Abstract
Genomic DNA of African trypanosomes contains a hypermodified thymidine residue termed base J (β-d-glucosyl-HOMedU). This modified base is localized primarily to repetitive DNA, namely the telomeres, and is implicated in the regulation of antigenic variation. The base is synthesized in a two-step pathway. Initially, a thymidine residue in DNA is hydroxylated by a thymidine hydroxylase (TH). This intermediate (HOMedU) is then glucosylated to form base J. Two proteins involved in J synthesis, JBP1 (J binding protein 1) and JBP2, contain a putative TH domain related to the family of Fe2+/2-oxoglutarate-dependent hydroxylases. We have previously shown that mutations in the TH domain of JBP1 kill its ability to stimulate J synthesis. Here we show that mutation of key residues in the TH domain of JBP2 ablate its ability to induce de novo J synthesis. While the individual JBP1 null and JBP2 null trypanosomes have reduced J levels, the deletion of both JBP1 and JBP2 generates a cell line that completely lacks base J but still contains glucosyl-transferase activity. Reintroduction of JBP2 in the J-null trypanosome stimulates HOMedU formation and site-specific synthesis of base J. We conclude that JBP2 and JBP1 are the TH enzymes involved in J biosynthesis.
INTRODUCTION
Base J is a modification of DNA found in all members of the kinetoplastid family as well as the closely related diplonema and euglena. Base J is primarily associated with repetitive DNA such as telomeres, 177, 50 and 70 bp repeats (1,2). In Trypanosoma brucei, J is developmentally regulated and found only in bloodstream stage parasites (3). This, along with its association with transcriptionally silent but not with active telomeric surface protein expression sites (3) has led to the hypothesis that the modified base functions to regulate antigenic variation, a mechanism which allows the parasite to persist in the mammalian host. However, the function of this modified base has yet to be elucidated.
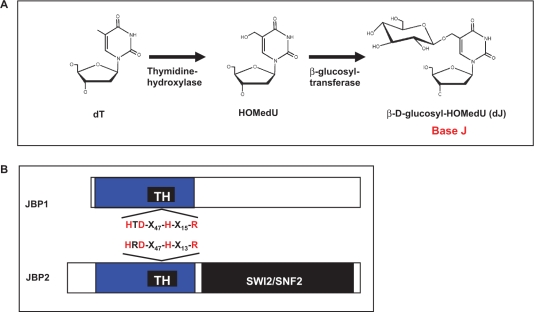
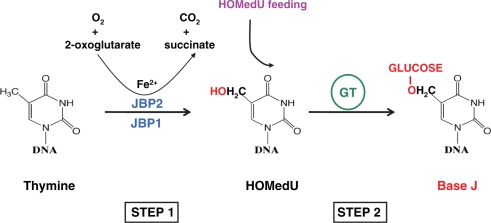
Whilst the function of J remains a mystery, much is known about its biosynthesis. J is synthesized in a two-step pathway (4) (Figure 1A). Step 1 involves the oxidation of thymidine residues, by a thymidine hydroxylase (TH), forming the intermediate HOMedU in DNA. This intermediate is then glucosylated by a glucosyl-transferase to form base J. Two key proteins involved in regulation of J synthesis have been identified; JBP1 (J binding protein 1) and JBP2 (5,6). JBP1 binds base J in double stranded DNA and stimulates additional J synthesis (5,7,8). JBP2 was identified in silico based on its N-terminal homology to JBP1 (34% identity, 45% similarity) (Figure 1B) and was shown to stimulate de novo J synthesis (6,9). Unlike JBP1, JBP2 does not bind to base J. However, the C-terminal domain of JBP2 contains a SWI2/SNF2 domain that may allow specific JBP2-chromatin interactions, thus regulating site specific de novo J synthesis. The SWI2/SNF2 domain of JBP2 contains the seven characteristic conserved motifs present in members of the family of ATPase/DNA helicase enzymes. The importance of the SWI2/SNF2 domain in JBP2 function was confirmed by mutation of the helicase motifs involved in ATP hydrolysis (6). Such mutations kill the ability of the protein to stimulate de novo J synthesis.
Figure 1.
Base J is synthesized in a two-step pathway. (A) Schematic model of the J-biosynthesis pathway. Step 1 involves the oxidation of thymidine (dT) residues in DNA forming the intermediate HOMedU. In step 2 the intermediate (HOMedU) is then glucosylated, by a glucosyl-transferase enzyme. This is then glucosylated, by a glucosyl-transferase enzyme, forming base J (dJ). The initial site-specific hydroxylation of thymidine, by TH, is thought to represent the key regulatory step of J biosynthesis. (B) Schematic diagram of the two proposed THs: JBP1 and JBP2. Shown in blue is the N-terminal region shared between both proteins. Within this region is the ∼70-amino-acid motif, indicated by the solid black box, which is related to the functional domain of members the Fe2+/2-oxoglutarate dependent hydroxylase family. Indicated above and below the putative TH motif (TH) for JBP2 and JBP1 is the amino acid signature shared among members of this hydroxylase family. The four key residues within this motif that have been implicated in catalysis by other members of this family of proteins (including JBP1), and which were mutated to alanine in JBP2 for the studies presented here, are highlighted in red. The C-terminal SWI2/SNF2 domain of JBP2 is shown in black.
Recently, we proposed that JBP1 and JBP2 are the TH enzymes that stimulate the formation of HOMedU during Step 1 of the J-synthesis pathway (9). We have shown that a region within the homologous N-terminal domain of JBP1 and JBP2 contains a motif that is conserved among members of the Fe2+/2-oxoglutarate dependent dioxygenase (hydroxylase) family of enzymes (Figure 1B). Four residues within this motif are critical for the coordination of Fe2+ and 2-oxoglutarate binding as well as functionality of this family of enzymes (10). Amino acid substitution of these residues within the putative hydroxylase region of JBP1 abolished its ability to stimulate J synthesis in the bloodstream form JBP1 null trypanosome (11). While it is tempting to hypothesize that both JBP1 and JBP2 are the TH enzymes, the comparative functional analysis of JBP2 has not been performed. Here we show that mutations within the proposed TH domain of JBP2 inhibit its ability to initiate de novo J synthesis. In addition, we demonstrate that the deletion of both JBP1 and JBP2 generates a bloodstream form trypanosome cell line that lacks the modified base (J null). This cell line still contains the glucosyl-transferase enzyme required for the Step 2 of the synthesis pathway since HOMedU feeding results in the formation of base J. Reexpression of JBP2 and JBP1 in the J null cell line rescues J synthesis. The expression of JBP2 alone stimulates site-specific de novo synthesis and concomitant expression of JBP1 in this cell line amplifies J synthesis within the specific regions of the genome seeded by JBP2. Furthermore, the reexpression of JBP2 in a J null background generates the intermediate HOMedU in genomic DNA. These results confirm the identity of JBP1 and JBP2 as the TH enzyme(s). A model is presented for the mechanism of JBP catalyzed thymidine hydroxylation and to explain why two enzymes are required to regulate J biosynthesis.
RESULTS
Mutation of the proposed hydroxylase domain of JBP2 inhibits de novo J synthesis
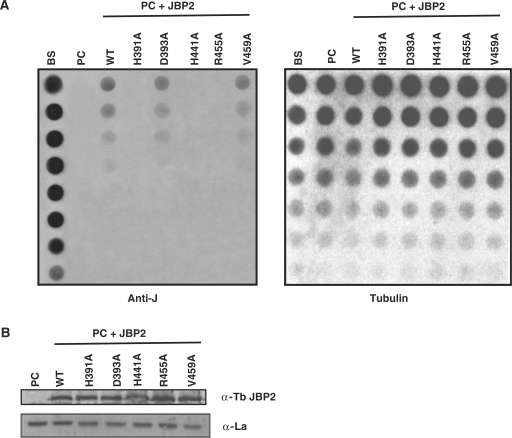
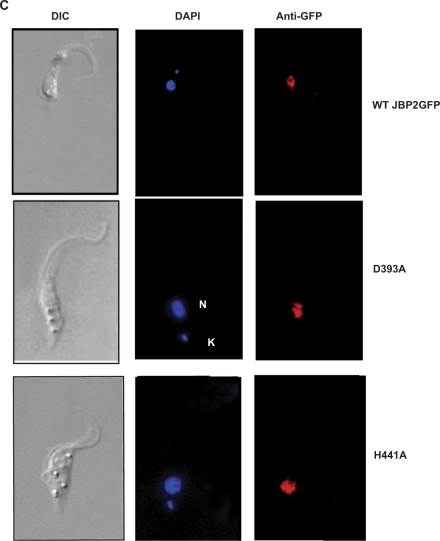
It was previously shown that alanine substitution of four key conserved residues critical for the function of Fe2+/2-oxoglutarate dependent hydroxylases in the N-terminal domain of JBP1 ablates the proteins ability to stimulate J synthesis in vivo (11). We decided to take a similar approach to address the importance of these residues in de novo synthesis by JBP2. Alanine substitutions were made in the four critical residues within the conserved HRDX47HX13R motif (Figure 1B). As a control, we made a valine to alanine substitution in a residue found outside of the proposed catalytic domain. We expressed either the wild type (WT) JBP2 or the mutant JBP2 proteins in insect stage trypanosomes, which do not contain the modified base and determined the J content of each cell line. J levels were assessed by DNA dot blot analysis using an antibody against base J. This analysis revealed that mutations made within the key catalytic domain ablated the ability of JBP2 to initiate de novo J synthesis (Figure 2A). Western blot analysis revealed that both WT and mutant JBP2 proteins are expressed at similar levels (Figure 2B), and immunofluorescence analysis showed that all localize to the nucleus (Figure 2C and data not shown). Therefore, the failure of the mutant proteins to function is not due to differential expression or localization within in the cell. While the aspartic acid mutation does not ablate de novo J synthesis in procyclic trypanosomes, it does cause a slight reduction in activity when compared with WT JBP2. We are uncertain as to why this alanine substitution does not ablate JBP2 function. However, other members of the Fe2+/2-oxoglutarate hydroxylase family exhibit flexibility in the importance of the Asp residue within the conserved motif in iron coordination and catalysis (12).
Figure 2.
Conserved residues with in the TH domain of JBP2 are critical for the protein to stimulate de novo J synthesis. (A) Dot-blot analysis of the JBP2 TH mutants. DNA was isolated from procyclic (insect stage) cells expressing either WT or mutant JBP2 and analyzed for J content by spotting DNA diluted onto a membrane and incubated with anti-J antisera. BS, WT bloodstream form cells; PC, WT procyclic cells; PC+JBP2, procyclic cells expressing JBP2 GFP; WT, procyclic cells expressing WT JBP2. H391A, D393A, H441A, R445A are procyclic cells expressing mutant versions of JBP2 as indicated in Figure 1. V459A is a control substitution of a residue outside the catalytic domain. (B) Expression of WT and mutant JBP2 in insect stage cells. Procyclic trypanosomes expressing either WT or mutant JBP2 were analyzed by western blot using anti-JBP2 antisera. Anti-La antisera was used as a loading control. Extract from untransfected procyclic cells is included as a negative control. (C) JBP2-GFP expression in procyclic trypanosomes was visualized microscopically by anti-GFP staining on formaldehyde fixed cells. DAPI staining reveals the localization of the nucleus (N) and the kinetoplast (K). WT JBP2, procyclic cells expressing WT protein; D391A, H441A are the corresponding mutant JBP2-GFP proteins.
The deletion of both JBP1 and JBP2 generates a cell line null for the modified base
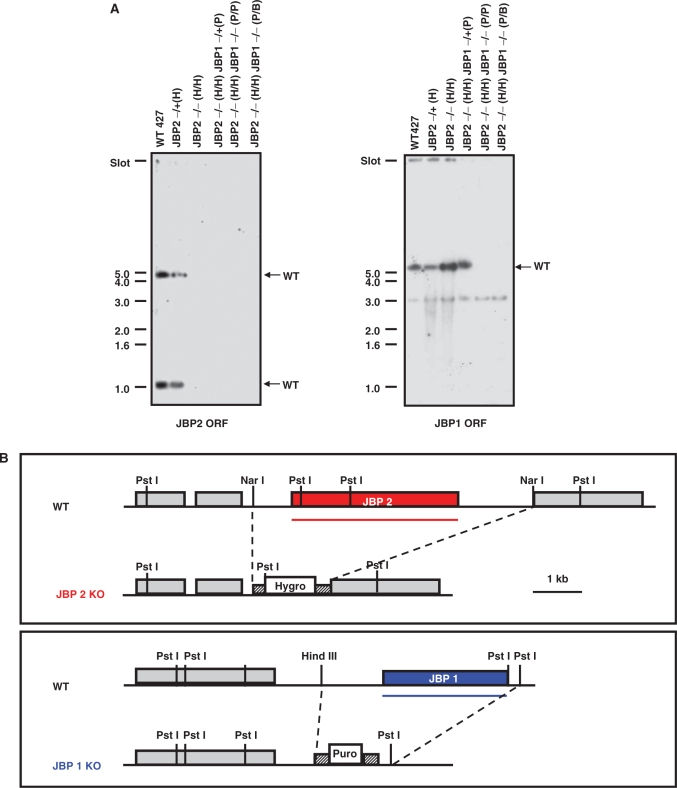
Cell lines null for either JBP1 or JBP2 have been generated and analyzed (9,13). While the reduction of base J levels in each of these cell lines indicated a role for JBP1 and JBP2 in J biosynthesis, presumably the activity of the remaining JBP maintained J synthesis to varying degrees. If JBP1 and JBP2 are the TH enzymes involved in J-biosynthesis, the deletion of both would be required to generate a trypanosome cell line completely lacking J synthesis. Therefore, we constructed a cell line in which both JBP1 and JBP2 were deleted from the trypanosome genome. To do this, we knocked out both alleles of JBP1 from the JBP2−/− cell line (see Methods section). Southern blot analysis confirmed deletion of both alleles of JBP1 and JBP2 from the genome of the resulting cell line (JBP2−/−/JBP1−/−) (Figure 3). J levels were quantified in the JBP2−/−JBP1−/− cell line by DNA dot blot analysis. As we have previously seen, the deletion of JBP1 or JBP2 alone cause a significant reduction in J levels in the genome (Figure 4). However, when both JBP1 and JBP2 are deleted, we generate a cell line that is null for the modified base (J null). The reintroduction of JBP2 in the J-null cell line stimulates de novo J synthesis. The expression of JBP1 in the presence of JBP2 acts to stimulate additional J synthesis (Figure 4). In contrast, reexpression of the JBP2 TH mutant (H441A) in the J null fails to stimulate J synthesis (data not shown).
Figure 3.
Southern blot analysis reveals the deletion of both JBP1 and JBP2. (A) DNA was isolated from WT bloodstream cells (WT427), and cells from which either one (JBP2−/+(H)) or both (JBP2−/− (H/H)) alleles of JBP2 were deleted as previously described (9). JBP1 was then deleted from this cell line by by LOH (JBP2−/− (H/H)/JBP1−/− (P/P)) and by conventional methods (JBP2−/− (H/H)/JBP1−/− (P/B)). DNA samples were digested with PstI and probed for the presence of the JBP2 (left) or JBP1 (right) open reading frame (ORF). A clear loss of a band correlating with JBP2 and JBP1 is apparent in cell lines from which all four JBP alleles were deleted, regardless of the method of generation. The 1 kb DNA size marker is indicated on the left. The WT gene fragments are indicated by the arrows on the right. A cross-reactive 3 kb band is visible on the JBP1 ORF hybridization. H, P and B refer to the hygomycin, puromycin and blasticidin resistance genes, respectively. (B) Map demonstrating area of JBP1 or JBP2 targeted by deletion constructs. Pst I, HindIII, Nar I restriction sites are indicated. Puro, puromycin resistance gene; Hygro, hygromycin resistance gene. The ORF probes used in panel A are indicated by the lines.
Figure 4.
JBP2 and JBP1 are essential for catalyzing the Step 1 of J biosynthesis in bloodstream trypanosomes. DNA was isolated from the indicated cell lines and analyzed for J content by anti-J DNA dot blot as in Figure 2. BS, WT bloodstream form cells; PC, procyclic cells; JBP2−/−, JBP2 knock out bloodstream; JBP1−/−, JBP1 knock out bloodstream; JBP2−/−/JBP1−/−, JBP2 and JBP1 knock out bloodstream; JBP2−/−/JBP1−/− + JBP2, JBP2 and JBP1 knock out bloodstream cells in which an ectopic JBP2 GFP is expressed; JBP2−/−/JBP1−/− + HOMedU, JBP2 and JBP1 knock out bloodstream cells feed with HOMedU; JBP2−/−/JBP1−/− + JBP2 + JBP1, JBP2 and JBP1 knock out bloodstream cells expressing both JBP2GFP and JBP1.
The requirement for Step 1 of the J-biosynthesis pathway can be bypassed (and therefore the hydroxylation of thymidine residues) by feeding cells a synthetic HOMedU, which is randomly incorporated into the genome during DNA replication (14). The ability of the J null to convert HOMedU into J (Figure 4) indicates that the cell still contains the glucosyl-transferase enzyme. Ultimately, the inability of the J-null cell line to synthesis J is due to ablation of the Step 1 of the biosynthesis pathway. This strongly implicates that both JBP1 and JBP2 play a pivotal role in the hydroxylation of thymidine residues in vivo.
The generation of the J-null cell line indicates that, in contrast to Leishmania (15) base J is not essential to T. brucei in vitro bloodstream form cultures. The J-null cell line has no significant growth rate or cell cycle defects when compared with WT trypanosomes (data not shown).
The reintroduction of JBP2 and JBP1 in the J-null cell line induces site-specific J synthesis
Previous analysis of JBP1 and JBP2 function in regulating J synthesis was done in the trypanosome life-stage (insect stage) that normally lacks base J (4). These studies indicated that JBP2 stimulated site-specific de novo base J in the genome and concomitant expression of JBP1 led to additional J synthesis in these specific regions. With the generation of the J-null cell line, we can confirm the localization of base J upon systematic expression of JBP2 and JBP1 in the native J-containing trypanosome life-stage. Therefore, we characterized the localization of base J in the J-null cell line expressing JBP2 and the null expressing both JBP proteins. To do this, genomic DNA was sonicated, immunoprecipitated with anti-J antisera and the purified J-DNA fragments were blotted and probed for regions of the genome known to contain J (177, 70 and 50 bp repeats and telomeres) as well as regions that are known to lack J (enolase). As expected, WT cells show high levels of J in the telomeric and subtelomeric repeats, and no J in the enolase gene (Figure 5). In contrast, when the J-null cells are fed HOMedU, which is then nonspecifically distributed throughout the genome (14), there is significant immunoprecipitation of all regions of the genome examined (Figure 5). As shown in Figure 5, the expression of JBP2 in the J-null cell line stimulates J synthesis in a site-specific manner as in WT cells, albeit to lower levels. The introduction of JBP1 in this cell line significantly amplifies the basal levels of J seeded at specific sites by JBP2.
Figure 5.
Reexpression of JBP2 and JBP1 in J-null bloodstream cells stimulates site-specific J synthesis. In order to localize J within the genome, DNA was isolated from the indicated cell lines, sonicated and J containing DNA fragments immunoprecipitated using J antisera. 10% of the input DNA (IN) and 100% of the immunoprecipitated DNA (IP) was blotted onto nitrocellulose. Blots were then hybridized with radio-labeled DNA probes corresponding to indicate regions of the trypanosome genome. (A) The percent IP was calculated for each region of the genome analyzed for the indicated cell line and values were normalized to background hybridization. ENO, enolase; TEL, telomere. The corresponding repeat regions were referred by 177, 70 and 50. Each value represents a mean between three and five hybridizations ± SEM. (B) Representative auto-radiographs of the hybridization are shown.
JBP2 induces the formation of HOMedU
All the data so far supports the hypothesis that JBP2 and JBP1 are the TH enzymes catalyzing Step 1 of the J-biosynthesis pathway. However, it is important to note that all our analysis to date has relied upon the detection of J as a read-out of biosynthesis. Clearly, this represents an endpoint of the J-biosynthesis pathway rather than a direct measure of the initial thymidine hydroxylation step. Therefore, we devised an anti-HOMedU immunoprecipitation assay to detect the formation of the intermediate in genomic DNA following JBP2 expression. To do this, DNA samples were digested to mononucleotides, postlalled labeled with 32P (16) and immunoprecipitated with anti-HOMedU antisera. The levels of HOMedU purified by the anti-HOMedU affinity resin were detected by scintillation counting. The specificity of the antibody was established by performing immunoprecipitation reactions on digested telomeric DNA oligos containing either four molecules of HOMedU, J or thymine. These control analyses indicate that the specificity of the antibody is significantly above any cross-reactivity with base J or unmodified DNA nucleotides (Figure 6A). As shown in Figure 6B, the expression JBP2 in the J-null cell line stimulates levels of HOMedU synthesis above the J-null background. Student's t-test indicates this difference is significant (P < 0.00001). Moreover, the expression of the TH mutant (JBP2-H319A) fails to stimulate HOMedU formation. As a positive control for the assay, high levels of HOMedU are detected in J-null cells grown in the presence of HOMedU. These data identifies JBP2 as an enzyme directly involved in the synthesis of HOMedU.
Figure 6.
JBP2 stimulates HOMedU formation in vivo. (A) Specificity of the HOMedU antibody was evaluated by immunoprecipitation assays using 32P-labeled mononucleotides derived from oligos containing HOMedU (HMU), base J (J) or thymidine (T) (see Methods section for oligo sequences). Postlabeled mononucleotides were incubated with α-HOMedU antibody conjugated to paramagnetic beads, washed to remove nonspecifically bound nucleotides followed by scintillation counting. Data are representative of three independent assays. Data are background subtracted and adjusted to results from unmodified DNA (T). (B) Anti-HOMedU immunoprecipitation analysis was used to examine HOMedU content of WT and the indicated mutant T. brucei cell lines. Genomic DNA was isolated from the indicated cell lines and analyzed for HOMedU content as in panel A. J-null (Null); J-null expressing WT JBP2GFP (Null+JBP2); J-null feed HOMedU (Null+HOMedU); J-null expressing JBP2 H441A mutant (Null+ JBP2 TH mutant). Each IP was repeated in triplicate and average ± SEM is shown.
DISCUSSION
Here we demonstrate that the JBP1 and JBP2 are ‘the’ key enzymes involved in the Step 1 of the J-biosynthesis pathway. The deletion of both proteins generates a J-null cell line. Upon reintroduction of both proteins, J synthesis is restored in a site-specific manner. Feeding of the J-null cell line with HOMedU results in J synthesis, indicating that the cell line still contains the glucosyl-transferase enzyme. Therefore deletion of JBP1 and JBP2 from the genome ablates Step 1 of the biosynthesis pathway but not Step 2 (Figure 7). Mutation of JBP2 at key residues conserved in members of the Fe2+/2-oxoglutarate dependent dioxygenases ablates the ability of JBP2 to stimulate de novo J synthesis. Previous analysis has revealed that the same mutations made in JBP1 allow the protein to bind J-DNA but inhibit its ability to propagate J synthesis (11). Furthermore, we can detect the formation of HOMedU in J-null cells expressing JBP2. Taken together, we believe that the evidence presented in this demonstrates that JBP1 and JBP2 are the two distinct TH enzymes involved in J biosynthesis.
Figure 7.
Proposed mechanism of JBP1 and JBP2 catalyzed thymidine hydroxylation. The two-step J-biosynthesis pathway in T. brucei. Indicated is the proposed mechanism of JBP2/JBP1 catalyzed hydroxylation of thymidine residues during the initial step of the pathway, as discussed in the text. Bypassing Step 1 (and therefore JBP catalyzed thymidine hydroxylation) can be achieved through feeding cells HOMedU, as indicated. The absence of base J in the genome of the J null cell, but its ability to convert incorporated HOMedU to J, indicates that only Step 1 of the synthesis pathway is ablated upon deletion of JBP1 and JBP2. The glucosyl transferase (GT) enzyme is still active in this cell line.
Previous to JBP1 and JBP2, no TH enzymes have been identified. However, thymine hydroxylase enzymes have been characterized in fungi including Neurospora crassa and Rhodotorula glutinis (17–21). These thymine 7-hydroxylase enzymes are involved in the thymine salvage pathway catalyzing the oxygenation of thymine to 5-hydroxymethyluracil coupled to oxidative decarboxylation of 2-oxoglutarate (20). Once thymine is converted to HOMedU, it can then be converted into uracil through a series of enzymatic steps, allowing the use of thymine as a pyrimidine source in the absence of a de novo synthesis pathway. As these enzymes are members of the Fe2+/2-oxoglutarate hydroxylase family, oxygen and 2-oxoglutarate are cosubstrates for the reaction and carbon dioxide and succinate the by products. Based on the functional analysis of JBP1 and JBP2, we believe that both proteins stimulate the hydroxylation of thymidine by a mechanism similar to the fungal enzymes, but at the DNA level rather than nucleotide level. Therefore, as proposed in Figure 7, JBP2 and JBP1 would convert thymidine into HOMedU in DNA using oxygen and 2-oxoglutarate as co-substrates, Fe2+ as a cofactor. The reaction would release carbon dioxide and succinate. The finding that both JBP1 and JBP2 bind chromatin (in a J-dependent and -independent manner, respectively) in vivo further supports this model (6).
Mutagenesis of the conserved residues involved in binding Fe2+ and 2-oxoglutarate strongly implicates JBP1 and JBP2 as members of the Fe2+/2-oxoglutarate family of hydroxylases. Regardless of the clear in vivo evidence, an in vitro DNA thymidine hydroxylation assay is needed to confirm this identity. To date, all attempts to detect the hydroxylation of thymidine in vitro has been unsuccessful. However, the recent isolation of recombinant JBP2 in the lab has opened up a number of new assays currently being pursued.
Previous analysis of insect-stage trypanosomes (which normally lack J) indicated that JBP1 and JBP2 regulate J synthesis (6). The analysis of JBP1 and JBP2 function in bloodstream form trypanosomes described here, mirrors that seen in insect stage cells (6). This data, along with the mutational analysis of JBP1 (11) indicates that these proteins regulate J biosynthesis by representing the key enzymes of the J-synthesis pathway. This finding raises the interesting question as to why there are two TH enzymes involved in J biosynthesis. We suggest that differences in DNA substrate specificity between JBP1 and JBP2 impart a distinct function for each in the biosynthesis pathway. JBP2 has been shown to bind to chromatin independent of base J (6). We propose that it recognizes specific regions of chromatin where it can then open up the chromatin via the C-terminal SWI2/SNF2 domain, allowing access of the TH domain to thymidine residues in double stranded DNA. JBP1, on the other hand, is able to bind J-DNA with high affinity (7,22). Once bound to regions of the genome containing base J, JBP1 can hydroxylate adjacent thymidine residues. The strong affinity of JBP1 for base J acts to maintain JBP1 at sites in the genome already seeded with J (by JBP2), allowing JBP1 to maintain and propagate J in a site-specific manner. In this way, JBP2-chromatin interactions provide the specificity of J-localization while JBP1, via the strong affinity for J-DNA, provides the stimulation and maintenance of J during replication at regions dictated by JBP2. While this separation of JBP1/JBP2 function in J biosynthesis is appealing, detailed analysis of the JBP2−/− suggests the model to be more complex. In the absence of JBP2, JBP1 is apparently unable to stably maintain J. The initial deletion of JBP2 from the bloodstream trypanosome resulted in a 5-fold reduction in J levels (9). However, as we show here, upon further growth (∼800 generations) J levels are 8-fold less than WT (Figure 4). Therefore, JBP1 may require ongoing chromatin remodeling by JBP2 to optimally propagate/maintain J within certain regions of the genome. The detailed analysis of JBP1 and JBP2 function in the bloodstream form trypanosome J-null cell line will help elucidate the contribution of each TH enzyme to the overall J-synthesis pathway.
A role for J in the regulation of antigenic variation has been proposed based on a number of key observations. J is developmentally regulated, being present only in the bloodstream form trypanosomes and not insect stage cells (which do not undergo antigenic variation) (23). Moreover, the association of J with silent and not transcriptionally active telomeric Variant Surface Glycoprotein (VSG) expression sites (3) suggests that the modified base plays a critical role in the repression of silent expression sites, ensuring monoallelic VSG gene transcription. Attempts to elucidate the function of J were performed on cell lines in which JBP1 or JBP2 had been deleted. However, despite causing a significant reduction in J levels, no clear phenotypic effect on VSG gene switching was measurable. It has not escaped our attention that the generation of a cell line which completely lacks the modified base is an invaluable tool in allowing us to address the biological function of base J in African trypanosomes including its potential role in the regulation of antigenic variation.
EXPERIMENTAL PROCEDURES
Enzymes and chemicals
Hygromycin, puromycin, blasticidin, neomycin and phleomycin were purchased from Research Diagnostic International. All restriction enzymes were purchased from New England Biolabs. Anti-GFP antibody, Alexa labeled goat anti-rabbit and Benchmark Pre-stained protein standard were purchased from Invitrogen. Prime-It II random primer labeling kit was purchased from Stratagene, α-32P-dATP and γ-32P-ATP was purchased from Perkin Elmer. Enhanced chemiluminescent (ECL) and Hybond-N+ were from Amersham. Goat anti-rabbit horseradish peroxidase (HRP) was purchased from Southern Biotec Inc. All other chemicals were purchased from Sigma Aldrich.
Trypanosome growth and cell culture
Both culturing and transfection of insect stage T. brucei 29-13 cells were carried out as described previously (6). Bloodstream form T. brucei cell line 221a of strain 427 were cultured as described previously (6). Bloodstream from transfections were carried out as previously described (24).
HOMedU feeding of bloodstream form trypanosomes
Cells were grown in HMI9 in the presence of 1mM HOMedU for 3 days. Cultures were started at low cell densities to ensure maximal incorportation.
Generation and analysis of T. brucei transfectants
The T. brucei JBP2−/− cell line was generated by loss of heterozygosity (LOH) as described (9). JBP1 was deleted from the JBP2−/− cell line using constructs containing either a blasticidin S deaminase (BSR) or a puromycin acetyltransferase (PAC) gene as described (13). Each knock out construct of weight 5 μg, digested with NotI and ApaI, was used to transform the JBP2−/− cell line. Transfectants were selected for resistance at 0.1 μg/ml puromycin or 2 μg/ml blasticydin. Two rounds of transfection were used to replace both JBP1 alleles. While LOH was also done to delete the second JBP1 allele, as previously described (9), these cell lines were not utilized in the studies shown here. The deletion of JBP1 from the genome was confirmed by southern blot analysis and hybridization with a full length JBP1 ORF probe.
The generation of J-null cells expressing JBP2 was performed by transfection with GFP-JBP2-Tub-phleo (6). The construct was digested by NotI and XhoI prior to transfection. Transfectants were selected with 2.5 μg/ml phleomycin. To generate cell lines expressing both JBP1 and JBP2, the null expressing JBP2 was subject to an additional round of transfection with JBP1-neo (8). This construct allows the expression of JBP1 from the ribosomal locus. Five microgram of the construct was linearized by AvaI digestion prior to transfection. Transfectants were selected at 2 μg/ml neomycin.
Mutagenesis
The mutations in JBP2 were made by site-directed mutagenesis using the Quik-Change site-directed mutagenesis kit (Stratagene) following the instruction of the manufacturer. The WT Tb GFP-JBP2-tub-phleo construct was used as a template. The different oligonucleotides used for the mutagenesis reaction as well as detailed maps of the expression vectors can be given upon request. Mutations were verified by sequencing using an ABI Prism 3700 DNA Analyzer (Applied Biosystems). The GFP-JBP2 constructs were linearized and transfected into the 29-13 insect stage cells as previously described (6).
Determination of the genomic level of J
To quantify the genomic J levels, we used the anti-J DNA immunoblot assay as described (3) on total genomic DNA, which was isolated as described (25). Briefly, serially diluted genomic DNA was blotted to nitrocellulose and incubated with anti-J antisera. Bound antibodies were detected by a secondary goat anti-rabbit antibody conjugated to HRP and visualized by ECL. The membrane was stripped and hybridized with a probe for the beta-tubulin gene to correct for DNA loading.
Western blotting
Proteins from 107 cell equivalents were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS page 8% gel), transferred to nitrocellulose and probed with anti-JBP2 as described (9). Bound antibodies were detected by a secondary goat anti-rabbit antibody conjugated the HRP and visualized by ECL.
Microscopy
The detection of WT and mutant GFP-JBP2 expressed in insect stage T. brucei was performed by anti-GFP immunofluorescence analysis. Cells were fixed in 4% formaldehyde, 0.04% gluteraldehyde for 20 min, and quenched using 10 mM ammonium chloride. Cells were then permeabilized in 0.1% NP40 and blocked for 20 min in normal goat serum. GFP was detected using anti-GFP (1 in 500 dilution), followed by secondary goat anti-rabbit (Alexa 594, 1 in 500 dilution). Cells were mounted in dapi-vecta shield. Images were acquired using an Axioobserver Z1 equipped with an Axiocam MRm camera controled by Axiovision, version 4.6, software.
Determination of the sequence distribution of J
Detection of HOMedU
Detection of HOMedU was performed by immunoprecipitation of kinase-labeled mononucleotides using a HOMedU specific goat-polyclonal antibody (ABCam). DNA digestion and postlabeling (via γ-32P-ATP and PNK) were carried out as previously described (16). Protein G coated paramagnetic beads (Dynabeads, Invitrogen) were washed according to the manufacturers recommendation prior to immunoprecipitation. For each immunoprecipitation, 25 μl of washed beads were incubated with 5 μl of HOMedU antiserum for 30 min at room temperature with agitation. Beads were washed (4 × 1 ml) in binding buffer (0.1 M NaPhosphate, 0.01% Tween-20 pH 8.0). Antibody coated beads were suspended in 250 μl of binding buffer, 32P-labeled mononucleotides were added, and the reaction was incubated at room temperature for 30 min with rocking. Beads were washed in binding buffer (6 × 1 ml) followed by scintillation counting. Oligos modified with HOMedU (H) (ACCCHAACCCHAACCCHAACCCHA), J (ACCCJAACCCJAACCCJAACCCJA) and an unmodified (thymidine) control (ACCCTAACCCTAACCCTAACCCTA) were used to examine specificity of the HOMedU polyclonal antibody. HOMedU content of WT and mutant T. brucei genomic DNA was evaluated as described for DNA oligos.
FUNDING
National Institutes of Health (grant number AI063523 to R.S.). Funding for open access charge: National Institutes of Health (063523).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We are grateful to John Harrington, Dilrukshi Ekanayake and Piet Borst for critical reading of the manuscript. Anti-J antisera and the JBP1-puran-neo construct were a gift from Piet Borst. The 70 bp probe was a gift from Richard McCulloch.
REFERENCES
- 1.van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres [see comments] Proc. Natl Acad. Sci. USA. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosal-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;109:133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, Borst P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 5.Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel G, van Boom J, van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Sabatini R, Meeuwenoord N, van Boom JH, Borst P. Recognition of base J in duplex DNA by J-binding protein. J. Biol. Chem. 2002;277:958–966. doi: 10.1074/jbc.M109000200. [DOI] [PubMed] [Google Scholar]
- 8.Toaldo CB, Kieft R, Dirks-Mulder A, Sabatini R, van Luenen HG, Borst P. A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1. Mol. Biochem. Parasitol. 2005;143:111–115. doi: 10.1016/j.molbiopara.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieft R, Brand V, Ekanayake DK, Sweeney K, DiPaolo C, Reznikoff WS, Sabatini R. JBP2, a SWI2/SNF2-like protein, regulates de novo telomeric DNA glycosylation in bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;156:24–31. doi: 10.1016/j.molbiopara.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 11.Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitson KS, Holmes SL, Ehrismann D, Hardy AP, Chowdhury R, Schofield CJ, McDonough MA. Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH) J. Biol. Chem. 2008;283:25971–25978. doi: 10.1074/jbc.M804999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross M, Kieft R, Sabatini R, Dirks-Mulder A, Chaves I, Borst P. J binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 2002;46:37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen F, Kieft R, Cross M, Borst P. Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Cell. Biol. 1998;18:5643–5651. doi: 10.1128/mcb.18.10.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genest PA, ter Riet B, Dumas C, Papadopoulou B, van Luenen HG, Borst P. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33:1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Leeuwen F, de Kort M, van der Marel GA, van Boom JH, Borst P. The modified DNA base beta-D-glucosylhydroxymethyluracil confers resistance to micrococcal nuclease and is incompletely recovered by 32P-postlabeling. Analytical Biochemistry. 1998;258:223–229. doi: 10.1006/abio.1998.2587. [DOI] [PubMed] [Google Scholar]
- 17.Thornburg LD, Lai MT, Wishnok JS, Stubbe J. A non-heme iron protein with heme tendencies: an investigation of the substrate specificity of thymine hydroxylase. Biochemistry. 1993;32:14023–14033. doi: 10.1021/bi00213a036. [DOI] [PubMed] [Google Scholar]
- 18.Thornburg LD, Stubbe J. Mechanism-based inactivation of thymine hydroxylase, an alpha-ketoglutarate-dependent dioxygenase, by 5-ethynyluracil. Biochemistry. 1993;32:14034–14042. doi: 10.1021/bi00213a037. [DOI] [PubMed] [Google Scholar]
- 19.Wondrack LM, Hsu CA, Abbott MT. Thymine 7-hydroxylase and pyrimidine deoxyribonucleoside 2′-hydroxylase activities in Rhodotorula glutinis. J Biol. Chem. 1978;253:6511–6515. [PubMed] [Google Scholar]
- 20.Holme E, Lindstedt G, Lindstedt S, Tofft M. 7-Hydroxylation of thymine in a Neurospora strain coupled to oxidative decarboxylation of 2-ketoglutarate. Biochim. Biophys. Acta. 1970;212:50–57. doi: 10.1016/0005-2744(70)90177-4. [DOI] [PubMed] [Google Scholar]
- 21.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr, Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim. Biophys. Acta. 2005;1723:256–264. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Grover RK, Pond SJ, Cui Q, Subramaniam P, Case DA, Millar DP, Wentworth P., Jr O-glycoside orientation is an essential aspect of base J recognition by the kinetoplastid DNA-binding protein JBP1. Angew Chem. Int. Ed. Engl. 2007;46:2839–2843. doi: 10.1002/anie.200604635. [DOI] [PubMed] [Google Scholar]
- 23.Gommers-Ampt J, Lutgerink J, Borst P. A novel DNA nucleotide in Trypanosoma brucei only present in the mammalian phase of the life-cycle. Nucleic Acids Research. 1991;19:1745–1751. doi: 10.1093/nar/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover L, Horn D. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO reports. 2006;7:93–99. doi: 10.1038/sj.embor.7400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernards A, Van der Ploeg LH, Frasch AC, Borst P, Boothroyd JC, Coleman S, Cross GA. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. doi: 10.1016/0092-8674(81)90391-3. [DOI] [PubMed] [Google Scholar]