, upper left is the fingers domain, where the mutations can be seen at the surface of the hinge point (knuckles) of the fingers. Primer and template DNA strands are rendered in stick mode (green) in the active site cleft.
In a previous study we found that an adjacent residue, Ile 707 renders the enzyme cold-sensitive when changed to Leu. Such Cs-mutants perform as highly specific ‘hot-start’ PCR enzymes. Remarkably, several neighboring codons implicated in the Cs-phenotype were mapped in this area as well, including the Glu 708 (moderately cold-sensitive when changed to Asp) (39).
At present it is unclear if this functional overlap of seemingly unrelated phenotypes has some functional relevance. Regardless, control tests with our best Cs-mutants (data not shown) and commercial hot-start enzymes (the present data) showed that neither the cold sensitivity nor the hot start feature alone is sufficient to overcome the blood or soil inhibition in PCR.
The novel mutant Taq and Klentaq enzymes should provide a simplified and efficient protocol for direct DNA amplification in crude blood and soil samples. Therefore, they could significantly facilitate some clinical and forensic PCR-based tests.
FUNDING
This work was supported by the National Institutes of Health (SBIR Grant 2R44GM07340102 to M.K.); and the US Department of Agriculture (SBIR Grant 2006-03071 to M.K.). Funding for open access charge: National Institutes of Health (SBIR Grant 2R44GM07340102 to M.K.).
ACKNOWLEDGEMENTS
We thank Dr Zh. Zhang and J. Hartman for critical reading of the manuscript and helpful comments. We also thank Dr W. Jack (New England Biolabs) for kindly providing the human MT gene primers and sequence.
Conflict of interest statement. None declared.
REFERENCES
- 1.Radstrom P, Knutsson R, Wolffs P, Lövenklev M, Löfström C. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol. Biotechnol. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 2.Lantz P-G, Al-Soud WA, Knutsson R, Hahn-Haegerald B, Radstrom P. Biotechnical use of the polymerase chain reaction for microbial analysis of biological samples. Biotechnol. Annu. Rev. 2000;5:87–130. doi: 10.1016/s1387-2656(00)05033-x. [DOI] [PubMed] [Google Scholar]
- 3.Altwegg M, Verhoef J. Amplification methods in diagnostic microbiology. J. Microbiol. Methods. 1995;23:1–2. [Google Scholar]
- 4.Al-Soud WA, Radstrom P. Effect of amplification facilitators on diagnostic PCR in the presence of blood, feces and meat. J. Clin. Microbiol. 2000;38:4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Soud AW, Radstrom P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frackman S, Kobs G, Simpson D, Storts D. Betaine and DMSO: enhancing agents for PCR. Promega Notes. 1998;65:27. [Google Scholar]
- 7.Topal MD, Sinha NK. Products of bacteriophage T4 genes 32 and 45 improve the accuracy of DNA replication in vitro. J. Biol. Chem. 1983;258:12274–12279. [PubMed] [Google Scholar]
- 8.Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Soud AW, Jönsson LJ, Radstrom P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Franchis R, Cross NC, Foulkes NS, Cox TM. A potent inhibitor of Taq polymerase copurifies with human genomic DNA. Nucleic Acids Res. 1988;16:10355–10366. doi: 10.1093/nar/16.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Soud WA, Radstrom P. Purification ands characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 13.Kuske CR, Banton KL, Adorada DL, Stark PC, Hill KK, Jackson PJ. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 1998;64:2463–2472. doi: 10.1128/aem.64.7.2463-2472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poussier S, Cheron J, Couteau A, Luisetti J. Evaluation of procedures for reliable PCR detection of Ralstonia solanacearum in common natural substrates. J. Microbiol. Methods. 2002;51:349–359. doi: 10.1016/s0167-7012(02)00111-2. [DOI] [PubMed] [Google Scholar]
- 15.Tsai Y-L, Olson BH. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson RJ, Blackwell B. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 2000;46:633–642. doi: 10.1139/w00-043. [DOI] [PubMed] [Google Scholar]
- 17.Yeates C, Gillings MR, Davison AD, Altavilla N, Veal DA. Methods for microbial DNA extraction from soil for PCR amplification. Biol. Proceed. Online. 1998;1:40–47. doi: 10.1251/bpo6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaMontagne MG, Michel FC, Jr, Holden PA, Reddy CA. Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J. Microbiol. Methods. 2001;49:255–264. doi: 10.1016/s0167-7012(01)00377-3. [DOI] [PubMed] [Google Scholar]
- 19.Watson RJ, Blackwell B. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 2000;46:633–642. doi: 10.1139/w00-043. [DOI] [PubMed] [Google Scholar]
- 20.Forbes BA, Hicks KE. Substance interfering with direct detection of mycobacterium tuberculosis in clinical specimens by PCR: effects of bovine serum albumin. J. Clin. Microbiol. 1996;34:2125–2128. doi: 10.1128/jcm.34.9.2125-2128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morata P, Queipo-Ortuno I, Colmenero J. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for diagnosis of human brucellosis. J. Clin. Microbiol. 1998;36:2443–2446. doi: 10.1128/jcm.36.9.2443-2446.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Soud AW, Lantz P-G, Bäckman A, Olcen P, Radstrom P. A sample preparation method which facilitates detection of bacteria in blood cultures by the polymerase chain reaction. J. Microbiol. Methods. 1998;32:217–224. [Google Scholar]
- 25.Klein A, Barsuk R, Dagan S, Nusbaum O, Shouval D, Galun E. Comparison of methods for extraction of nucleic acid from hemolytic serum for PCR amplification of hepatitis B virus DNA sequences. J. Clin. Microbiol. 1997;35:1897–1899. doi: 10.1128/jcm.35.7.1897-1899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cattaneo C, Graig OE, James NT, Bolton H. Comparison of three DNA extraction methods on bone and blood stains up to 43 years old and amplification of three different gene sequences. J. Forensic Sci. 1997;42:1126–1135. [PubMed] [Google Scholar]
- 27.Bourke MT, Scherczinger CA, Ladd C, Lee HC. NaOH treatment to neutralize inhibitors of Taq polymerase. J. Forensic Sci. 1999;44:1046–1050. [PubMed] [Google Scholar]
- 28.Harry M, Gambier B, Bourezgui Y, Garnier-Sillam E. Evaluation of purification procedures for DNA extracted from organic rich samples: interference with humic substances. Analusis. 1999;27:439–441. [Google Scholar]
- 29.Lebuhn M, Effenberger M, Garces G, Gronauer A, Wilderer PA. Evaluating real-time PCR for the quantification of distinct pathogens and indicator organisms in environmental samples. Water Sci. Technol. 2004;50:263–270. [PubMed] [Google Scholar]
- 30.Moreira D. Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res. 1998;26:3309–3310. doi: 10.1093/nar/26.13.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneegurt MA, Dore SY, Kulpa CF. Direct extraction of DNA from soils for studies in microbial ecology. Curr. Issues Mol. Biol. 2003;5:1–8. [PubMed] [Google Scholar]
- 32.Burgmann H, Pesaro M, Widmer F, Zeyer J. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods. 2001;45:7–20. doi: 10.1016/s0167-7012(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 33.Kramvis A, Bukovzer S, Kew MC. Comparison of hepatitis B virus DNA extractions from serum by the QIAamp blood kit, Genereleaser, and the phenol-chloroform method. J. Clin. Microbiol. 1996;34:2731–2733. doi: 10.1128/jcm.34.11.2731-2733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ong JL, Loakes D, Jaroslawski S, Too K, Holliger P. Directed evolution of DNA polymerase, RNA polymerase and reverse transcriptase activity in a single polypeptide. J. Mol. Biol. 2006;361:537–550. doi: 10.1016/j.jmb.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 35.Vichier-Guerre S, Ferris S, Auberger N, Mahiddine K, Jestin JL. A population of thermostable reverse transcriptases evolved from Thermus aquaticus DNA polymerase I by phage display. Angew. Chem. Int. Ed. Engl. 2006;45:6133–6137. doi: 10.1002/anie.200601217. [DOI] [PubMed] [Google Scholar]
- 36.Schönbrunner NJ, Fiss EH, Budker O, Stoffel S, Sigua CL, Gelfand DH, Myers TW. Chimeric thermostable DNA polymerases with reverse transcriptase and attenuated 3′-5′ exonuclease activity. Biochemistry. 2006;45:12786–12795. doi: 10.1021/bi0609117. [DOI] [PubMed] [Google Scholar]
- 37.Gloeckne RC, Sauter KB, Marx A. Evolving a thermostable DNA polymerase that amplifies from highly damaged templates. Angew. Chem. Int. Ed. Engl. 2007;46:3115–3117. doi: 10.1002/anie.200603987. [DOI] [PubMed] [Google Scholar]
- 38.Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 39.Kermekchiev MB, Tzekov A, Barnes W. Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR. Nucleic Acids Res. 2003;31:6139–6147. doi: 10.1093/nar/gkg813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai UJ, Pfaffle PK. Single-step purification of a thermostabile DNA polymerase expressed in Escherichia coli. BioTechniques. 1995;19:780–784. [PubMed] [Google Scholar]
- 41.Barnes WM. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- 42.Nath K, Sarosy JW, Hahn J, Di Como CJ. Effects of ethidium bromide and SYBR green I on different polymerase chain reaction systems. J. Biochem. Biophys. Methods. 2000;42:15–29. doi: 10.1016/s0165-022x(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 43.Monis PT, Giglio S, Saint CP. Comparison of SYTO9 and SYBR Green I for real time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 2005;340:24–34. doi: 10.1016/j.ab.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 44.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 45.Gudnason H, Dufva M, Bang DD, Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rees WA, Yager TD, Korte J, von Hippel PH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 47.Zipper H, Buta C, Lämmle K, Brunner H, Bernhagen J, Frank Vitzthum F. Mechanisms underlying the impact of humic acids on DNA quantification by SYBR Green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res. 2003;31:e39. doi: 10.1093/nar/gng039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutlovic D, Gamulin S, Definis-Gojanovic M, Gugic D, Andjelinovic S. Interaction of humic acids with human DNA: proposed mechanisms and kinetics. Electrophoresis. 2008;7:1467–1472. doi: 10.1002/elps.200700699. [DOI] [PubMed] [Google Scholar]
- 49.Byrnes J, Downey K, Esserman L, So A. Mechanism of hemin inhibition of erythroid cytoplasmic DNA polymerase. Biochemistry. 1975;14:796–799. doi: 10.1021/bi00675a023. [DOI] [PubMed] [Google Scholar]
- 50.Korolev S, Nayal M, Barnes WM, DiCera E, Waksman G. Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 25 Å resolution: structural basis for thermostability. Proc. Natl Acad. Sci. USA. 1995;92:9264–9268. doi: 10.1073/pnas.92.20.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Korolev S, Waksman G. Chrystal structure of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levene M, Korlach J, Turner S, Foquet M, Craighead H, Webb W. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 53.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–382. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]