Abstract
microRNAs (miRNAs) play integral roles in diverse processes including tumorigenesis. miRNA gene loci are often found in close conjunction, and such clustered miRNA genes are transcribed from a common promoter to generate polycistronic primary transcript. The primary transcript (pri-miRNA) is then processed by two RNase III proteins to release the mature miRNAs. Although it has been speculated that the miRNAs in the same cluster may play related biological functions, this has not been experimentally addressed. Here we report that the miRNAs in two clusters (miR-106b∼93 ∼ 25 and miR-222 ∼ 221) suppress the Cip/Kip family members of Cdk inhibitors (p57Kip2, p21Cip1 and p27Kip1). We show that miR-25 targets p57 through the 3′-UTR. Furthermore, miR-106b and miR-93 control p21 while miR-222 and miR-221 regulate both p27 and p57. Ectopic expression of these miRNAs results in activation of Cdk2 and facilitation of G1/S phase transition. Consistent with these results, both clusters are abnormally upregulated in gastric cancer tissues compared to the corresponding normal tissues. Ectopic expression of miR-222 cluster enhanced tumor growth in the mouse xenograft model. Our study demonstrates the functional associations between clustered miRNAs and further implicates that effective cancer treatment may require a combinatorial approach to target multiple oncogenic miRNA clusters.
INTRODUCTION
microRNAs (miRNAs) are noncoding RNAs of ∼22 nt that function as post-transcriptional regulators. By base-pairing with the complementary sites in the 3′ untranslated region (UTR) of mRNA, miRNAs can control the mRNA stability and the efficiency of translation (1). The latest release of miRBase (Release 12.0) annotates 695 miRNA loci in the human genome (2). Bioinformatic analyses predict that miRNAs may control more than 30% of human protein-coding genes. miRNAs and their targets constitute complex regulatory networks. Each miRNA is predicted to suppress over hundreds of targets. Conversely, each target mRNA can be controlled by multiple miRNAs. Efforts have been made to identify miRNA targets for individual miRNAs using both computational and experimental approaches.
The genes of miRNAs are often organized in clusters in the genome. Approximately 40% of total human miRNA loci are located in <3 kb from the adjacent miRNA locus (3). Expression analyses showed strong positive correlations among the closely located miRNAs, indicating that they may be controlled by common regulatory element(s). In fact, experimental evidence demonstrated that clustered miRNA loci form an operon-like gene structure and that they are transcribed from a common promoter (4,5). Following transcription, the polycistronic primary transcript (pri-miRNA) gets processed by two RNase III proteins, Drosha and Dicer, to liberate the mature forms of the miRNAs (6,7). Interestingly, these gene organizations are often evolutionarily conserved. For instance, the let-7∼miR-100∼lin-4 cluster is found in most animals (2). Other interesting examples include the miR-106b∼93 ∼ 25 cluster and the miR-222 ∼ 221 cluster, which are found in all vertebrate genomes with minor variations. Expression patterns as well as the conservation of clustered miRNAs led to the hypothesis that the clustered miRNAs may have related functions. However, it has not been experimentally examined whether the operonic organization of miRNA loci have any functional significance.
Dysregulation of miRNAs is often associated with human malignancy (8,9). By targeting oncogenes and tumor suppressor genes, miRNAs themselves can function in various pathways in the development of a tumor. Identification of oncogenic miRNAs opens a new window for anticancer treatment. Proof-of-principle studies have shown that the inhibition of oncogenic miRNAs such as miR-21 leads to the regression of tumors in vivo (10). For effective anti-miRNA strategy, it is crucial to correctly determine the oncogenic miRNAs that play key roles in the development/maintenance of the tumor of interest. Because it is expected that multiple miRNAs contribute to tumor development and that certain miRNAs may have cooperative and/or redundant functions, it would be important to understand the functional relationships between miRNAs, particularly those in clusters.
In addressing this issue, we chose to study miRNA clusters that are upregulated in gastric cancer. Gastric cancer is the second most common malignancy and is responsible for ∼10% of tumor-related deaths worldwide (11). Particularly high incidences and mortality rates are observed in eastern Asia (11). Although the molecular pathology of gastric cancer remains fragmentary, it is established that gastric cancer cells typically show abnormal levels of cell-cycle regulators.
Transitions between cell-cycle phases are mediated by cyclin-dependent kinases (Cdks) and their modulators. Cdks can be controlled by Cdk inhibitors (CKIs) that bind to Cdks (12,13). In mammalian cells, two families of Cdk inhibitors are responsible for regulating different Cdks. Members of the Ink4 family (p15Ink4b, p16Ink4a, p18Ink4c and p19Ink4d) bind to Cdk4 and 6/cyclin D complexes, thereby inhibiting progression through the G1 restriction point. Members of the Cip/Kip family (p21Cip1, p27Kip1 and p57Kip2) share homology in their N-terminal regions and affect the complexes of Cdk2, 4 and 6 with cyclin A, D and E. The Cip/Kip family proteins block the progression through all stages of G1/S, thereby functioning as a ‘brake of cell cycle’. The Cip/Kip proteins are also involved in many aspects of cellular physiology, including apoptosis, transcriptional regulation, cell fate determination, cell migration and cytoskeletal dynamics (12).
Here we study miRNA clusters that are abnormally upregulated in gastric cancer. We found that miR-106b cluster and miR-222 cluster are upregulated in cancer and modulate cell cycle by targeting the Cip/Kip family proteins (p21Cip1, p27Kip1 and p57Kip2).
MATERIALS AND METHODS
Microarray
Total RNA was extracted from gastric-cancer patient tissue using Trizol reagent (Invitrogen). RNA quality was checked by Bioanalyzer RNA Nano kit (Agilent). Agilent Human miRNA Microarray 8 × 15k (G4470A) platform and Agilent Microarray Scanner were used for miRNA profiling (Agilent). GeneSpring GX 7.3 software (Agilent) was used to analyze the microarray result.
Tissue specimens
Tumors and the corresponding normal tissues were obtained from patients with primary gastric adenocarcinoma at the Seoul National University Hospital, Seoul, Korea. After surgical removal, the tissues were frozen immediately in liquid nitrogen and stored at –80°C. Ethical approval was obtained from the Seoul National University Hospital Institutional Review Board for this study, and informed consent was obtained from all patients.
Cell culture
Human gastric cancer cell lines SNU-638, AGS and MKN-28, and breast cancer cell line MCF7 were obtained from a Korean cell line bank. The human uterine cervix adenocarcinoma cell line HeLa was grown in 10% FBS (Invitrogen) in DMEM. SNU-638, AGS, MKN-28 and MCF7 were cultured in RPMI1640 supplemented with 10% FBS. These cells were maintained at 37°C under an atmosphere of 5% CO2.
DNA constructs
The fragment containing the 3′-UTR region of human p27 cDNA was amplified by PCR using the primers of 5′-ACA GCT CGA ATT AAG AAT ATG TTT CC-3′ (forward) and 5′-ACT TGG CTC AGT ATG CAA CCT TTT AAG-3′ (reverse). It was cloned into pGEM-T easy vector (Promega) and subsequently transferred into the pGL3_CMV vector (modified from pGL3 basic vector; Promega) at XbaI site. The nucleotide sequences of the plasmid were confirmed by sequencing. To amplify the 3′-UTR region of human p57, the primers of 5′-GCT CTA GAG CCA ATT TAG AGC CCA AAG AG-3′ (forward) and 5′-GGA ATT CTT TGC ACT GAG TTT CAG CAG AG-3′ (reverse) were used. And to clone the 3′-UTR region of human p21, the primers of 5′-TCT AGA CAC AGG AAG CCT GCA GTC CT-3′ (forward) and 5′-CCG CGG AGC ACC TGC TGT ATA TTC AGC-3′ (reverse) were used.
Transfection
Plasmids and RNA oligonucleotides were transfected into each cell using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
RNA preparation and northern blot
Total RNA was prepared using Trizol (Invitrogen), treated with DNase I (Takara), and then reverse-transcribed using an oligo-dT primer (Invitrogen). Reverse transcription was carried out using the Superscript II kit (Invitrogen) according to the manufacturer's protocol. For northern blot, total RNA was resolved on 15% urea–polyacrylamide gel and transferred to a Zetaprobe-GT-membrane (Bio-Rad). Oligonucleotides complementary to each miRNA were endlabeled using T4 polynucleotide kinase (Takara) and used as probes. BAS 2500 Image Analysis System (Fujifilm) and Multi Gauge V3.0 software (Fujifilm) were used for quantitation.
Real-time PCR
RNA quantifications were conducted by real-time PCR using the comparative Ct method with the Applied Biosystems 7300 Real-Time PCR system. Real-time PCR reaction was conducted by using the SyBR green 2X master mix (Applied Biosystems), and the PCR primers were 5′-TCC GGC TAA CTC TGA GGA CAC-3′ (forward) and 5′-TGT TTT GAG TAG AAG AAT CGT CGG T-3′ (reverse) for the amplification of p27 mRNA. To amplify the p57 mRNA, primer set 5′-ATT TTC ATC GCG GAC TCT GG-3′ (forward) and 5′-TAA GAG AGA CAG CGA AAG CGC-3′ (reverse) were used. For the amplification of p21 mRNA, primer set 5′-AGC AGA GGA AGA CCA TGT GGA C-3′ (forward) and 5′-TTT CGA CCC TGA GAG TCT CCA G-3′ (reverse) were used. For normalization, human GAPDH amplification kit was used (Applied Biosystems).
Western blot analysis
Thirty micrograms of protein were used for western blotting. Antimouse p21 antibody (Cell signaling), antimouse p27 antibody (BD Biosciences) and antirabbit p57 antibody (Abcam) were used for western blot experiments. Normalization was done using mouse monoclonal anti-GAPDH or rabbit polyclonal antitubulin antibody.
Luciferase assay
For luciferase assays, firefly luciferase reporter plasmids were transiently transfected with siRNA into HeLa or SNU-638 cells. Renilla luciferase construct was co-transfected for normalization of transfection efficiency. Twenty-four hours before transfection, cells were plated in 24-well dishes at 8 × 104 cells/well. Twenty-four hours post-transfection, cells were harvested and dual-luciferase assay (Promega) was carried out according to the manufacturer's protocol. Results were normalized against the value of Renilla luciferase activity. All assays were performed more than three times.
Mutagenesis
Site-directed mutagenesis was carried out using the Quick-Change Site-Directed Mutagenesis kit (Stratagene) to confirm that the 3′-UTR of each gene is directly regulated by miRNAs. The sequences of the primer sets used for mutagenesis are given in the Supplementary Data.
FACS analysis
For cell-cycle analysis, attached cells were trypsinized, washed with PBS and fixed in 70% ethanol at 4°C. After fixation, ethanol was removed and PI-staining solution [propidium iodide (PI, Sigma) and 50 μg/ml RNase A in 0.1% BSA solution] was added. After 30 min of incubation, cell-cycle profile was analyzed by Becton Dickinson FACSCalibur. Modfit LT (ver. 3.1) software was used to analyze the distribution of cell cycle.
Tumor formation in nude mice
Tumorigenicity of miR-222 ∼ 221 was investigated after the injection of stably transfected cells (5 × 106) into the right hind legs of 5-week old athymic nude mice subcutaneously. Before injection, the cell suspension was mixed with the same amount of matrigel (250 μl) (BD matrigel matrix solution [BD biosciences]) for effective attachment in each mouse. All cells were injected simultaneously and tumor formation in nude mice was monitored over a 4-week period. Four weeks later, the mice were sacrificed to measure tumor mass. The tumor volume was calculated by the formula V = 0.5 × L × W2 (14).
RESULTS
miRNA expression profiling in gastric cancer
In order to identify the miRNA clusters that may be involved in the tumorigenesis of gastric cancer, we carried out miRNA expression profiling in gastric cancer patient tissues. Several miRNAs were found to be enriched in tumor tissues compared to their normal counterparts (Supplementary Table 1 and Figure 1A). Previous studies also reported the enhanced expression of these miRNAs in various tumors including stomach, colon and prostate cancer (15,16). We selected 56 miRNAs, whose levels were increased more than 1.5-fold in tumor samples compared to normal tissues. We excluded the miRNAs whose absolute expression value in microarray was very low, because the P-values of these miRNAs were unacceptable. More than half of the miRNAs (30/56) belong to miRNA clusters (Supplementary Table 1 and Figure 1A). Average fold of miRNA upregulation was highest among the members of the miR-17, miR-222 and miR-106b clusters. Certain members of the miR-106a cluster were also increased in tumor samples, but miR-363, which belongs to miR-106a cluster, was decreased in the tumors, implicating a post-transcriptional regulation of miR-363 processing (Figure 1A). Both the miR-17 and miR-106a cluster contains six miRNA hairpin structures that are highly similar to those in the miR-106b cluster in sequences, indicating that these miRNA clusters are paralogs of the miR-106b cluster but have undergone additional gene duplication events.
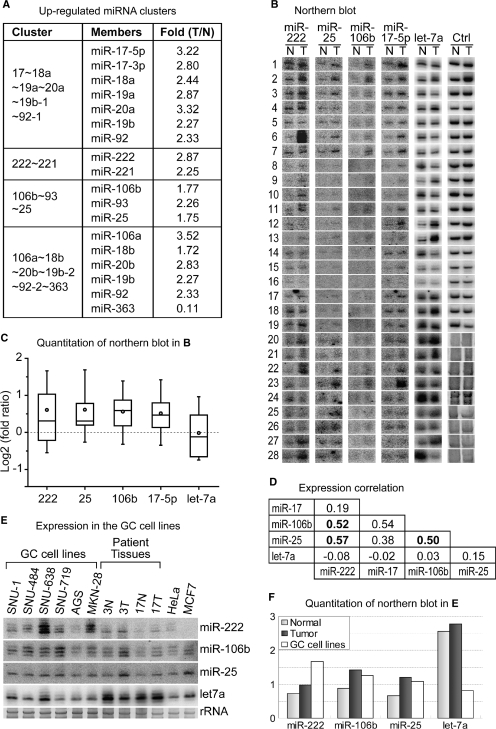
Figure 1.
Expression of miRNA clusters in gastric cancer tissues. (A) Selected results from microarray experiments. The table presents the miRNA clusters whose members are upregulated in tumor samples. (B) Northern blotting. Total RNA was extracted from frozen tissue samples from gastric cancer patients. Both normal (left, N) and tumor (right, T) tissues from the same donor were analyzed side by side. Control RNAs (tRNA for sample No. 1-19 or rRNA for sample No. 20-28) demonstrate equal loading of RNA. (C) Statistical analysis of northern blotting. The band intensity from northern blots was measured (Supplementary Table 2) and the intensity ratio (tumor/normal) was used to calculate Log2 value. A box plot was drawn for each miRNA. The circles indicate the mean values. Boxes and whiskers range from 25% to 75% and from 10% to 90%, respectively. (D) Expression correlation among miRNAs. Correlation coefficient between each miRNA pair was calculated based on the results from northern blotting. The array of intensity ratio (tumor/normal) from each miRNA was grouped to calculate the correlation coefficients. (E) Enhanced expression of miR-222, miR-106b and miR-25 in gastric cancer cells. Thirty micrograms of total RNA were used for northern analysis. The average levels of miR-222, miR-106b and miR-25 in gastric cancer (GC) cell lines are higher than those in normal stomach tissues. (F) Quantitation of northern blotting in (E).
To confirm the expression profiling data, northern blot experiments were carried out using the tissues from 28 gastric cancer patients. We determined the levels of miR-17 from the miR-17 ∼ 18a∼19a∼20a∼19b-1 ∼ 92-1 cluster, miR-106b and miR-25 from the miR-106b∼93 ∼ 25 cluster and miR-222 from the miR-222 ∼ 221 cluster. These miRNAs were upregulated in the gastric tumor samples compared to the normal counterparts (Figure 1B). In contrast, the expression pattern of let-7a did not show a meaningful correlation. The northern blot results were quantitated and normalized against tRNA or rRNA signals (Supplementary Table 2). For statistical analysis, we calculated the log2 value of fold ratio and drew a box plot for each miRNA (Figure 1C). miR-222, miR-25, miR-106b and miR-17-5p are upregulated in gastric cancer. On the contrary, the expression level of let-7a did not change significantly. Correlation coefficient between each miRNA pair was calculated using the quantitated northern blot signals (Figure 1D). Interestingly, the correlation coefficient values between miR-222 and members of the miR-106b cluster (miR-106b or miR-25) were very high; even higher than the coefficient between miR-106b and miR-25, which belong to the same cluster. The correlation between miR-17 and miR-222 was relatively low. Thus, the miR-222 cluster may be more closely associated with the miR-106b cluster than the miR-17 cluster in gastric cancer. Let-7a miRNA has nearly no correlation with other miRNAs we tested. For further study, we focused on the miR-222 ∼ 221 cluster and the miR-106b∼93 ∼ 25 cluster, because these clusters are highly increased in tumor samples and their expression patterns correlate most highly with each other.
To examine whether these miRNA clusters show overlapping expression patterns in gastric cancer cells, we measured miRNA levels in six different gastric cancer cell lines, and compared them with those in normal stomach tissues. These gastric cancer cell lines are homogenous in cell population. We selected representative patient tissues, and using the expression level in these samples, we normalized average expression level of each miRNA in the whole patient samples. Northern blotting data indicate that the cell lines express higher levels of miR-222, miR-106b and miR-25 compared to the normal tissues (Figure 1E). Thus, our result suggests that these miRNAs may indeed be co-expressed in gastric cancer cells.
To investigate the function of these miRNAs in cancer, we searched for the target genes using miRNA target prediction programs (17). The target genes were annotated to each miRNA, and their GO terms were analyzed (http://cgap.nci.nih.gov/Genes/GOBrowser/). We found that the GO terms of ‘cell cycle’, ‘cell division’, ‘cell proliferation’, ‘apoptosis’ and ‘replication’ were overpresented (Supplementary Table 3). This was interesting because these terms are associated with the tumor development process. When we compared the putative target genes for these miRNAs, we found that several protein families are predicted to be regulated by both the miR-106b and miR-222 clusters (Supplementary Table 3). Among them, the Cip/Kip family was most intriguing, because all three members (p21Cip1, p27Kip1 and p57Kip2) from this family were predicted to be regulated by the miR-106b and miR-222 clusters.
The miR-106b∼93 ∼ 25 cluster regulates p57 as well as p21
miR-106b and miR-93 are identical in the seed sequences and similar in the 3′-half (Supplementary Figure 1), suggesting that they may have similar functions. In contrast, miR-25 has no sequence similarity to miR-106b or miR-93 and, therefore, is expected to have a separate function. Interestingly, miR-25 has a putative binding site on p57 mRNA while miR-106b/93 is predicted to control a related Cdk inhibitor, p21 (Supplementary Figure 2).
To validate the prediction, we transfected miRNA duplexes into HeLa or MCF7 cells, and carried out western blot analysis (Figure 2). As predicted, the protein level of p57 was decreased in the miR-25-transfected sample (Figure 2A, lane 2). We also tested the effect of miR-106b on p21 by transfecting the miRNA duplex into MCF7 cells. Figure 2B shows that p21 is downregulated by miR-106b. Similar results were obtained when HeLa cells were used for transfection (data not shown).
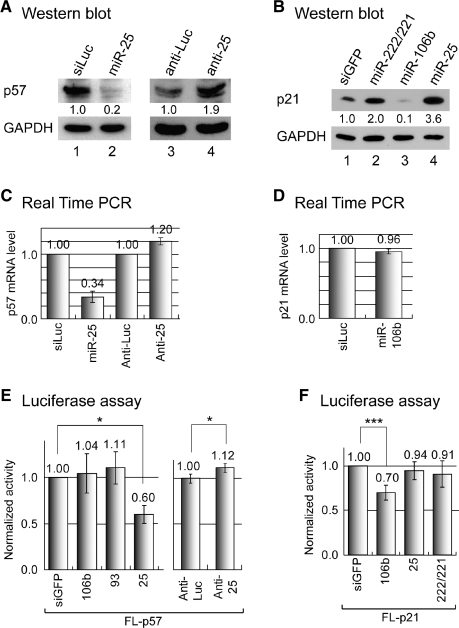
Figure 2.
Regulation of p57 and p21 by miR-106b∼93 ∼ 25 cluster. (A) Western blot analysis of p57 protein. HeLa cells were transfected with miR-25 or control siRNA (siLuc) duplex (30 nM). To inhibit miRNA function, either control (anti-Luc) or miR-25 inhibitor (anti-25) was used (200 nM). Because the p57 level is usually low, 1 day after the transfection of RNA duplexes, dexamethasone (100 nM) was used in HeLa cells for 16 h in lanes 1 and 2. In the case of lanes 3 and 4, dexamethasone (100 nM) was treated for 4 h after 36 h of transfection. Band intensity of p57 was measured using densitometer and normalized against that of GAPDH. (B) Western blot analysis of p21. A total of 30 nM of small RNA duplexes were transfected into MCF7 cells. Two days later, western blot was carried out. (C) Quantitative RT-PCR to measure the p57 mRNA level after the transfection of miR-25 duplex (left) or anti-miR-25 (right). The same cells as in (A) were used. (D) Quantitative RT-PCR to measure the p21 mRNA level after the transfection of miR-106b into HeLa cells. (E) p57 is directly regulated by miR-25 through the 3′-UTR. The reporter containing the 3′-UTR of p57 (FL-p57) was co-transfected with miRNA duplexes or antisense inhibitors into HeLa cells. Luciferase assay was carried out 24 h post-transfection (n = 3, mean±SD). (F) p21 is directly regulated by miR-106b through the 3′-UTR. The reporter containing the 3′-UTR of p21 (FL-p21) was transfected into HeLa cells along with miRNA duplexes (n = 5, mean±SD). Paired one-tail T-test was used to calculate the P-value (*P < 0.1, ***P < 0.001).
We also tested the effect of miRNA inhibition on the target proteins. Antisense inhibitors used in this assay are complementary to mature miRNA throughout the length and contain modified 2′-O-methyl ribose groups. When we introduced anti-miR-25 into HeLa cells, the protein level of p57 was increased (Figure 2A, lanes 3 and 4), confirming the repression of p57 by endogenous miR-25.
We also measured the mRNA levels of p57 and p21 after miRNA overexpression or inhibition. The p57 mRNA level was reduced by miR-25 duplex while anti-miR-25 treatment elevated the p57 mRNA level (Figure 2C). Thus, p57 is likely to be suppressed by miR-25 through both translational inhibition and mRNA decay. The p21 mRNA level was only slightly reduced by miR-106b treatment (Figure 2D), indicating that repression of p21 may be mainly due to translational inhibition.
To examine if these effects are mediated by the 3′-UTR binding of miRNAs, we carried out reporter assays using the luciferase constructs harboring the 3′-UTR region of the target genes. miR-25 can specifically suppress the expression of the reporter that contains the 3′-UTR of p57 (FL-p57) (Figure 2E, left). When anti-miR-25 was transfected, the expression level of the p57 reporter plasmid was increased (Figure 2E, right). We also examined the regulation of p21 by using the reporter harboring the 3′-UTR of p21 mRNA (FL-p21) (Figure 2F). There are two predicted target sites for miR-106b/93 in the 3′-UTR of p21 (Supplementary Figure 2). Transfection of miR-106b or miR-93 decreased reporter expression (Figure 2F and data not shown). Our data are consistent with a recent report that showed the suppression of p21 by miR-106b (18,19).
The miR-222 ∼ 221 cluster regulates both p27 and p57
miR-222 and miR-221 are closely related to each other in sequences. Their loci are in close vicinity to each other in the genome, suggesting that the cluster was created by the duplication of a common ancestral miRNA. We noticed potential target sites of miR-222 and miR-221 in the 3′-UTR of the p27 and p57 mRNAs. These sites are conserved in almost all vertebrates. To validate this prediction, we transfected synthetic miRNA duplexes into HeLa cells that express relatively low levels of miR-222 and miR-221 (Figure 1E). When the mixture of miR-222 and miR-221 (miR-222/221) was transfected, the protein levels of p27 and p57 were decreased as determined by western blotting (Figure 3A). Because the p57 level is low in HeLa cells, dexametasone was added to the medium to increase the p57 level so that the silencing effect can be seen more clearly (Figure 3A, lanes 3 and 4) (20). miR-222 and miR-221 showed comparable effects when they were transfected separately (Supplementary Figure 3). Even a small amount of miRNA (0.5 nM of miR-222 plus 0.5 nM of miR-221) was sufficient to induce significant silencing of p27 (data not shown). These data independently confirm the previous reports on the suppression of p27 and p57 by the miR-222 family (21–25).
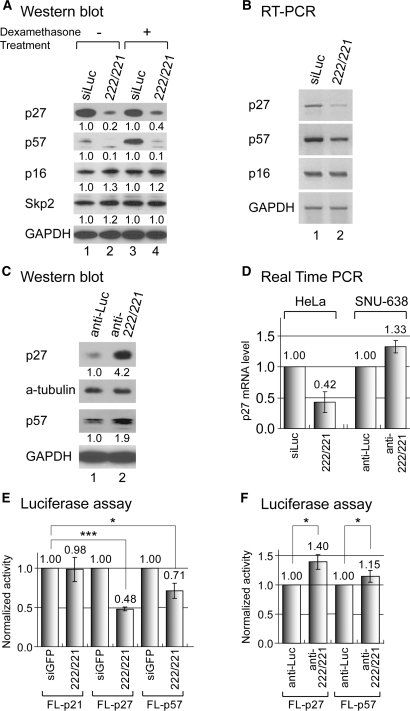
Figure 3.
Regulation of p27 and p57 by miR-222 ∼ 221 cluster. (A) Western blot analysis shows that p27 and p57 proteins are down-modulated by miR-222 ∼ 221. A total 30 nM of small RNA duplexes were transfected into HeLa cells. For dexamethasone treatment (lanes 3 and 4), HeLa cells were transfected with RNA duplexes and one day later, 100 nM of dexamethasone was treated for another 16 h. (B) RT-PCR demonstrates that p27 and p57 are suppressed by miR-222 ∼ 221 at the mRNA level. Total RNA was extracted from the same samples as in (A). (C) Western blot analysis. p27 and p57 are upregulated in the presence of the inhibitors of miR-222 and miR-221. SNU-638 cells were transfected with a mixture of antisense inhibitors against miR-222 and miR-221 (200 nM). (D) RT-PCR. The same samples as in (B) and (C) were used for quantitative real-time RT-PCR to measure p27 mRNA level. Endogenous GAPDH level was measured for normalization. (E and F) p27 and p57 are directly regulated by miR-222 ∼ 221 through the 3′-UTR. The reporters containing the 3′-UTR of p27 or p57 were transfected into HeLa (E) or SNU-638 (F) cells. At the same time, small RNA duplex [(E), 30 nM] or 2′-O-methyl RNA inhibitors [(F), 200 nM] were co-transfected, and luciferase expression was assayed 24 h-post-transfection (n = 3, mean±SD). Paired one-tail T-test was used to calculate the P-value (*P < 0.1, ***P < 0.001).
The level of p16Ink4a, which belongs to an unrelated family of Cdk inhibitors, was not affected by miR-222 ∼ 221, indicating that these effects are specific to the Cip/Kip family Cdk inhibitors. Because both p27 and p57 are the natural substrates of ubiquitin ligase Skp2/Csk1 (26,27), we also checked the level of Skp2 (Figure 3A). There was no detectable difference at the level of Skp2 protein, indicating that the silencing is unlikely to be a result of proteolysis mediated by Skp2.
Downregulation was also observed at the mRNA levels as determined by RT-PCR (Figure 3B). The mRNA levels of p27 and p57 were decreased by miR-222/221, indicating that miR-222/221 may elicit not only translational repression but also mRNA destabilization of p27 and p57.
Then, to address the question whether the miRNA inhibitor can reverse the suppression of p27 and p57, we introduced the mixture of two inhibitors (anti-miR-222 and anti-miR-221) into gastric cancer cell line SNU-638, which expresses relatively high levels of miR-222 and miR-221 (Figure 1E). When anti-miR-222/221 were transfected, p27 and p57 proteins were upregulated specifically (Figure 3C).
We also quantitatively measured the mRNA level of p27 in HeLa and SNU-638 cells by real-time PCR (Figure 3D). As expected, the p27 mRNA level was decreased when the cells were transfected with miR-222/221 while it was increased in the presence of anti-miR-222/221. Notably, a previous study reported that miR-222/221 does not affect the p27 mRNA level (22). The difference may be due to the experimental condition or differences in cell types.
Next, the reporter plasmid containing the 3′-UTR of the target gene was co-transfected with synthetic miRNA duplexes (Figure 3E) or antisense inhibitors (Figure 3F). When miR-222 and miR-221 duplexes were introduced into HeLa cells, the expressions of the p27 reporter (FL-p27) as well as the p57 reporter (FL-p57) were reduced (Figure 3E). When the antisense inhibitors of miR-222 ∼ 221 were transfected into SNU-638 cells, the luciferase expression was reproducibly elevated as expected (Figure 3F).
Because our data show that p57 is regulated by both miR-25 and miR-222/221, we co-transfected these miRNAs and determined the expression of p57 reporter (Figure 4A). When these miRNAs were co-transfected, the reporter expression was slightly more suppressed compared to the individual miRNA-transfected samples. This result suggests that multiple miRNAs may play redundant roles and may have additive effects to some extent.
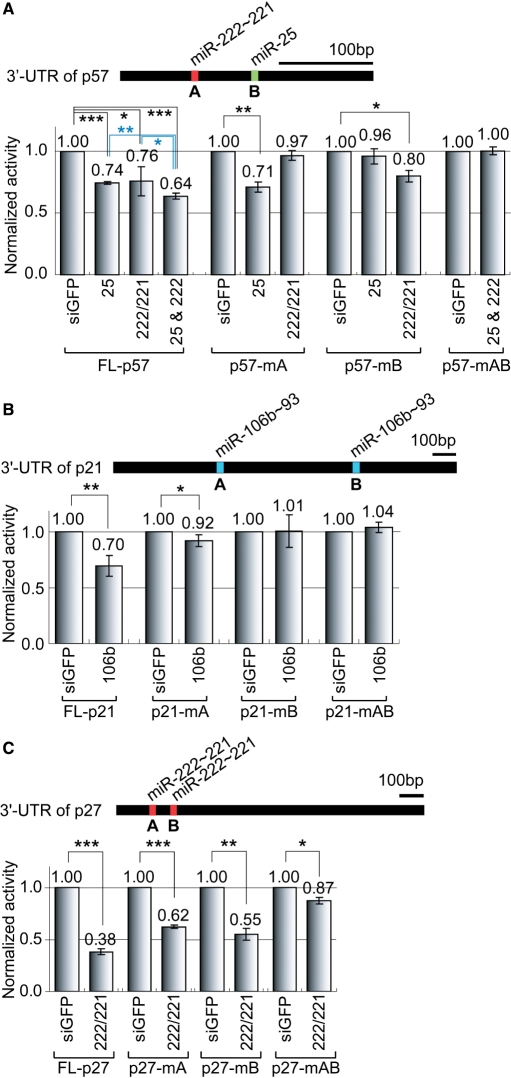
Figure 4.
Direct regulation of Cip/Kip family proteins by miRNAs through their 3′-UTR sequences. (A) The 3′-UTR of p57 contains a single binding site for miR-25 and a single binding site for miR-222 ∼ 221. The same experiment as in Figure 2E was conducted, except that 1 nM of RNA duplexes were used to confirm the additive effect in this experiment (n = 3, mean±SD). (B) The 3′-UTR of p21 contains two binding sites for miR-106b. We generated mutations in the putative target sites, and a similar experiment as in (A) was conducted (n = 4, mean±SD). (C) The 3′-UTR of p27 harbors two putative binding sites for miR-222 ∼ 221. We tested the effects of mutations in the target sites as in (B) (n = 3, mean±SD). Paired one-tail T-test was used to calculate the p-value (*P < 0.1, **P < 0.01, ***P < 0.001).
To confirm that the predicted target sites are responsible for the silencing, we carried out site-directed mutagenesis on the target sites (Figure 4). We generated mutations at the ‘seed’-sequences of the target sites, which are known to be critical for the binding of miRNA to its target mRNA. The 3′-UTR of p57 contains one putative target site for miR-222 ∼ 221 and one target site for miR-25 (Figure 4A). When the miR-222/221 site (site A) was mutated, the reporter failed to respond to miR-222/221. When we introduced a mutation into the miR-25 target site (site B), the reporter also became resistant to miR-25 treatment. Thus, these data indicate that the predicted sites are indeed responsible for the regulation of p57.
The 3′-UTR of p21 possesses two predicted target sites for miR-106b/93 (Figure 4B). When we mutated any one of the target sites, the silencing effect was diminished, suggesting that both target sites are required for the suppression of p21 (Figure 4B and data not shown).
There are two predicted target sites for miR-222/221 in the 3′-UTR of the p27 mRNA (Figure 4C). When we mutated the target sites individually, the silencing was partially relieved. When we introduced double mutations to both target sites, the silencing was nearly abolished, indicating that these two sites mediate miR-222 ∼ 221-induced suppression.
Cell-cycle regulation by the miRNA clusters
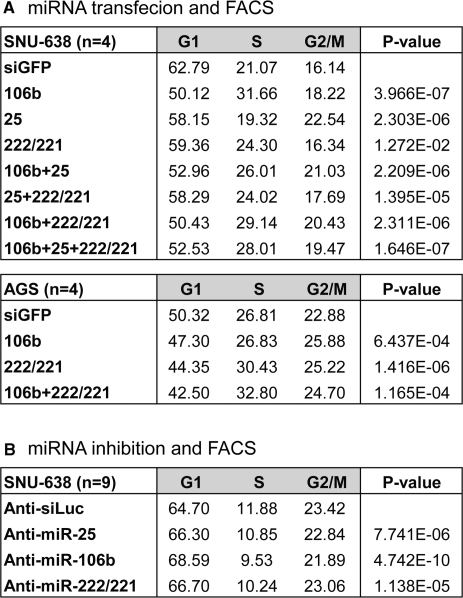
Because miR-106b and miR-222 clusters downregulate the Cdk inhibitors, it is expected that they may induce the activation of Cdk. As expected, the activity of Cdk2, as measured by phosphorylation of histone H1, was markedly increased in the miR-222/221-transfected sample, indicating that Cdk2 was stimulated as a result of miRNA transfection (Supplementary Figure 4). Because it is well established that the Cip/Kip inhibition and the Cdk2 activation is a prerequisite for G1/S transition, one would expect that cell-cycle profile may be influenced by miR-106b and miR-222 cluster. To test this, we transfected synthetic miRNAs into gastric cancer cell lines SNU-638 and AGS. Two days later, the cell-cycle profile was analyzed by FACS. The proportion of G1 phase fell in cells transfected with miR-106b or miR-25 (Figure 5A). Transfection of miR-222/221 also resulted in a decrease in G1 proportion (Figure 5A). We also transfected the mixture of half or quarter amounts of miR-106b, miR-25 or miR-222/221 to SNU-638 or AGS cells. When multiple miRNAs are co-transfected simultaneously, the proportion of G1 phase was decreased more than the average value of that from individual miRNAs (Figure 5A). Interestingly, slight but reproducible additive effect was observed in AGS cells suggesting the effect may be cell type dependent. Thus, although these miRNAs appear to have largely redundant functions in cell-cycle control, co-expression of these miRNAs may have significant additive effects in vivo, ensuring the G1/S transition.
Figure 5.
Effects of miR-106b or miR-222 clusters on cell cycle. FACS analysis was performed using the cells transfected with either miRNA or miRNA inhibitors. (A) Thirty nanomoles of control siRNA (siGFP), miR-106b, miR-25, miR-222/221 or combinations of these miRNAs were transfected into SNU-638 or AGS cells. Two days later, cell-cycle profile was analyzed (n = 4, mean). (B) Antisense inhibitors of siLuc, miR-25, miR-106b or miR-222/221 were transfected into SNU-638 cells. Two days later, cell-cycle profile was analyzed by FACS (n = 9, mean). For each treatment, paired one-tail T-test was used to calculate the P-value.
We also analyzed the effect of anti-miRNA oligonucleotides on cell cycle. Cell numbers in G1 phase was reproducibly increased by anti-miR-25, anti-miR-106b or anti-miR-222/221 (Figure 5B). Together, our data indicate that miR-106b and miR-222 clusters facilitate the cell-cycle progression by silencing Cdk inhibitors.
The high levels of miR-106b and miR-222 clusters in cancer cells may keep the expression of Cdk inhibitors under strong suppression, thereby paving the way for tumor progression. miR-222 ∼ 221 is highly expressed in many tumors and is induced by serum treatment (Supplementary Figure 5). When AGS and MKN-28 gastric cancer cells were depleted of serum for 24 h and re-added with serum, miR-222 ∼ 221 was induced while the p27 level was reduced. This result suggests that the miR-222 cluster may be activated by mitogenic stimuli.
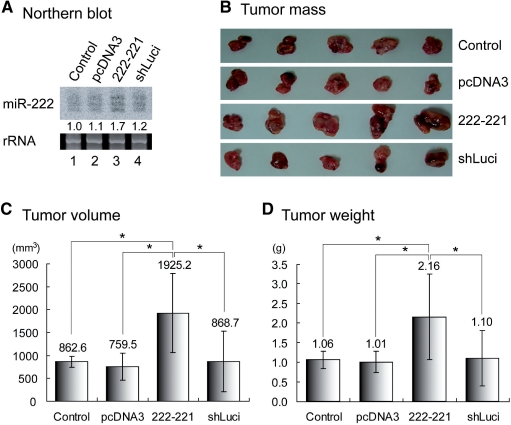
It was previously shown that the miR-17 ∼ 18a∼19a∼20a∼19b-1 cluster enhances tumor formation in transgenic mice (28). Because the miR-17 cluster is highly similar to the miR-106b family, the miR-106b cluster is likely to have a similar tumorigenic activity. However, the miR-222 cluster has not yet been examined for tumorigenic effects using a solid tumor model (29). To address this issue, we carried out tumor xenograft experiments using nude mice (Figure 6). For this, the gastric cancer cell line AGS was introduced with the miR-222 ∼ 221-expression plasmid. The stable cell line that overexpresses miR-222 ∼ 221 was selected and injected into nude mice. Two control cell lines were used for comparison: the cell line transfected with pcDNA3 null vector and the other cell line transfected with the vector expressing shRNA against luciferase gene. Four weeks later, the volume and weight of the tumor were measured. Mice injected with miR-222 ∼ 221-expressing cells developed significantly bigger tumor mass compared to the mice with the control cell lines (Figure 6). These data indicate that high levels of miR-222 ∼ 221 may enhance the proliferation of cancer cells in vivo.
Figure 6.
Effect of miR-222 overexpression on tumor formation in nude mice. (A) The expression level of miR-222 in each stable cell line was measured by northern blot analysis. Ethidium bromide staining of rRNA was used for RNA loading control. (B) Stable cell lines were injected at the same time and the tumor formation in nude mice was monitored over a 4-week period. Four weeks later, the mice were sacrificed to obtain tumor masses. Average value of (C) tumor volume and (D) tumor weight were measured and shown with standard deviation (n = 5). Paired one-tail T-test was used to calculate the P-value (*P < 0.1).
DISCUSSION
One of the hallmarks of cancer is its rapid proliferation. Overexpression of miRNAs that repress the inhibitors of Cdk would help cancer cells to overcome the cell-cycle checkpoint. Our study demonstrates that two miRNA clusters (miR-106b and miR-222 clusters) exert their oncogenic functions by co-operatively suppressing three related Cdk inhibitors (p57, p21 and p27) (Figure 7). We also show that miR-222/221 can stimulate gastric tumor growth in vivo in a mouse xenograft model. Thus these miRNAs may contribute to the development of human tumors including gastric cancer. Although gastric cancer is a major health burden throughout the world, the molecular pathology remains poorly understood. Analyses of miRNA regulation will provide opportunities to develop new means for diagnosis and treatment of gastric cancer.
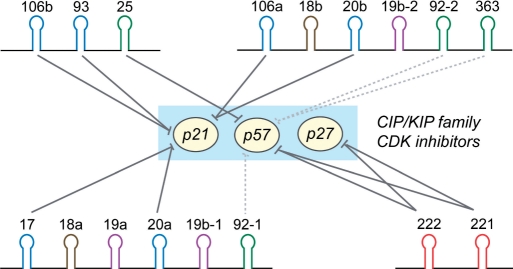
Figure 7.
Schematic model of the regulation of Cip/Kip family proteins by the miR-222 cluster and the miR-106b cluster. The miR-17 and miR-106a clusters are the paralogs of miR-106b cluster. miR-106 paralogs were recently shown to target p21 (18,19). The suppression of p27 and p57 by the miR-222 family was recently described by other groups (21–25). Solid lines indicate the experimentally confirmed inhibition from previous and our studies (see the text). The probable inhibition inferred from miRNA sequences are indicated with dotted lines.
There exist multiple paralogs of the miR-106b cluster in the human genome while the miR-222 cluster does not have an apparent paralog. The miR-106b paralogs are likely to play similar roles in facilitating cell proliferation (Supplementary Figure 1). The miR-17 cluster on chromosome 13 caused B-cell lymphoma in transgenic mice when overexpressed with c-myc (28). The miR-17 cluster contains three additional components (miR-19a, miR-19b-1 and miR-18a) that have slightly different seed sequences from miR-17/miR-20/miR-106/miR-93. Interestingly, the miR-17-related miRNAs were recently shown to control several key molecules in cell-cycle control including cyclin D1 (30). The two embryonic stem-cell-specific miRNA clusters (the miR-302b∼302c∼302a∼302d∼367 cluster on chromosome 4 and the miR-371 ∼ 372 ∼ 373 cluster on chromosome 19) also contain miRNAs that are similar to miR-106b/93 and miR-25 (31), which may contribute to the rapid proliferation of stem cells by downregulating p21 and p57. Recent studies reported the functions of miR-106b in targeting p21 and that of miR-222/221 in controlling p27 and p57 (18,19,21–25). Our study independently confirms and extends these observations. We show that p27 or p57 is suppressed not only at the protein level but also at the mRNA level. In addition, we confirm that the silencing of p27 and p57 by miR-222/221 is not attributable to Skp2-mediated proteolysis.
We further find that miR-25 directly targets p57. miR-25 and its paralogs (miR-92, miR-367, and miR-32) may collaborate with the miRNAs in the same cluster (such as miR-106b, miR-93, miR-17, miR-20 and miR-302) so as to effectively control Cdk inhibitors. So our study provides strong evidence that miRNA clusters (operons) contain functionally related miRNAs (cistrons). The first two cistrons of the miR-106b cluster (mir-106b and miR-93) control p21 while the last cistron of the operon (miR-25) targets p57. Perhaps miRNA operons have evolved to coordinate the expression of the functionally related miRNAs. Transcription from the common promoter can ensure the generation of individual components in a co-regulated manner.
Our study also shows that separate clusters can be co-expressed and have related activities. While the miR-106b and miR-222 clusters are separately located in the genome, their expression patterns are highly similar and they play a similar role in cell cycle control by inhibiting three Cdk inhibitors. It would be of interest to understand how these two clusters are co-regulated. It would also be interesting to see if miRNA operons cooperate with each other to enhance disease phenotype. Because multiple miRNAs play related roles during tumorigenesis, effective anticancer therapy will require combinatorial approaches targeting multiple oncogenic miRNAs. Thus, it is of high importance to identify functionally related miRNAs and to investigate the cooperative and/or redundant activities of oncogenic miRNAs. Understanding of the miRNA gene structures and regulatory mechanisms will provide the basis for such combinatorial therapeutic approaches.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Creative Research Initiatives from the Ministry of Education, Science and Technology of Korea (R16-2007-073-01000-0); The National R&D Program for Cancer Control by the Ministry of Health, Welfare and Family affairs of Korea (0720090); and a BK21 Research Studentship (to Y.K.K. and S.Y.P.). Funding for open access charges: The National Creative Research Initiatives from the Ministry of Education, Science and Technology of Korea (R16-2007-073-01000-0).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the members of our laboratory for their helpful discussions and technical assistance. We offer particular thanks to Jung Hyun Lee and Jinuk Choi for their assistance in FACS experiments. We are also grateful to Dr Deog Soo Hwang and Dr Hong-Duk Youn for the antibodies, and Dr Yong-Keun Jung, Kyung Yong Lee, and Ho-June Lee for their advice.
REFERENCES
- 1.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell. Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 11.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 12.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev. Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 14.Cao ZA, Daniel D, Hanahan D. Sub-lethal radiation enhances anti-tumor immunotherapy in a transgenic mouse model of pancreatic cancer. BMC Cancer. 2002;2:11. doi: 10.1186/1471-2407-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Samuelsson MK, Pazirandeh A, Davani B, Okret S. p57Kip2, a glucocorticoid-induced inhibitor of cell cycle progression in HeLa cells. Mol. Endocrinol. 1999;13:1811–1822. doi: 10.1210/mend.13.11.0379. [DOI] [PubMed] [Google Scholar]
- 21.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 22.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 24.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 25.Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell. 2004;6:661–672. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 27.Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc. Natl Acad. Sci. USA. 2003;100:10231–10236. doi: 10.1073/pnas.1831009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 30.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell. Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.