Abstract
Antimonials remain the first line drug against the protozoan parasite Leishmania but their efficacy is threatened by resistance. We carried out a RNA expression profiling analysis comparing an antimony-sensitive and -resistant (Sb2000.1) strain of Leishmania infantum using whole-genome 70-mer oligonucleotide microarrays. Several genes were differentially expressed between the two strains, several of which were found to be physically linked in the genome. MRPA, an ATP-binding cassette (ABC) gene known to be involved in antimony resistance, was overexpressed in the antimony-resistant mutant along with three other tandemly linked genes on chromosome 23. This four gene locus was flanked by 1.4 kb repeated sequences from which an extrachromosomal circular amplicon was generated in the resistant cells. Interestingly, gene expression modulation of entire chromosomes occurred in the antimony-resistant mutant. Southern blots analyses and comparative genomic hybridizations revealed that this was either due to the presence of supernumerary chromosomes or to the loss of one chromosome. Leishmania parasites with haploid chromosomes were viable. Changes in copy number for some of these chromosomes were confirmed in another antimony-resistant strain. Selection of a partial revertant line correlated antimomy resistance levels and the copy number of aneuploid chromosomes, suggesting a putative link between aneuploidy and drug resistance in Leishmania.
INTRODUCTION
The protozoan parasite Leishmania is the etiological agent of a group of diseases termed leishmaniasis. Human leishmaniasis has a prevalence of 12 million cases, an estimated population of 350 million at risk and an incidence of 2 million new cases annually (1). No effective vaccine is yet available against this parasite and its control relies primarily on chemotherapy. Pentavalent antimony containing compounds such as sodium stibogluconate (Pentostam) and N-methylglucamine (Glucantime) remain the mainstay against all forms of Leishmania infections (1), but their efficacy is threatened by antimony-resistant parasites now described on a frequent basis in several endemic regions (2–4). Other drugs include pentamidine and amphotericin B but severe side effects and high cost have limited their widespread use. The aminoglycoside paromomycin is now more frequently used (1,5,6) and the first oral drug miltefosine shows promising efficacy despite evidence suggesting that resistance could develop rapidly (7). Since there are few new drugs in the pipeline (5,8–11) and resistance to first-line drug(s) has a significant therapeutic impact on this important parasitic disease, a better understanding of the molecular and biochemical mechanisms leading to resistance is warranted.
The mechanisms involved in antimony resistance in Leishmania are partly understood, at least in in vitro selected antimony-resistant strains. These are mainly associated with a reduced activation or an altered transport of the drug. Pentavalent antimony (SbV) is believed to be a prodrug that requires biological reduction to its trivalent form (SbIII) in either the host macrophages and/or the parasites to acquire antileishmanial activity [reviewed in (5,8)] and a decreased SbV reduction rate has been reported in sodium stibogluconate-resistant amastigotes (12). The uptake of SbIII in Leishmania has been shown to be mediated by aquaglyceroporins and downregulation of aquaglyceroporin 1 (AQP1) was associated with an increased SbIII resistance (13,14). Furthermore, an increased level of trypanothione (TSH), the main cellular thiol in Leishmania (15), has been observed in mutants selected for antimony resistance (16–21) and could help to restore thiol redox potential which is perturbed following antimony accumulation (22,23). Increased levels of TSH also allow for the formation of thiol adducts of antimony that are either detoxified by their transport inside an intracellular organelle by the ATP-binding cassette (ABC) transporter MRPA or extruded from the parasite by an unidentified plasma membrane efflux system. The increased expression of MRPA is often due to the amplification of its gene in antimony-resistant strains (16,24,25). Other genes and proteins including a heat-shock protein 70 (26), a heat-shock protein 83 (27), a protein of the leucine-rich repeats (LRRs) superfamily (28), an amplified DNA from field isolates (29), a tryparedoxin peroxidase (30) and a very large polypeptide (31) were also associated with an increased tolerance toward antimony in some parasites. Several, but not all, of the antimony resistance mechanisms identified while studying in vitro selected mutants were also found in resistant field isolates (2,22,32–36).
Resistance to antimonials seems to implicate several mutational events co-existing within the same cell (8). Accordingly, the simultaneous analysis of the whole genome could provide useful information about the complex events leading to resistance to this class of drugs. Small targeted DNA microarrays have already shown their usefulness in studying drug resistance mechanisms in Leishmania (17,37,38). In order to study gene expression modulation associated with antimony resistance on a full-genomic scale, we have carried out a RNA expression profiling experiment comparing an antimony-sensitive (WT) and an antimony-resistant (Sb2000.1) strain of Leishmania infantum using recently generated whole-genome 70-mer oligonucleotide microarrays for Leishmania (39,40). These experiments pinpointed specific mechanisms and novel changes in ploidy (supernumerary chromosomes and monosomy) that were correlated to resistance.
MATERIALS AND METHODS
Cell lines
Leishmania infantum promastigotes were grown at 25°C in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum and 10 μg/ml hemin. The L. infantum wild-type (WT) strain (MHOM/MA/67/ITMAP-263), the L. infantum Sb2000.1 mutant (37) and the L. infantum Sb4000.4 mutant (38) selected for SbIII resistance have been described previously. The L. infantum Sb400.2 and Sb1000.3 mutants were selected from a cloned parental population using a stepwise selection until they were resistant to 400 and 1000 μM SbIII, respectively. The L. infantum Sb2000.1 mutant was grown in the absence of SbIII for 30 passages in order to obtain a (partial) revertant line. Leishmania promastigotes were transfected by electroporation as reported previously (41). Growth curves were obtained by measuring absorbance at 600 nm of a 200-μl culture aliquot at 72 h using an automated microplate reader.
DNA manipulations
Total DNA was isolated using DNAzol reagent (Invitrogen) as recommended by the manufacturer. For quantitative Southern blots, the genomic DNA was digested with BamHI and EcoRV and migrated overnight at 30 V in 0.7% agarose gels. Southern blots, hybridizations and washes were performed following standard protocols (42) and all probes were obtained by PCR. Densitometric analyses of Southern blots were performed using ImageQuant 5.2 (Molecular Dynamics). The pSPαNEOα-MRPA construct has been described previously (43). The LinJ32_V3.2190 inactivation cassette was generated using the folowing PCR primers: primer 1, 5′-GCTCTAGAAGCTCCTGCGGTTTGCCTAC and primer 2, 5′-CCGTTATTGTGCCGACTGCCGGATCCGGGTGCGGAGTATGAAGGATG to amplify a region of 600-bp upstream of the gene and primer 3, 5′-GGCAGTCGGCACAATAACGG; and primer 4, 5′-CCCAAGCTTGGGTGTCACGCAGCTCCTTG in order to amplify a DNA fragment of 600-bp downstream of the gene. The upstream and downstream fragments were fused by PCR using primers 1 and 4 and the resulting DNA fragment was cloned in the pGEM-T-easy vector leading to the pGEM-UP/DOWN-32 vector. The XbaI, BamHI and HindIII restriction sites are underlined in the primers. An α-NEO-α cassette was isolated from the vector pSP72 α-NEO-α by digestion with BamHI and cloned in a unique BamHI restriction site located between the upstream and downstream fragments in pGEM-UP/DOWN-32 leading to the pGEM-KO32NEO plasmid. The LinJ32_V3.2190 inactivation cassette was isolated from pGEM-KO32NEO by an XbaI-HindIII digest and transfected in L. infantum WT, Sb2000.1 and Sb2000.1rev as previously described. The integration of the inactivation cassette at the LinJ32_V3.2190 locus was confirmed by PCR using the primers 5′- CAATACGCACGATGCACAGG and 5′-ACAACGTCGAGCACAGCTGC, and by Southern blots. Genomic DNA labeling was performed as previously described (40).
Microarray design
The recent completion of the genome sequencing of L. major (44) and L. infantum (45) has allowed the generation of multispecies high-density oligonucleotide microarrays as previously described (39,40).
RNA isolation and labeling
Total RNA was isolated from 108 Leishmania cells during the mid-log phase of growth using the RNeasy Plus Mini Kit (Qiagen) as described by the manufacturer. The RNAs were treated with RNase-free DNAse I Turbo (Ambion) to avoid any genomic contamination. The quality and quantity of RNA were assessed using RNA 6000 Nano Assay chips on a Bioanalyzer 2100 (Agilent technologies). The major criterion for RNA integrity was the presence of three clear ribosomal peaks (18S, 24Sα and 24Sβ) and the absence of RNA degradation. Ten micrograms of RNA were converted to aminoallyl-dUTP incorporated cDNA using random hexamers (Roche) and the SuperScript III Rnase H-Reverse Transcriptase (Invitrogen). Aminoallyl-dUTP incorporated cDNAs were thereafter coupled to Alexa Fluor 555 or Alexa Fluor 647 following manufacturer recommendations. Fluorescent cDNAs were purified with MinElute Spin Column (Qiagen) and quantified spectrophotometrically.
Microarrays hybridization
Pre-hybridization and hybridization were performed as previously described (39,40). Hybridizations were performed in five replicates using five independent RNA extractions and dye swapping.
Microarray data acquisition and analysis
Detection of the Alexa Fluor 555 and Alexa Fluor 647 signals was performed on a G2565CA DNA microarray scanner (Agilent technologies) at a 5-μm resolution. The signal intensity data were extracted from the primary scanned images using the GenePix Pro 6.0 software (Axon Technologies). Raw data were imported and normalization and statistical analyses were performed in R 2.2.1 using the LIMMA (Linear Models for Microarray Data) 2.7.3 package (46–48) as previously described (40). Multiple testing corrections were done using the false discovery rate method with a threshold P-value of 0.05. Only genes statistically significant with an absolute log2-transformed expression ratio greater than 0.58 were considered as differentially expressed. GeneSpring GX 3.1 was used for the generation of the chromosome map of expression ratios. The entire dataset was deposited to GEO under the reference number series GSE9949.
Real-time RT-PCR
Three independent RNA preparations were used for each real-time PCR experiment. First-strand cDNA was synthetized from 10 μg of RNA using random hexamers (Roche) and the SuperScript III RNase H-Reverse Transcriptase (Invitrogen) according to the manufacturer recommendations. Equal amounts of cDNA were run in triplicate and amplified in 15 μl reactions containing 7.5 μl of 2X Universal PCR Master Mix (Applied Biosystems), 10 nM Z-tailed forward primer, 100 nM reverse primer, 250 nM Amplifluor Uniprimer probe (Chemicon) and 1 µl cDNA target. Mixtures were incubated at 50°C for 2 min, at 95°C for 4 min and then cycled 55 times at 95°C for 15 s and at 55°C for 30 s using an Applied Biosystems Prism 7900 Sequence Detector at the Gene Quantification Core Laboratory of the Centre de Génomique de Québec (https://genome.ulaval.ca/qrtpcr). No-template controls were used as recommended. Amplification was normalized to the LinJ18_V3.0630 and LinJ36_V3.0850 genes, for which a highly stable expression was noted in several conditions by different microarrays experiments, before quantities of target genes were calculated according to standard curves using the REST 2005 software (Corbett Life Science). Primers were designed using Primer Express 2.0 (Applied Biosystems).
RESULTS
Gene expression profiling in L. infantum antimony-resistant mutants
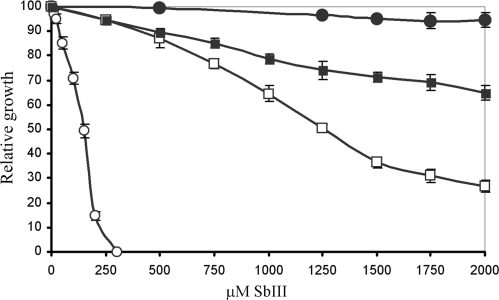
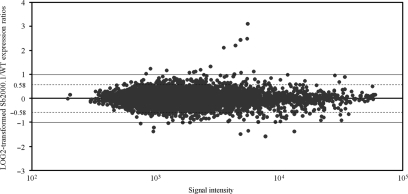
The completion of the L. major (44) and L. infantum (45) genome sequences allowed the generation of full genome 70-mer oligonucleotides DNA microarrays suitable for genome-wide expression profiling analyses in L. major and L. infantum (39,40). The arrays were used to study gene expression modulation associated with antimony resistance in the L. infantum Sb2000.1 mutant. This mutant is highly resistant to SbIII compared to its parental sensitive WT strain (Figure 1). The Sb2000.1 mutant has already been studied with small targeted arrays of 50 genes (37,38) but not at the whole genome level. The log2-transformed gene expression ratios of Sb2000.1 compared to its parental strain are shown in Figure 2. The majority of the spots aligned along the line of best fit, suggesting that genes represented by these spots were equally expressed in both samples. Nonetheless, when applying a cutoff of at least 2-fold differential expression (P < 0.05), 24 genes were significantly modulated at the RNA level in the Sb2000.1 mutant (Table 1). These included the gene coding for the ABC transporter MRPA whose amplification in Sb2000.1 has previously been reported (37,38). When the cutoff was adjusted from a 2-fold to a 1.5-fold difference in mRNA abundance (P < 0.05), a total of 84 genes were found to be significantly modulated in the Sb2000.1 mutant (Table 1).
Figure 1.
Antimony susceptibilities in L. infantum. Growth of L. infantum promastigote cells in the presence of increasing concentrations of SbIII was monitored at 72 h by OD mesurements at 600 nm. The average of three experiments is shown. Leishmania infantum WT (open circles); L. infantum Sb2000.1 (filled circles); L. infantum Sb2000.1rev P30 (open squares); L. infantum Sb2000.1rev P30 transfected with pSPαNEOα-MRPA (filled squares).
Figure 2.
Gene expression profiling of antimony-resistant L. infantum promastigotes. Plot of log2-transformed Sb2000.1/WT expression ratios as a function of hybridization intensities. An average of five independent experiments is presented. External lines and dashed lines indicate 2- and 1.5-fold differences, respectively.
Table 1.
Genes significantly modulated in L. infantum Sb2000.1 as determined by microarrays*
| GeneDB V3.0 systematic ID | Gene description | Fold difference |
|---|---|---|
| Downregulated in L. infantum Sb2000.1 | ||
| LinJ02_V3.0520 | Hypothetical protein, unknown function | −1.72 |
| LinJ03_V3.0360 | Hypothetical protein, conserved | −2.08 |
| LinJ05_V3.0280 | Protein tyrosine phosphatase, putative | −1.73 |
| LinJ05_V3.0830 | Methylthioadenosine phosphorylase, putative | −3.01 |
| LinJ06_V3.1280 | Hypothetical protein, conserved | −2.60 |
| LinJ06_V3.1340 | Protoporphyrinogen oxidase-like protein | −1.74 |
| LinJ06_V3.1350 | Hypothetical protein, unknown function | −2.00 |
| LinJ09_V3.0100 | RNA-binding protein 5-like protein | −1.70 |
| LinJ09_V3.0580 | LRR protein, putative | −1.70 |
| LinJ09_V3.0650 | Cyclin 1, putative | −1.78 |
| LinJ09_V3.0690 | Hypothetical protein, conserved | −2.00 |
| LinJ09_V3.0970 | Calmodulin, putative | −1.89 |
| LinJ09_V3.0980 | Calmodulin, putative | −1.89 |
| LinJ09_V3.1050 | Hypothetical protein, conserved | −1.69 |
| LinJ12_V3.0010 | Surface antigen protein 2, putative | −1.88 |
| LinJ12_V3.0020 | Surface antigen protein 2, putative | −1.75 |
| LinJ12_V3.0040 | Surface antigen protein 2, putative | −1.88 |
| LinJ12_V3.0080 | No description | −1.88 |
| LinJ12_V3.0110 | Surface antigen protein, putative | −1.99 |
| LinJ12_V3.0120 | HGPRT,hypoxanthine-guanine phosphoribosyltransferase | −1.77 |
| LinJ12_V3.0130 | XRPT, xanthine phosphoribosyltransferase | −1.87 |
| LinJ12_V3.0290 | Hypothetical protein, unknown function | −1.72 |
| LinJ12_V3.0350 | 3′-nucleotidase/nuclease, putative | −1.68 |
| LinJ12_V3.0680 | Surface antigen protein 2, putative | −1.75 |
| LinJ12_V3.0690 | Surface antigen protein 2 precursor | −1.88 |
| LinJ17_V3.0120 | Receptor-type adenylate cyclase a | −1.80 |
| LinJ18_V3.1640 | Hypothetical protein, conserved | −1.76 |
| LinJ23_V3.1830 | Hypothetical protein, unknown function | −1.79 |
| LinJ23_V3.1930 | Hypothetical protein, conserved | −1.98 |
| LinJ26_V3.0120 | Adenine phosphoribosyltransferase | −1.85 |
| LinJ29_V3.0260 | Oxidase-like protein | −2.81 |
| LinJ30_V3.2870 | Hypothetical protein, conserved | −1.73 |
| LinJ31_V3.0610 | Amino acid transporter aATP11, putative | −1.72 |
| LinJ31_V3.0430 | Calpain-like cysteine peptidase, putative | −1.87 |
| LinJ31_V3.1240 | Vacuolar-type proton translocating pyrophosphatase 1 | −1.94 |
| LinJ31_V3.2380 | 3′-nucleotidase/nuclease precursor, putative | −2.59 |
| LinJ32_V3.0240 | Dynein light chain, flagellar outer arm, putative | −1.79 |
| LinJ32_V3.0360 | Hypothetical protein, conserved | −1.70 |
| LinJ32_V3.0690 | Centrin, putative | −1.79 |
| LinJ32_V3.1760 | Hypothetical protein, conserved | −2.57 |
| LinJ32_V3.2020 | Hypothetical protein, conserved | −1.96 |
| LinJ32_V3.2030 | Hypothetical protein, conserved | −1.72 |
| LinJ32_V3.2150 | COP-coated vesicle membrane protein p24 precursor | −1.84 |
| LinJ32_V3.2400 | Hypothetical protein, unknown function | −2.33 |
| LinJ32_V3.3100 | Nucleoside diphosphate kinase b | −1.69 |
| LinJ32_V3.3110 | Nucleoside diphosphate kinase b | −1.69 |
| LinJ32_V3.3350 | Hypothetical protein, conserved | −1.79 |
| LinJ32_V3.4080 | Hypothetical protein, conserved | −1.71 |
| LinJ34_V3.2150 | Hypothetical protein, unknown function | −1.87 |
| LinJ35_V3.1050 | Hypothetical protein, unknown function | −2.06 |
| LinJ36_V3.0430 | Hypothetical protein, conserved | −1.71 |
| Upregulated in L. infantum Sb2000.1 | ||
| LinJ01_V3.0470 | α/β-hydrolase-like protein | 2.24 |
| LinJ01_V3.0540 | Long chain fatty acid CoA ligase, putative | 1.77 |
| LinJ11_V3.0020 | Hypothetical protein, conserved | 1.89 |
| LinJ11_V3.0030 | Hypothetical protein, conserved | 1.69 |
| LinJ11_V3.0040 | ABC transporter, putative | 1.75 |
| LinJ11_V3.0080 | Hypothetical protein, conserved | 1.68 |
| LinJ11_V3.0130 | NGG1 interacting factor 3-like protein | 1.72 |
| LinJ11_V3.0170 | Hypothetical protein, conserved | 1.71 |
| LinJ11_V3.0180 | Hypothetical protein, conserved | 2.13 |
| LinJ11_V3.0210 | Acidocalcisomal pyrophosphatase | 1.99 |
| LinJ11_V3.0240 | Proteasome alpha 7 subunit, putative | 2.08 |
| LinJ11_V3.0520 | Nucleoside transporter-like protein | 2.49 |
| LinJ11_V3.0550 | Amino acid permease/transporter, putative | 1.78 |
| LinJ11_V3.0560 | Protein kinase, putative | 1.71 |
| LinJ11_V3.0570 | Hypothetical protein, conserved | 2.09 |
| LinJ11_V3.0580 | Tetratricopeptide repeat (TPR) protein | 1.86 |
| LinJ11_V3.0590 | Hypothetical protein, conserved | 1.83 |
| LinJ11_V3.0680 | Hypothetical protein, conserved in leishmania | 1.88 |
| LinJ11_V3.0690 | Hypothetical protein, conserved in leishmania | 1.88 |
| LinJ11_V3.0730 | Hypothetical protein, conserved | 1.95 |
| LinJ14_V3.1420 | δ-6 fatty acid desaturase, putative | 1.69 |
| LinJ14_V3.1450 | Myo-inositol-1-phosphate synthase | 2.27 |
| LinJ17_V3.0390 | Hypothetical protein, conserved | 1.76 |
| LinJ19_V3.0870 | Pteridine transporter, putative | 1.70 |
| LinJ20_V3.1740 | Aminoacylase, putative | 2.35 |
| LinJ23_V3.0270 | Hypothetical protein, conserved | 4.30 |
| LinJ23_V3.0280 | Terbinafine resistance locus protein (yip1) | 8.50 |
| LinJ23_V3.0290 | Multidrug resistance protein, putative | 5.39 |
| LinJ23_V3.0300 | Argininosuccinate synthase, putative | 5.59 |
| LinJ23_V3.0700 | Hypothetical protein | 1.92 |
| LinJ25_V3.0740 | Eukaryotic initiation factor 5a, putative | 1.91 |
| LinJ25_V3.0750 | Eukaryotic initiation factor 5a, putative | 1.91 |
| LinJ25_V3.0760 | Eukaryotic initiation factor 5a, putative | 1.91 |
| LinJ25_V3.1040 | Hypothetical protein, conserved | 2.02 |
| LinJ25_V3.1210 | ATPase β subunit, putative | 1.80 |
| LinJ25_V3.1680 | Hypothetical protein, conserved | 1.69 |
| LinJ25_V3.1700 | Hypothetical protein, conserved | 1.71 |
| LinJ25_V3.2260 | Hypothetical protein, conserved | 1.68 |
| LinJ25_V3.2360 | Hypothetical protein, conserved | 1.69 |
| LinJ25_V3.2580 | ATPase β subunit, putative | 1.96 |
| LinJ29_V3.1320 | Hypothetical protein, conserved | 1.73 |
| LinJ29_V3.2940 | Hypothetical protein, conserved | 1.88 |
| LinJ31_V3.0750 | Hypothetical protein, conserved | 2.16 |
| LinJ33_V3.3260 | Hypothetical protein, conserved | 1.77 |
| LinJ33_V3.3390 | h1 histone-like protein | 2.15 |
*P < 0.05.
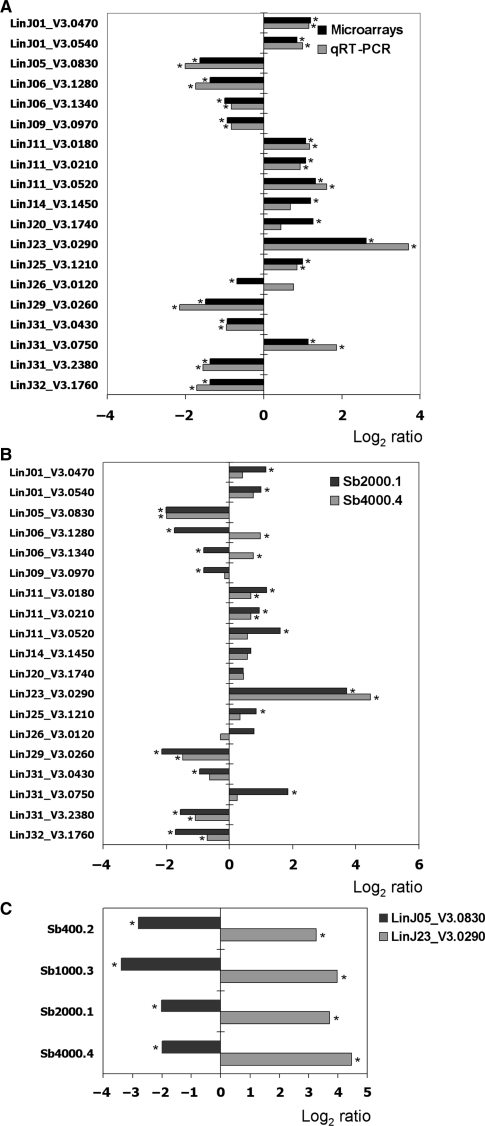
DNA microarrays data were confirmed by real-time quantitative RT-PCR (qRT-PCR) experiments performed on a set of 21 selected genes (19 test genes and 2 control genes) and the two techniques correlated, with only few discrepant results (Figure 3A). Indeed, whereas LinJ26_V3.0120 was found downregulated using DNA microarrays (−1.6-fold), it was found overexpressed in the qRT-PCR experiments (+1.7-fold), although not at a significant level. Furthermore, whereas LinJ14_V3.1450 and LinJ20_V3.1740 were significantly upregulated using DNA microarrays, their overexpression was determined as not significant in the qRT-PCR experiments. The remaining genes showed similar significant differences in mRNA abundance whether they were analyzed by qRT-PCR or DNA microarrays. Genes whose expressions are modulated in several independent resistant mutants would make good candidate drug resistance genes. Therefore, in order to link specific gene expression modulation with antimony resistance, the expression of the same set of selected genes was assayed by qRT-PCR in another independently selected antimony-resistant line, the L. infantum Sb4000.4 mutant. Out of the 16 genes significantly modulated in Sb2000.1 by qRT-PCR, 7 were also differentially expressed in Sb4000.4 (Figure 3B). The other nine genes were either not modulated at a significant level or were inversely regulated in Sb4000.4. With its 4-fold decreased expression, LinJ05_V3.0830 was the gene that showed the highest fold difference in both mutants, apart from LinJ23_V3.0290 (MRPA) (Figure 3A and B). This is the first report of LinJ05_V3.0830 being downregulated in antimony-resistant mutants of Leishmania and interestingly this gene was also shown to be highly downregulated in two other independently selected SbIII-resistant L. infantum strains, the Sb400.2 and Sb1000.3 mutants (Figure 3C). Since the expression of seven genes was similarly modulated in two independent mutants, we performed transfection experiments to test whether any of these genes could be involved in antimomy resistance. The upregulated and downregulated genes were transfected in L. infantum WT and Sb2000.1, respectively. However, under our experimental conditions, we did not observe a change in susceptibility to SbIII except for cells transfected with LinJ23_V3.0290 (MRPA) (data not shown and Figure 1), suggesting that if the genes tested are involved in resistance it is through indirect means.
Figure 3.
Validation of DNA microarrays expression data by quantitative real-time RT-PCR (qRT-PCR). (A) The mean log2-transformed Sb2000.1/WT ratios of selected genes from microarrays expression data (black bars) are compared to qRT-PCR data (grey bars). (B) The mean log2-transformed Sb2000.1/WT (black bars) and Sb4000.4/WT (grey bars) qRT-PCR expression ratios of a group of selected genes. (C) The mean log2-transformed qRT-PCR expression ratios of LinJ05_V3.0830 and LinJ23_V3.0290 in different L. infantum antimony-resistant mutants compared to the parental WT strain. The qRT-PCR data were normalized with the LinJ18_V3.0630 and LinJ36_V3.0850 genes, for which a highly stable expression was noted in several conditions by different microarrays experiments. An asterix beside the bars indicates a significant fold difference (P < 0.05).
Extrachromosomal circular amplification of a gene locus on chromosome 23
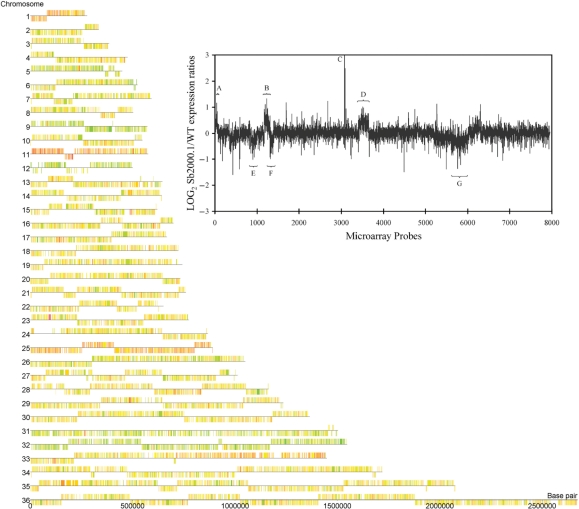
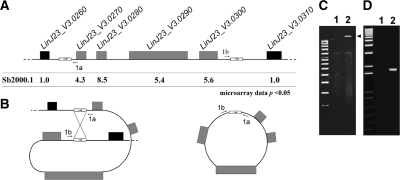
The data generated from the microarray hybridizations can be illustrated by a chromosome map representing gene expression levels on a genomic scale (Figure 4). The expression ratios of every gene on a particular chromosome is represented by a color code with overexpressed genes shown as orange to red features, downregulated genes shown as pale to bright green features and unmodulated genes shown as yellow features. This representation enabled the identification of a locus on chromosome 23 that was clearly overexpressed in Sb2000.1 (Figure 4, red locus on chromosome 23). A close examination of the expression data derived from the microarrays allowed to precisely define the genes located on this overexpressed locus. Indeed, the overexpressed genomic region comprises the genes LinJ23_V3.0270, LinJ23_V3.0280, LinJ23_V3.0290 (MRPA) and LinJ23_V3.0300 (Figure 5A), which were all substantially upregulated in Sb2000.1, while the genes located on each side of this four genes locus (LinJ23_V3.0260 and LinJ23_V3.0310) were not differentially expressed. Alkaline lysis allowed the recovery of a circular amplicon from Sb2000.1 (Figure 5C) that hybridized with a MRPA probe (data not shown) indicating that the increased gene expression observed by DNA microarrays at the MRPA locus resulted from the circular amplification of the small overexpressed genomic region. Generation of circular amplicons in Leishmania is often due to homologous recombination between direct repeats (24,25,49,50) and a close examination of the sequences bordering the amplified locus indeed revealed the presence of a 1389-bp repeated sequence of 100% nucleotide identity on each side of the locus amplified (Figure 5A). These direct repeats were also found on chromosome 23 of L. major and L. braziliensis (data not shown). To ascertain that the extrachromosomal circular amplicon was generated from homologous recombination between these direct repeats, a pair of PCR primers was designed to amplify a DNA fragment of 1.8 kb only from an extrachromosomal circular DNA template created by the recombination of the repeats and not from the chromosomal locus (Figure 5A). As expected, amplification of the 1.8-kb DNA fragment was observed from total DNA of L. infantum Sb2000.1 while no amplification was observed from total DNA of L. infantum WT (Figure 5D). Sequencing of the PCR fragment confirmed that the circle was formed by homologous recombination between the repeated sequences (data not shown).
Figure 4.
Chromosome map of L. infantum Sb2000.1/WT gene expression modulation. DNA microarrays data were analyzed with the GeneSpring GX3.1 software to illustrate the Sb2000.1/WT expression ratios on a chromosome map of L. infantum. Orange to red features indicate genes overexpressed in Sb2000.1, whereas pale to bright green features indicate genes downregulated in Sb2000.1. Yellow features indicate genes equally expressed in both samples. (Insert) Log2-transformed Sb2000.1/WT expression ratios plotted as a function of the chromosomal location of every probes represented on the full-genome microarrays from chromosome 1 (left end) to chromosome 36 (right end). Probes are plotted by coordinates along each chromosome. Vertical bars represent the log2-transformed expression ratio of individual genes. Whereas most of the genes represented by the microarrays do not show a modulated expression, some chromosomes are entirely overexpressed or downregulated at the RNA level. (A) chromosome 1; (B) chromosome 11; (C) overexpressed locus on chromosome 23; (D) chromosome 25; (E) chromosome 9; (F) chromosome 12; (G) chromosome 32. The plot represents the average values of five independent hybridizations.
Figure 5.
Extrachromosomal circular amplification of a genomic region encoding the MRPA gene on chromosome 23 of Leishmania. (A) Genomic organization of the MRPA locus in L. infantum with the Sb2000.1/WT microarrays expression ratios indicated underneath. Direct repeats of 1.4 kb are indicated by small boxes and the approximate position of PCR primers 1a and 1b is indicated. (B) Model for the formation of the extrachromosomal circular DNA generated through homologous recombination between the direct repeats. (C) Isolation of a circular DNA amplicon by standard alkaline lysis preparation. The arrow head indicates the presence of the extrachromosomal circular DNA. (D) PCR amplification of a 1.8 kb DNA fragment with primers 1a and 1b to support the model shown in Figure 5B. For (C) and (D), L. infantum wild type (lane 1); L. infantum Sb2000.1 (lane 2).
Antimony resistance selection leads to aneuploidy
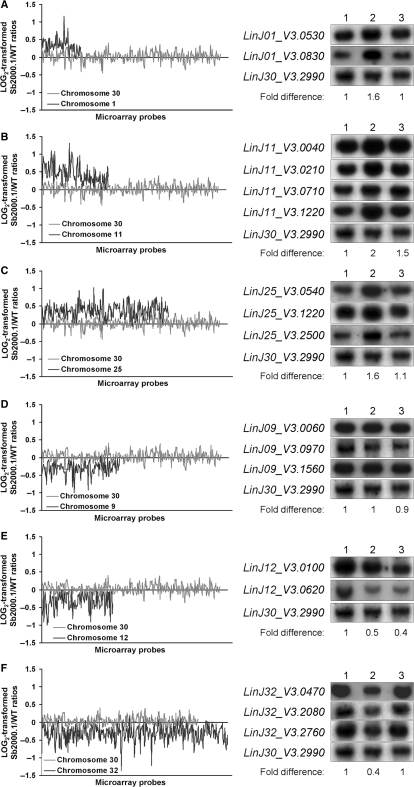
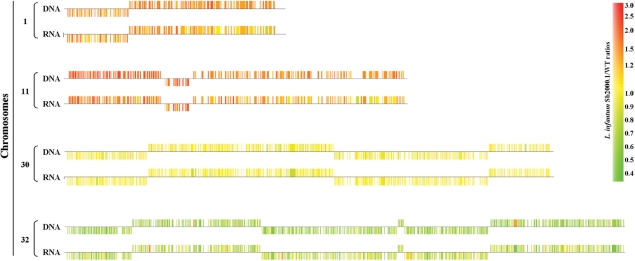
The expression of whole chromosomes seemed to be modulated in L. infantum Sb2000.1, as suggested by the chromosome map of gene expression ratios (Figure 4). Indeed, whereas genes from chromosomes 1, 11 and 25 were mostly overexpressed (predominantly colored in red), those of chromosomes 9, 12 and 32 were mostly downregulated (predominantly colored in green). A close analysis of the normalized expression data generated by the microarray experiments confirmed that most genes located on these chromosomes were modulated by a factor ranging from 1.5 to 2 in mRNA abundance (data not shown). Plotting the gene expression data as a function of microarray probes ordered by chromosomal location for the entire genome (insert in Figure 4) or for selected chromosomes (Figure 6) confirmed the uniform gene expression modulation of these chromosomes. Although a number of hypotheses could explain this phenomenon, one favored explanation for the modulated expression of entire chromosomes would be a concomitant-specific change in the number of allele for these chromosomes. This contention received support from quantitative Southern blot hybridizations since the DNA contents of chromosomes 1, 11 and 25 were, respectively, increased by 1.6-, 2- and 1.6-fold in Sb2000.1 (Figure 6A, B and C, lanes 1 and 2), while those of chromosomes 12 and 32 (Figure 6E and F, lanes 1 and 2) were decreased by 2-fold in this mutant. Conversely, whereas the microarray data indicated that the gene expression of the entire chromosome 9 was decreased in L. infantum Sb2000.1, its DNA content was unchanged compared to L. infantum WT (Figure 6D, lane 1 and 2). Interestingly, the DNA content of chromosome 11 was also increased in the Sb4000.4 antimony-resistant mutant (Figure 6B, lane 3) and the DNA content of the entire chromosome 12 was also decreased in this independently selected strain (Figure 6E, lane 3). As a control, chromosome 30, whose gene expression was unchanged in the microarray data, showed equal DNA contents in L. infantum WT, Sb2000.1 and Sb4000.4 (Figure 6A–F). The 2–4 chromosome-specific probes used for DNA hybridizations were physically located far apart from each other and the data supported a change in ploidy for the entire chromosomes. This was further confirmed by comparative genomic hybridization experiments (CGH) that revealed a change in ploidy for several chromosomes in L. infantum Sb2000.1. Indeed, the DNA copy number correlated well with the mRNA profile and changes were constant along the whole chromosome length (Figure 7). Statistical analysis of the median signal difference for each chromosome also confirmed whole chromosome copy number changes whether the microarrays were hybridized to cDNA or genomic DNA (data not shown).
Figure 6.
Chromosome aneuploidy in L. infantum Sb2000.1 and Sb4000.4 antimony-resistant mutants. Log2-transformed Sb2000.1/WT expression ratios of the upregulated chromosome 1 (A), chromosome 11 (B) and chromosome 25 (C) and of the downregulated chromosome 9 (D), chromosome 12 (E) and chromosome 32 (F) plotted as a function of the location of microarray probes on each chromosomes. For each plot, the log2-transformed gene expression ratios of chromosome 30, which was equally expressed in both samples, are shown as a control (grey line). Quantitative Southern blot hybridizations of digested genomic DNA extracted from L. infantum WT (lane 1), Sb2000.1 (lane 2) and Sb4000.4 (lane 3) were performed to correlate gene expression modulation of entire chromosomes with the chromosome DNA copy number. The hybridization signal of LinJ30_V3.2990 was used for normalization. The hybridization signals were quantified using ImageQuant 5.2 (Molecular Dynamics) and the fold differences in DNA copy number of Sb2000.1 compared to WT are found below each blot.
Figure 7.
Comparison of relative hybridization data between RNA expression profiling and CGH. RNA- or genomic DNA-derived probes were prepared from L. infantum Sb2000.1 and its sensitive parent strain and hybridized to DNA microarrays. A subset of whole chromosome comparisons showing the correlation between RNA and DNA hybridization data is shown. Examples shown are: chromosomes 1 and 11 showing increased RNA and DNA levels, chromosome 32 showing decreased RNA and DNA levels between the two strains and chromosome 30 where either RNA or DNA remained unchanged. The expression ratio of each gene is indicated as orange to red features for overexpressed genes, pale to bright green features for downregulated genes and yellow features for non-modulated genes.
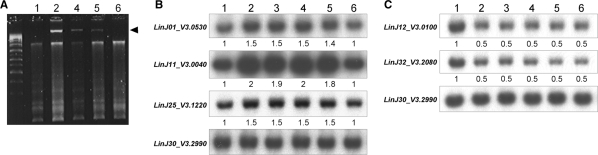
A correlation between aneuploidy and drug resistance has already been described in yeast (51). In order to verify if the aneuploidy observed in Sb2000.1 and Sb4000.4 could be associated with antimony resistance, a partial revertant line of Sb2000.1 (Sb2000.1rev) was generated by successive passages in the absence of antimony. The SbIII resistance level of the revertant line decreased in cells that have grown for passages in the absence of drug but never completely reverted to the parental wild-type level (Figure 1). The decreased resistance after 30 passages in the absence of SbIII was correlated with the concomitant loss of the extrachromosomal amplicon carrying MRPA (Figures 1 and 8A) and with the regression of the aneuploidy of chromosomes 1, 11 and 25 to wild-type ploidy (Figures 1 and 8B). Interestingly, resistance in cells grown for 10 passages without drug were not less resistant than the parent mutant (data not shown) but also their polyploidy was not changed (Figure 8). Furthermore, the transfection of MRPA into the revertant line at passage 30 only partially restored the resistance to antimony observed in the parental mutant (Figure 1), an observation that further circumstantially links the antimony resistance levels and the supernumerary chromosomes. Finally, the L. infantum Sb2000.1 mutant and its revertant strain appeared haploid for chromosomes 12 and 32, a genotype that remained stable for 30 passages without antimony (Figure 8C) and that correlated with the remaining SbIII resistance levels observed in L. infantum Sb2000.1revP30 (Figure 1). Despite their partial haploid genome, the Sb2000.1 mutant and its revertant line divided at the same rate as wild-type cells (data not shown). To further confirm the monosomy of chromosome 32, we tested whether a single round of allelic inactivation would lead to a null mutant in Sb2000.1 and Sb2000.1rev. We chose LinJ32_V3.2190, a gene coding for an unclassified ABC protein (38). A single NEO-containing cassette deleting LinJ32_V3.2190 (Figure 9A) was electroporated in wild-type cells (Figure 9, lane 2), in the Sb2000.1 mutant (Figure 9, lane 4) and in its revertant (Figure 9, lane 6). PCR analysis of the integration confirmed that the cassette has integrated at the right locus (Figure 9B). Southern blot analysis indicated that the integration of the NEO cassette in the WT strain led to haploid parasites with one intact allele remaining for the ABC protein gene (note the decrease in intensity of 50% between lane 1 and lane 2 in Figure 9C). However, the ABC protein gene was deleted after a single round of inactivation with the NEO cassette in the Sb2000.1 mutant and its revertant (Figure 9C, lanes 4 and 6), a situation usually not observed as Leishmania is a diploid organism. These LinJ32_V3.2190 null mutant parasites grew as their control parent cells and their susceptibility to antimonials was unchanged (data not shown).
Figure 8.
Stability of the extrachromosomal circular amplicon and of chromosome aneuploidy in the revertant strain L infantum Sb2000.1rev. (A) Extrachromosomal circular amplicon isolated by alkaline lysis. The arrow head indicates the presence of the extrachromosomal circular DNA. (B) Quantitative Southern blot hybridizations of digested genomic DNA hybridized with probes specific to chromosomes 1, 11, 25 and 30 (as a control). (C) Quantitative Southern blot hybridizations of digested genomic DNA hybridized with probes specific to chromosomes 12, 32 and 30 (as a control). For (B) and (C), L. infantum WT (lane 1), L. infantum Sb2000.1 (lane 2), L. infantum Sb2000.1 grown for 5 (lane 3), 10 (lane 4), 20 (lane 5) and 30 (lane 6) passages in the absence of SbIII. The hybridization signals of LinJ30_V3.2990 located on a chromosome equally expressed between L. infantum WT and Sb2000.1 were used for normalization of Southern blot hybridization signals. Fold differences in hybridization intensities between L. infantum Sb2000.1 and WT are indicated below the Southern blots.
Figure 9.
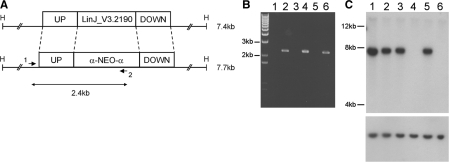
Monosomy of chromosome 32 in L. infantum Sb2000.1 and L. infantum Sb2000.1rev. (A) Genomic organization of the LinJ32_V3.2190 region. The LinJ32_V3.2190 upstream and downstream fragments that were used to carry out gene disruption are shown. The primers 1 and 2 used to confirm the integration of the inactivation cassette at the LinJ32_V3.2190 locus are shown. ‘H’ represents HindIII restriction sites. (B) PCR amplification to confirm the integration of the inactivation cassette. (C) Southern blot of L. infantum genomic DNA digested with HindIII and hybridized to a LinJ32_V3.2190 probe (top) or a LinJ30_V3.2990 probe as a control (bottom). For (B) and (C), L. infantum WT (lane 1), L. infantum WT with one LinJ32_V3.2190 allele disrupted (lane 2), L. infantum Sb2000.1 (lane 3), L. infantum Sb2000.1 with one LinJ32_V3.2190 allele disrupted (lane 4), L. infantum Sb2000.1rev P30 (lane 5), L. infantum Sb2000.1rev P30 with one LinJ32_V3.2190 allele disrupted (lane 6).
DISCUSSION
Microarrays monitoring gene expression are now being used for the study of various aspects of Leishmania. The initial studies have dealt with a limited number of genes but genome-wide surveys are now being reported (39,40,52–54). Gene expression studies using whole-genome 70-mer oligonucleotide microarrays revealed the complexity of the genotype associated with antimony resistance in the L. infantum Sb2000.1 mutant. Indeed, several genes showed a significant difference in expression compared to the parental sensitive WT strain, some of which were part of an extrachromosomal amplicon while others were located on aneuploid chromosomes. The set of differentially regulated genes between L. infantum WT and Sb2000.1 included two ABC protein-coding genes, LinJ11_V3.0040/ABCH1 and LinJ23_V3.0290/MRPA. The genome-wide survey reported here revealed that the overexpression of ABCH1 is due to an increase in the number of copies of chromosome 11. Furthermore, the microarrays were useful to map precisely the amplified region encoding MRPA and three other genes on chromosome 23. Homologous recombination events that occured between direct repeats that bordered this amplified locus were responsible for the generation of the circular amplicon isolated from Sb2000.1. Gene amplification through homologous recombination between repeated sequences is a common mechanism of drug resistance in Leishmania (24,25,49,50,55,56) and is probably a consequence of the lack of transcriptional control in trypanosomatid parasites (57). Indeed, gene-specific expression modulation happened only for a limited number of genes and genes with a substantial variation in RNA levels were part of an amplicon generated by DNA recombination events. Accordingly, repeated sequences may flank key drug resistance genes to allow their amplification under conditions where an increase of a specific protein is required. Consistent with this proposal, the direct repeats flanking the MRPA locus on chromosome 23 are conserved in L. infantum, L. major and L. braziliensis, despite 15 million years of divergence (58).
It is plausible that Leishmania could resort to aneuploidy to modulate the expression of relevant genes in the absence of direct or inverted repeats in the vicinity of a resistance gene or if more than one gene on a particular chromosome is required to confer resistance. Indeed, analysis of our microarrays data showed that gene expression modulation of entire chromosomes (e.g chromosomes 1, 11, 12, 25 and 32) was associated with a concomitant change in ploidy in the Sb2000.1 antimony-resistant strain (Figures 6 and 7). The only exception was the minor decrease in gene expression of chromosome 9 that was not correlated to a change in copy number. This phenomenon was previously observed in a L. major methotrexate-resistant strain (40) and may involve some epigenetic mechanisms that remain to be elucidated. Supernumerary chromosomes have already been described when attempting to inactivate essential genes in Leishmania (59,60) and drug-resistant parasites could also increase the copy number of chromosomes carrying genes relevant to resistance as recently demonstrated in methotrexate-resistant Leishmania (40). In support for this hypothesis, whole-chromosome aneuploidy and specific segmental aneuploidy have already been observed for chromosomes encoding drug resistance genes in azole-resistant strains of Candida albicans (51,61) and in drug-resistant carcinomatous lung cancer cells (62). The decreased expression of genes present on chromosomes 12 and 32 resulted from the loss of one copy of their chromosome and led to a partial haploid mutant. While segmental haploidy could be induced by telomere-mediated chromosome fragmentation (63), this is the first example of drug-induced haploidy in Leishmania. The antimony-related metalloid arsenite is known to induce several mitotic abnormalities such as derangement of the spindle apparatus and alteration of chromosome segregation (64) and it is possible that haploidy, coming from the loss of specific chromosomes, could be associated with antimony resistance in Leishmania, as previously described for ovarian carcinoma cell lines selected for resistance to microtubules stabilizing agents (65). The mutant strain Sb2000.1 which is haploid for two chromosomes had no growth defect as promastigotes and also infected macrophages similarly to wild-type parasites (37).
A role for aneuploidy in Leishmania drug resistance is supported by the presence of similar changes in ploidy for two chromosomes in the independently selected Sb2000.1 and Sb4000.4 antimony-resistant strains (Figure 6) and by the correlation observed between SbIII resistance levels and the copy number of aneuploid chromosomes in Sb2000.1 (Figures 1 and 8). Accordingly, given that supernumerary chromosomes have also been observed in a L. major strain selected for methotrexate resistance (40), it is possible that alterations in the number of copies of specific chromosomes could be associated with gene expression changes implicated in drug resistance in Leishmania. Obviously, further work will be required to identify which genes, if any, could be associated with antimony resistance in Sb2000.1. One potential candidate could be LinJ11_V3.0710, a gene located on chromosome 11 that is supernumerary in the two tested SbIII-resistant mutants and which encodes an anion-transporting ATPase homologous to the bacterial ArsA protein. Although ArsA homologs were not thought to be involved in metal resistance in eukaryotes, it has recently been reported that downregulation of the ArsA homolog in Caenorhabditis elegans (asna-1) resulted in sensitization of the worm to antimony and to the related metalloid arsenite (66). However, transfection of this gene did not reveal a role in SbIII resistance in Leishmania (our unpublished data), and thus other candidates must be studied.
Finally, the LinJ05_V3.0830 gene encoding a methylthioadenosine phosphorylase (MTAP) was downregulated several folds in many independent L. infantum SbIII-resistant mutants (Figure 3). A direct link with antimony resistance is still missing but the downregulation of this gene might be a good biomarker candidate for predicting SbIII resistance, at least in L. infantum parasites. The genes flanking LinJ05_V3.0830 were not differentially expressed between L. infantum WT and Sb2000.1 (data not shown) and this suggests that its specific downregulation could be mediated by changes in mRNA stability (67,68). MTAP is part of the methionine recycling pathway and downregulation of its homologue in yeast (meu1) is associated with an increase in ornithine decarboxylase (ODC) activity (69). ODC is frequently overexpressed (19,33) along with GSH1 (16,33) in metal-resistant Leishmania mutants in order to increase the synthesis of TSH, a key player of antimony resistance in trypanosomatid parasites. Interestingly, ODC (LinJ12_V3.0100) is located on chromosome 12 which has lost a copy in Sb2000.1 and Sb4000.4. Accordingly, downregulation of LinJ05_V3.0830 may be linked to the decreased polyamine pool generated by the haploidy of chromosome 12. Further experimental work will be required to test this hypothesis but it is interesting to note that a correlation between expression of specific genes and aneuploidy has already been described in Saccharomyces cerevisiae (70).
Overall, the whole-genome expression profiling experiments enabled the identification of several genotypic changes associated with antimony resistance in Leishmania. Although few genes identified in this study displayed a dramatic modulation in expression, the antimony-resistant phenotype may occur from the synergistic interactions of several genes. With the exception of MRPA, the expression of several genes known to be involved in antimonials resistance (see ‘Introduction’ section) was not modulated in the Sb2000.1 mutant, which suggests that several mechanisms can lead to resistance.
The arrays also allowed the identification of two novel phenomenons, including partial haploidy and supernumerary chromosomes. Further work is required to isolate the putative resistance genes located on aneuploid chromosomes and should also reveal how widespread these changes in chromosome copy number in Leishmania isolates are.
FUNDING
CIHR group grant (to J.C. and M.O.) and operating grants (to M.O.); CIHR studentship (to P.L. and F.R.); Canada Research Chair in Medical Genomics (to J.C.) and Burroughs Wellcome Fund Scholar in Molecular Parasitology and holds the Canada Research Chair in Antimicrobial Resistance (M.O.). Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 2.Hadighi R, Mohebali M, Boucher P, Hajjaran H, Khamesipour A, Ouellette M. Unresponsiveness to glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Med. 2006;3:e162. doi: 10.1371/journal.pmed.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lira R, Sundar S, Makharia A, Kenney R, Gam A, Saraiva E, Sacks D. Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J. Infect. Dis. 1999;180:564–567. doi: 10.1086/314896. [DOI] [PubMed] [Google Scholar]
- 4.Rojas R, Valderrama L, Valderrama M, Varona MX, Ouellette M, Saravia NG. Resistance to antimony and treatment failure in human Leishmania (Viannia) infection. J. Infect. Dis. 2006;193:1375–1383. doi: 10.1086/503371. [DOI] [PubMed] [Google Scholar]
- 5.Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jha TK, Olliaro P, Thakur CP, Kanyok TP, Singhania BL, Singh IJ, Singh NK, Akhoury S, Jha S. Randomised controlled trial of aminosidine (paromomycin) v sodium stibogluconate for treating visceral leishmaniasis in North Bihar, India. Br. Med. J. 1998;316:1200–1205. doi: 10.1136/bmj.316.7139.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Victoria FJ, Gamarro F, Ouellette M, Castanys S. Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J. Biol. Chem. 2003;278:49965–49971. doi: 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- 8.Ouellette M, Drummelsmith J, Papadopoulou B. Leishmaniasis: drugs in the clinic, resistance and new developments. Drug Resist. Updat. 2004;7:257–266. doi: 10.1016/j.drup.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Sundar S, Rai M. Treatment of visceral leishmaniasis. Expert Opin. Pharmacother. 2005;6:2821–2829. doi: 10.1517/14656566.6.16.2821. [DOI] [PubMed] [Google Scholar]
- 10.Berman J. Current treatment approaches to leishmaniasis. Curr. Opin. Infect. Dis. 2003;16:397–401. doi: 10.1097/00001432-200310000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Croft SL, Coombs GH. Leishmaniasis–current chemotherapy and recent advances in the search for novel drugs. Trends Parasitol. 2003;19:502–508. doi: 10.1016/j.pt.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Shaked-Mishan P, Ulrich N, Ephros M, Zilberstein D. Novel Intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J. Biol. Chem. 2001;276:3971–3976. doi: 10.1074/jbc.M005423200. [DOI] [PubMed] [Google Scholar]
- 13.Gourbal B, Sonuc N, Bhattacharjee H, Legare D, Sundar S, Ouellette M, Rosen BP, Mukhopadhyay R. Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J. Biol. Chem. 2004;279:31010–31017. doi: 10.1074/jbc.M403959200. [DOI] [PubMed] [Google Scholar]
- 14.Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol. Microbiol. 2005;57:1690–1699. doi: 10.1111/j.1365-2958.2005.04782.x. [DOI] [PubMed] [Google Scholar]
- 15.Fairlamb AH, Cerami A. Metabolism and functions of trypanothione in the Kinetoplastida. Annu. Rev. Microbiol. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]
- 16.Grondin K, Haimeur A, Mukhopadhyay R, Rosen BP, Ouellette M. Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenite-resistant Leishmania tarentolae. EMBO J. 1997;16:3057–3065. doi: 10.1093/emboj/16.11.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimond C, Trudel N, Brochu C, Marquis N, El Fadili A, Peytavi R, Briand G, Richard D, Messier N, Papadopoulou B, et al. Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Res. 2003;31:5886–5896. doi: 10.1093/nar/gkg806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haimeur A, Brochu C, Genest P, Papadopoulou B, Ouellette M. Amplification of the ABC transporter gene PGPA and increased trypanothione levels in potassium antimonyl tartrate (SbIII) resistant Leishmania tarentolae. Mol. Biochem. Parasitol. 2000;108:131–135. doi: 10.1016/s0166-6851(00)00187-0. [DOI] [PubMed] [Google Scholar]
- 19.Haimeur A, Guimond C, Pilote S, Mukhopadhyay R, Rosen BP, Poulin R, Ouellette M. Elevated levels of polyamines and trypanothione resulting from overexpression of the ornithine decarboxylase gene in arsenite-resistant Leishmania. Mol. Microbiol. 1999;34:726–735. doi: 10.1046/j.1365-2958.1999.01634.x. [DOI] [PubMed] [Google Scholar]
- 20.Legare D, Papadopoulou B, Roy G, Mukhopadhyay R, Haimeur A, Dey S, Grondin K, Brochu C, Rosen BP, Ouellette M. Efflux systems and increased trypanothione levels in arsenite-resistant Leishmania. Exp. Parasitol. 1997;87:275–282. doi: 10.1006/expr.1997.4222. [DOI] [PubMed] [Google Scholar]
- 21.Mukhopadhyay R, Dey S, Xu N, Gage D, Lightbody J, Ouellette M, Rosen BP. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc. Natl Acad. Sci. USA. 1996;93:10383–10387. doi: 10.1073/pnas.93.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandal G, Wyllie S, Singh N, Sundar S, Fairlamb AH, Chatterjee M. Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology. 2007;134:1679–1687. doi: 10.1017/S0031182007003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyllie S, Cunningham ML, Fairlamb AH. Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J. Biol. Chem. 2004;279:39925–39932. doi: 10.1074/jbc.M405635200. [DOI] [PubMed] [Google Scholar]
- 24.Grondin K, Papadopoulou B, Ouellette M. Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res. 1993;21:1895–1901. doi: 10.1093/nar/21.8.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouellette M, Hettema E, Wust D, Fase-Fowler F, Borst P. Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J. 1991;10:1009–1016. doi: 10.1002/j.1460-2075.1991.tb08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brochu C, Haimeur A, Ouellette M. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite leishmania. Cell Stress Chaperones. 2004;9:294–303. doi: 10.1379/CSC-15R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vergnes B, Gourbal B, Girard I, Sundar S, Drummelsmith J, Ouellette M. A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol. Cell Proteomics. 2007;6:88–101. doi: 10.1074/mcp.M600319-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Genest PA, Haimeur A, Legare D, Sereno D, Roy G, Messier N, Papadopoulou B, Ouellette M. A protein of the leucine-rich repeats (LRRs) superfamily is implicated in antimony resistance in Leishmania infantum amastigotes. Mol. Biochem. Parasitol. 2008;158:95–99. doi: 10.1016/j.molbiopara.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Singh N, Singh RT, Sundar S. Novel mechanism of drug resistance in kala azar field isolates. J. Infect. Dis. 2003;188:600–607. doi: 10.1086/377133. [DOI] [PubMed] [Google Scholar]
- 30.Wyllie S, Vickers TJ, Fairlamb AH. Roles of trypanothione S-transferase and tryparedoxin peroxidase in resistance to antimonials. Antimicrob. Agents Chemother. 2008;52:1359–1365. doi: 10.1128/AAC.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhury K, Zander D, Kube M, Reinhardt R, Clos J. Identification of a Leishmania infantum gene mediating resistance to miltefosine and SbIII. Int. J. Parasitol. 2008;38:1411–1423. doi: 10.1016/j.ijpara.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Decuypere S, Rijal S, Yardley V, De Doncker S, Laurent T, Khanal B, Chappuis F, Dujardin JC. Gene expression analysis of the mechanism of natural Sb(V) resistance in Leishmania donovani isolates from Nepal. Antimicrob. Agents Chemother. 2005;49:4616–4621. doi: 10.1128/AAC.49.11.4616-4621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee A, Padmanabhan PK, Singh S, Roy G, Girard I, Chatterjee M, Ouellette M, Madhubala R. Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J. Antimicrob. Chemother. 2007;59:204–211. doi: 10.1093/jac/dkl494. [DOI] [PubMed] [Google Scholar]
- 34.Goyeneche-Patino DA, Valderrama L, Walker J, Saravia NG. Antimony resistance and trypanothione in experimentally selected and clinical strains of Leishmania panamensis. Antimicrob. Agents Chemother. 2008;52:4503–4506. doi: 10.1128/AAC.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittal MK, Rai S, Ashutosh, Ravinder, Gupta S, Sundar S, Goyal N. Characterization of natural antimony resistance in Leishmania donovani isolates. Am. J. Trop. Med. Hyg. 2007;76:681–688. [PubMed] [Google Scholar]
- 36.Singh N, Almeida R, Kothari H, Kumar P, Mandal G, Chatterjee M, Venkatachalam S, Govind MK, Mandal SK, Sundar S. Differential gene expression analysis in antimony-unresponsive Indian kala azar (visceral leishmaniasis) clinical isolates by DNA microarray. Parasitology. 2007;134:777–787. doi: 10.1017/S0031182007002284. [DOI] [PubMed] [Google Scholar]
- 37.El-Fadili K, Messier N, Leprohon P, Roy G, Guimond C, Trudel N, Saravia NG, Papadopoulou B, Legare D, Ouellette M. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob. Agents Chemother. 2005;49:1988–1993. doi: 10.1128/AAC.49.5.1988-1993.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leprohon P, Legare D, Girard I, Papadopoulou B, Ouellette M. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryot. Cell. 2006;5:1713–1725. doi: 10.1128/EC.00152-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ubeda JM, Legare D, Raymond F, Ouameur AA, Boisvert S, Rigault P, Corbeil J, Tremblay MJ, Olivier M, Papadopoulou B, et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11:3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Papadopoulou B, Roy G, Dey S, Rosen BP, Ouellette M. Contribution of the Leishmania P-glycoprotein-related gene ltpgpA to oxyanion resistance. J. Biol. Chem. 1994;269:11980–11986. [PubMed] [Google Scholar]
- 44.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 47.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 49.Grondin K, Roy G, Ouellette M. Formation of extrachromosomal circular amplicons with direct or inverted duplications in drug-resistant Leishmania tarentolae. Mol. Cell. Biol. 1996;16:3587–3595. doi: 10.1128/mcb.16.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beverley SM. Gene amplification in Leishmania. Annu. Rev. Microbiol. 1991;45:417–444. doi: 10.1146/annurev.mi.45.100191.002221. [DOI] [PubMed] [Google Scholar]
- 51.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol. Biochem. Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol. Biochem. Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Saxena A, Lahav T, Holland N, Aggarwal G, Anupama A, Huang Y, Volpin H, Myler PJ, Zilberstein D. Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol. Biochem. Parasitol. 2007;152:53–65. doi: 10.1016/j.molbiopara.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Chang KP. The 63-kilobase circular amplicon of tunicamycin-resistant Leishmania amazonensis contains a functional N-acetylglucosamine-1-phosphate transferase gene that can be used as a dominant selectable marker in transfection. Mol. Cell. Biol. 1992;12:4112–4122. doi: 10.1128/mcb.12.9.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chow LM, Wong AK, Ullman B, Wirth DF. Cloning and functional analysis of an extrachromosomally amplified multidrug resistance-like gene in Leishmania enriettii. Mol. Biochem. Parasitol. 1993;60:195–208. doi: 10.1016/0166-6851(93)90131-g. [DOI] [PubMed] [Google Scholar]
- 57.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lukes J, Mauricio IL, Schonian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Tintaya KW, Jirku M, Chocholova E, et al. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc. Natl Acad. Sci. USA. 2007;104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cruz AK, Titus R, Beverley SM. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc. Natl Acad. Sci. USA. 1993;90:1599–1603. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Calvillo S, Stuart K, Myler PJ. Ploidy changes associated with disruption of two adjacent genes on Leishmania major chromosome 1. Int. J. Parasitol. 2005;35:419–429. doi: 10.1016/j.ijpara.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 61.Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d’Enfert C, Berman J, Sanglard D. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell. 2007;6:1889–1904. doi: 10.1128/EC.00151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doubre H, Cesari D, Mairovitz A, Benac C, Chantot-Bastaraud S, Dagnon K, Antoine M, Danel C, Bernaudin JF, Fleury-Feith J. Multidrug resistance-associated protein (MRP1) is overexpressed in DNA aneuploid carcinomatous cells in non-small cell lung cancer (NSCLC) Int. J. Cancer. 2005;113:568–574. doi: 10.1002/ijc.20617. [DOI] [PubMed] [Google Scholar]
- 63.Tamar S, Papadopoulou B. A telomere-mediated chromosome fragmentation approach to assess mitotic stability and ploidy alterations of Leishmania chromosomes. J. Biol. Chem. 2001;276:11662–11673. doi: 10.1074/jbc.M009006200. [DOI] [PubMed] [Google Scholar]
- 64.Yih LH, Ho IC, Lee TC. Sodium arsenite disturbs mitosis and induces chromosome loss in human fibroblasts. Cancer Res. 1997;57:5051–5059. [PubMed] [Google Scholar]
- 65.Wang Y, O’Brate A, Zhou W, Giannakakou P. Resistance to microtubule-stabilizing drugs involves two events: beta-tubulin mutation in one allele followed by loss of the second allele. Cell Cycle. 2005;4:1847–1853. doi: 10.4161/cc.4.12.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tseng YY, Yu CW, Liao VH. Caenorhabditis elegans expresses a functional ArsA. FEBS J. 2007;274:2566–2572. doi: 10.1111/j.1742-4658.2007.05791.x. [DOI] [PubMed] [Google Scholar]
- 67.McNicoll F, Muller M, Cloutier S, Boilard N, Rochette A, Dube M, Papadopoulou B. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 2005;280:35238–35246. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- 68.Bringaud F, Muller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, Papadopoulou B, Ghedin E. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subhi AL, Diegelman P, Porter CW, Tang B, Lu ZJ, Markham GD, Kruger WD. Methylthioadenosine phosphorylase regulates ornithine decarboxylase by production of downstream metabolites. J. Biol. Chem. 2003;278:49868–49873. doi: 10.1074/jbc.M308451200. [DOI] [PubMed] [Google Scholar]
- 70.Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]