Abstract
Short-patch repair of DNA single-strand breaks and gaps (SSB) is coordinated by XRCC1, a scaffold protein that recruits the DNA polymerase and DNA ligase required for filling and sealing the damaged strand. XRCC1 can also recruit end-processing enzymes, such as PNK (polynucleotide kinase 3′-phosphatase), Aprataxin and APLF (aprataxin/PNK-like factor), which ensure the availability of a free 3′-hydroxyl on one side of the gap, and a 5′-phosphate group on the other, for the polymerase and ligase reactions respectively. PNK binds to a phosphorylated segment of XRCC1 (between its two C-terminal BRCT domains) via its Forkhead-associated (FHA) domain. We show here, contrary to previous studies, that the FHA domain of PNK binds specifically, and with high affinity to a multiply phosphorylated motif in XRCC1 containing a pSer-pThr dipeptide, and forms a 2:1 PNK:XRCC1 complex. The high-resolution crystal structure of a PNK–FHA–XRCC1 phosphopeptide complex reveals the basis for this unusual bis-phosphopeptide recognition, which is probably a common feature of the known XRCC1-associating end-processing enzymes.
INTRODUCTION
Short-patch repair of DNA single-strand breaks and gaps (SSBs) is coordinated by the multi-domain XRCC1 protein, which acts as a ‘scaffold’, recruiting and organising the DNA polymerase (Polβ) and DNA ligase (Lig3α) that fill in the missing nucleotides and seal the phosphodiester backbone (1,2). Facile error-free repair of SSBs depends on the availability of a free 3′-hydroxyl on one side of the gap, and a 5′-phosphate group on the other, which respectively permit template-directed DNA synthesis and ATP-dependent ligation. However, in many circumstances the DNA damage event that caused the gap, leaves end-chemistries that cannot be extended or ligated, and end-processing is required before SSB repair can be completed (3).
Polynucleotide kinase 3′ phosphatase (PNK or PNKP) is a bifunctional DNA end-processing enzyme that facilitates SSB repair by phosphorylating 5′-hydroxyls and/or dephosphorylating 3′-phosphates at the margins of DNA single-strand gaps (4,5). PNK interacts, via its N-terminal forkhead-associated domain (FHA), with a segment of XRCC1 (between the central and C-terminal BRCT domains) that is multiply phosphorylated by casein kinase 2 (CK2) (6,7). PNK is also involved in non-homologous end-joining repair of double strand breaks (DSBs) via its interaction with a similar phosphorylated segment C-terminal to the coiled-coil region of the structurally unrelated XRCC4 scaffold protein (8,9). FHA domains are implicated in protein–protein interactions in a wide and diverse range of cellular proteins, and have been found to bind preferentially to amino-acid sequences containing a single phospho-threonine (pThr) residue.
Although previous studies had indicated that the PNK–FHA domain has the conventional specificity for phospho-threonine (9,10), the tight clustering of the identified CK2 phosphorylation sites within XRCC1, suggests that the situation might be more complicated. To gain some insight into this we have analysed the binding of PNK to phosphorylated XRCC1 protein biochemically, and performed a quantitative calorimetric analysis of the interaction of the PNK–FHA domain with a series of phosphorylated XRCC1 peptides. We show here that, contrary to previous studies, the FHA domain of PNK binds specifically, and with high affinity to a bis-phosphorylated target sequence in XRCC1 containing vicinal pThr-pSer residues. The structural basis for this is revealed by a high-resolution crystal structure of the human PNK–FHA domain binary complex with a phosphorylated XRCC1-derived peptide. The distinctive characteristics of the PNK–FHA are also found on the FHA domains of two other DNA repair end-processing enzymes, APTX (aprataxin) and APLF (aprataxin/PNK-like factor), defining a distinct subset of FHA domains specifically adapted to bis-phosphorylation-specific binding to the DNA repair scaffold proteins, XRCC1 and XRCC4.
MATERIALS AND METHODS
Antibodies
The phospho-XRCC1 (pSer518/pSer519/pThr523) antibody used throughout this study was purchased from Bethyl Laboratories, Montgomery, TX, USA (Catalogue number: A300-059A).
Peptides
All peptides were synthesized and purified in-house by the Peptide Synthesis Facility, ICR.
Expression vectors
pTWO-E is a modified pET-17b (Novagen, Merck Chemicals, Nottingham, UK) vector engineered to encode an N-terminal, 3C-Protease cleavable, His6 affinity tag.
pTWO-B is a modified pFASTBAC1 (Invitrogen, Paisley, UK) that encodes an N-terminal, 3C-Protease cleavable, His6 affinity tag.
pTHREE-E is a modified pGEX-6P-1 (GE Healthcare, UK Ltd, Little Chalfont, UK) vector engineered to contain the multiple cloning site of pET-17b and encodes an N-terminal 3C-Protease cleavable, GST affinity tag.
Gene synthesis
A synthetic gene encoding the N-terminal region of human PNK (amino acids 1–139, including the FHA domain) was constructed using recursive PCR (11). Juniper—an in-house synthetic gene design program—selected the sequences of the required overlapping oligonucleotides. By choosing an average length of 70 bases, eight oligonucleotides were needed for gene construction, the sequences of which are shown below:
‘Forward’ oligos—5′–3′:
1. CATATGGGTGAAGTTGAAGCTCCAGGTCGT CTGTGGCTGGAATCCCCGCCGGGCGAAGC TCCGCCGATC
2. GGGTCGTGGTCCGCTGACTCAGGTTACCGA CCGTAAATGCTCCCGTACTCAGGTAGAACT GGTTGCTGACCC
3. GCGTTAACCCGTCCACCACCGGCACCCAGG AACTGAAACCGGGTCTGGAAGGTTCCCTG GGTGTTGGTGACA
4. ACCCTGCGTTGGGAAGAAACCCGTACCCCG GAATCCCAGCCGGACACCCCGCCAGGTACC CCGCTGGTTTCC
‘Reverse’ oligos—5′–3′:
5. CAGCGGACCACGACCCAGAACCAGAGCCT GACCGTCGGACGGCAGGAAGATCGGCGG AGCTTCGC
6. GTGGACGGGTTAACGCCCAGCTGTTTAACA GCAACGGTGCGGGTTTCCGGGTCAGCAA CCAGTTCT
7. TTCTTCCCAACGCAGGGTCAGCGGGTGCAG ACCGTTAACCAGGTACAGGGTGTCACCAAC ACCCAGGG
8. GAATTCTCATCAACGTTTTTTCGGCAGTTCA GCGTCACGTTTTTCGTCCTGGGAAACCAGC GGGGTAC
The resulting product was blunt-end ligated into the vector pCR-Blunt (Invitrogen Ltd, Paisley, UK). Several clones were analysed and sequenced in-house, before sub-cloning the DNA encoding amino acids 1–109 into the expression vectors pTWO-E and pTHREE-E.
Protein expression and purification
PNK–FHA
The pTWO-E/PNK–FHA construct was transformed into Escherichia coli strain Rosetta2(DE3) pLysS (Merck Chemicals, Nottingham, UK). Cells were cultured in shaker flasks, containing LB media [supplemented with 50 µg/ml carbenicillin (Melford Laboratories, Ipswich, UK) and 34 µg/ml chloramphenicol (Sigma-Aldrich, Dorset, UK)], with growth conditions of 37°C and 220 r.p.m. Cell density was monitored until an OD of 0.5 was achieved when protein expression was induced by the addition of IPTG to a final concentration of 1 mM. Cells were harvested after 3 h by centrifugation, at 5000 g for 10 min at 4°C, and the resulting pellet stored at −80°C until required.
The cell pellet resulting from 5 l of expression culture was resuspended, on ice, in 45 ml of buffer A: 50 mM HEPES/HCl pH 7.5, 200 mM NaCl, 10% v/v glycerol, 10 mM imidazole, supplemented with protease inhibitors. Cells were lysed by sonication, then clarifed by high-speed centrifugation at 40 000 g, for 1 h at 4°C. The resulting supernatant was filtered through 4 μm filter.
Ten millilitres of Talon (Takara Bio Europe) resin was added to the filtered supernatant and incubated, with mixing, at 4°C for 30 min. The unbound fraction was removed by passing the slurry through an empty gravity-flow chromatography column. The retained resin was washed with 200 ml of buffer A. The retained protein was eluted by the addition of buffer B: 50 mM HEPES/HCl pH 7.5, 200 mM NaCl, 10% v/v glycerol, 300 mM imidazole. Fractions containing the protein were identified by SDS–PAGE, pooled, then concentrated to a volume of 10 ml using Vivaspin 20 (5000 MWCO) concentrators (Sartorius Stedim Biotech, Goettingen, Germany), before being applied to a Superdex 75 gel filtration column, pre-equilibrated with buffer C: 20 mM HEPES/HCl pH 7.5, 250 mM NaCl, 1 mM DTT, 1 mM EDTA. Fractions containing the protein were again identified by SDS–PAGE, pooled, and then incubated with 3C-Protease (PreScission protease, GE Healthcare) overnight, at 4°C to remove the affinity-tag when required.
One millilitre of Glutathione Sepharose resin (GE Healthcare) then added to the sample and then incubated with mixing, at 4°C for 30 min, to remove the 3C-Protease. The resin was removed by passing the slurry through a gravity-flow chromatography column. The protein was again concentrated, and as before, applied to a Superdex 75 gel filtration column (GE Healthcare). Fractions containing the purified protein were identified, and then concentrated to 15 mg/ml, where it was either stored at 4°C for immediate use, or snap-frozen at −80°C for long-term storage.
XRCC1ΔN
A full-length clone of human XRCC1 was obtained by PCR using Human MTC Panel I (Takara Bio Europe, Saint-Germain-en-Laye, France) as a template.
Sub-cloning of the DNA encoding amino acids 218–633 into the vectors pTWO-E and pTWO-B produced the XRCC1ΔN constructs for expression in E. coli and Sf9 insect cells, respectively. Recombinant baculovirus expressing XRCCΔN was generated from the pTWO-B donor vector, following the manufacturer's recommended protocol (Invitrogen).
Biochemical and biophysical experiments
Isothermal titration calorimetry
Heats of interaction were measured on a VP-ITC microcalorimeter (MicroCal Europe, Milton Keynes, UK) with a cell volume of 1.458 ml. Purified PNK–FHA was dialysed overnight in assay buffer (20 mM HEPES/HCl pH 7.5, 250 mM NaCl, 1 mM EDTA and 1 mM TCEP).
For the PNK–FHA/XRCC1 phosphopeptide interactions, 28 injections of 10 μl of 400 µM peptide were injected into 40 μM PNK–FHA at 10°C.
The heat of dilution was determined in a separate experiment by injecting peptide into the sample buffer. Data corrected for the heat of dilution were fitted using a non-linear least square curve-fitting algorithm (Microcal Origin) with three floating variables: stoichiometry, binding constant and change in enthalpy of interaction. All peptides were dissolved in assay buffer. Concentrations were determined by UV spectroscopy using a ND-1000 spectrophotometer (NanoDrop Products, Wilmington, DE, USA) and a molar extinction coefficient of 1440 M−1 cm−1 at 274 nm (due to the single tyrosine residue present in each peptide sequence).
Crystallization, data collection and refinement
PNK–FHA
Crystallization trials of PNK–FHA were carried out at 15 mg/ml, in hanging drop experiments, at 20°C using MDL Structure Screen I. Crystals were obtained in condition 32 (0.1 M Tris–HCl pH 8.5, 2.0 M ammonium sulphate). No further optimization of the condition was required, as crystals diffracted in-house to a resolution of 2.2 Å. The crystals were cryo-protected by swiping through successive buffers that contained increasing amounts of glycerol, to a final concentration of 20% v/v.
Data to 1.4 Å were collected from a single crystal at 100 K at the ESRF, Grenoble, France. Images were integrated using MOSFLM (12), and reduced/scaled using programs of the CCP4 suite (13). The protein crystallized in the spacegroup P21 with cell dimensions of a = 31.81 Å, b = 40.05 Å, c = 37.51 Å and β = 90.96. A single molecule comprised the asymmetric unit.
Initial phases were calculated by molecular replacement [Phaser (14)] using the solution structure of the murine PNK FHA domain (PDB: 1UJX) as a search model. The calculated difference maps were then used to build a molecular model, both automatically by ARP/wARP (15) and manually using Coot (16). Iterative cycles of refinement [REFMAC5 (17)] and manual intervention produced the current model which consists of 775 protein atoms, 144 solvent atoms and two sulphate ions with R = 0.150 and Rfree = 0.183 for 5% of the data omitted from refinement. Coordinates have been deposited in the Protein Databank (PDB: 2BRF).
Statistics for the data collection and refinement are given in Table S1 of the Supplementary Data.
PNK–FHA/XRCC1 phosphopeptide complex
The single-point mutant L99E was generated by using a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) following the manufacturer's recommended protocol. The mutant protein was expressed and purified exactly as the wild-type protein.
Several of the XRCC1-derived phosphopeptides described in this study were used in co-crystallization trials, however reproducible hits were only obtained with the triple-phosphorylated YAGpSpTDENpTDSE peptide.
Protein and peptide were combined at a 2:1 ratio, then incubated at 4°C for 1 h, prior to crystallization screening in hanging drop experiments at 20°C. Initial hits were obtained in condition 44 of the PEGS II suite (Qiagen Ltd, West Sussex, UK): 0.1 M HEPES pH 7.5, 0.2 M CaCl2, 30% w/v PEG 4000.
This was optimized, again in hanging drops at 20°C, to mixing 1 μl of the protein/peptide complex (15 mg/ml, with respect to protein, in 20 mM HEPES/NaOH pH 7.5, 250 mM NaCl, 1 mM EDTA, 1 mM TCEP) with 1 μl of precipitant containing 0.1 M Tris–HCl pH 8.5, 0.25 M CaCl2, 28% w/v PEG 4000 and 0.2 M NDSB-221.Crystals were cryo-protected, for data collection, by a rapid swipe through successive buffers containing increasing amounts of glycerol, to a final concentration of 20% v/v.
Diffraction data to 1.85Å were collected from a single crystal at 100 K on station I04 at the Diamond Light Source, Didcot, UK. Data were processed and scaled as before using the software packages MOSFLM and Scala. The protein crystallized in the spacegroup P3 with cell dimensions of a = 56.90 Å, b = 56.90 Å, c = 62.77 Å. Two molecules of PNK–FHA comprised the asymmetric unit.
Phases were calculated by molecular replacement using the crystal structure of PNK–FHA as a search model. Difference electron density corresponding to the bound peptide was readily observed in the initial maps. A molecular model was built, both automatically by ARP/wARP and manually using Coot. Iterative cycles of refinement (REFMAC5) and manual intervention produced the current model which consists of 1508 protein atoms, 286 solvent atoms and six calcium ions with R = 0.18 and Rfree = 0.24 for 5% of the data omitted from refinement. Coordinates have been deposited in the Protein Databank (PDB: 2W3O).
Statistics for the data collection and refinement are given in Table S1 of the Supplementary data.
RESULTS
Expression and purification of phosphorylated XRCC1
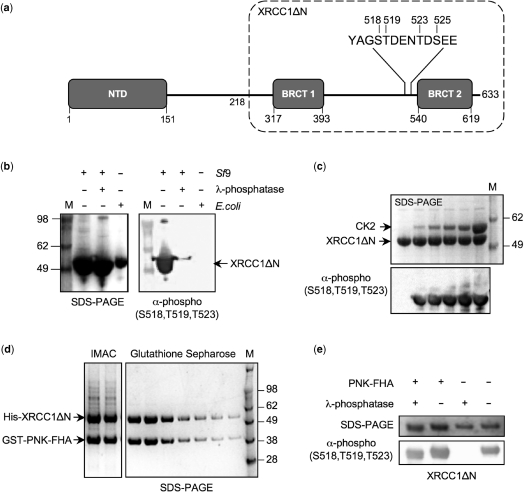
We had previously characterized in our laboratory a construct expressing a truncated form of human XRCC1 that was resistant to proteolytic degradation—XRCC1ΔN (amino acids 218–633)—containing both BRCT domains and the intervening linker region (Figure 1A). As for full-length XRCC1 (6), the truncated protein encoded by this construct was phosphorylated when purified from insect cells (Figure 1B); confirmed by western blot with a polyclonal antibody that simultaneously detects phosphorylation sites (pS518/pT519/pT523) within the linker region connecting the two BRCT domains of XRCC1 (α-pXRCC1). The reactivity to this antibody was readily lost on incubation with λ-phosphatase. In contrast, XRCC1ΔN expressed in and purified from E. coli, showed no inherent reactivity to the antibody, but developed a signal on incubation with maize CK2 (Figure 1C) confirming previous observations that these sites are targets for CK2 phosphorylation (6).
Figure 1.
PNK–FHA protects CK2-phosphorylated XRCC1 from dephosphorylation. (a) Schematic representation of the domain structure of human XRCC1. The predicted amino-acid boundaries for each domain are indicated, with a dotted box delineating the extent of the XRCC1ΔN expression construct used in this study. The positions of the known phosphorylation sites within the linker region connecting the two C-terminal BRCT domains are also shown. (b) XRCC1ΔN expressed in insect cells (Sf9) is phosphorylated, as detected by a phospho-specific polyclonal antibody (Bethyl Laboratories, A300-059A). Incubation with λ-phosphatase readily removes the detected sites of phosphorylation. In contrast, XRCC1ΔN expressed in E.coli is not phosphorylated. One hundred micrograms of purified XRCC1ΔN protein was incubated with 50 μg of λ-phosphatase for a period of 1 h at room temperature, before being analysed by SDS–PAGE. (c) Incubation of the E.coli expressed XRCC1ΔN with recombinant maize CKII reproduces the phosphorylation observed in Sf9 cells. Sixty micrograms of purified XRCC1ΔN was incubated with increasing amounts of CKII (ranging from 0 to 95 μg), for a period of 2 h at room temperature. (d) Cell lysates from insect cells expressing His6-tagged XRCC1ΔN and E.coli expressing GST-tagged PNK–FHA were mixed, then applied to two sequential affinity columns (Talon/IMAC and Glutathione Sepharose). The SDS–PAGE analyses of the eluates from each step are shown. (e) The presence of the PNK–FHA domain protects, at least, the three phosphorylation sites detected by the polyclonal antibody in XRCC1 from the enzymatic action of λ-phosphatase. SDS–PAGE gels were stained with SimplyBlue SafeStain (Invitrogen, Paisley, UK).
Affinity capture of XRCC1ΔN by the FHA domain of PNK
To test the functionality of the XRCC1ΔN construct in binding PNK, we expressed the FHA domain of human PNK (PNK–FHA: amino acids 1–109) as a GST-fusion in E. coli, and incubated cell lysates containing PNK–FHA and XRCC1ΔN (from insect cells) together. Both proteins co-eluted in successive IMAC and Glutathione Sepharose affinity capture steps (Figure 1D), confirming that the PNK–XRCC1 interaction does not require the N-terminal part of XRCC1. A purified PNK–FHA–XRCC1ΔN complex could then be obtained with further chromatographic steps, following proteolytic removal of the GST and His6 affinity tags.
PNK–FHA protects XRCC1 phosphorylation sites
The binding site for PNK has previously been localized to the ∼150 amino-acid segment linking the two BRCT domains in XRCC1, which contains the major documented CK2 phosphorylation sites pSer518, pThr519, pThr523 and pSer525 (6,7), the first three of which are detected by the α-pXRCC1 phospho-specific antibody.
To confirm that these sites are directly involved in the interaction with PNK, we analysed the ability of the purified PNK–FHA domain to protect the phosphorylation sites in insect cell-purified XRCC1ΔN from the enzymatic action of λ-phosphatase (Figure 1E). Isolated XRCC1ΔN was rapidly dephosphorylated as before, as judged by loss of reactivity to the α-pXRCC1 antibody.
However, within the XRCC1ΔN–PNK–FHA complex, the sites detected by the antibody were protected from the enzymatic activity of λ-phosphatase, although the electrophoretic mobility of XRCC1ΔN was slightly altered; suggesting that some additional phosphorylation sites were removed.
The experiment confirms the interaction of PNK–FHA with this particular region of XRCC1, and suggesting that all three phosphorylation sites might be involved in the PNK–FHA interaction.
Isothermal titration calorimetry of PNK–FHA interaction with defined XRCC1 phospho-peptides
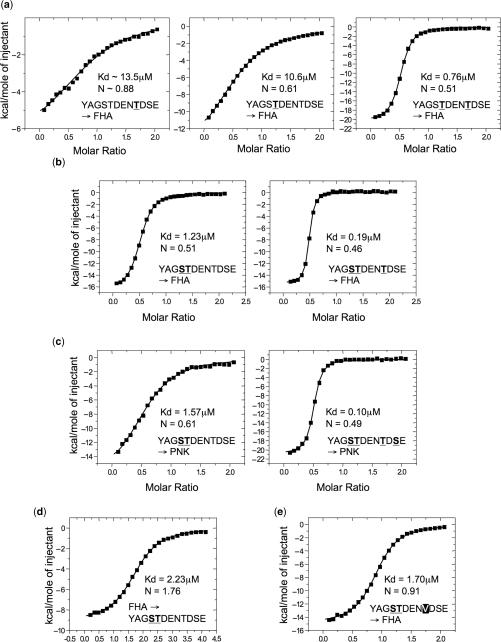
As at least three of the documented CK2 sites (pSer518, pThr519 and pThr523) appeared to be involved in PNK recruitment, we sought to determine the contribution to PNK–FHA-binding provided by each XRCC1 phosphorylation site, individually and in combination, using isothermal titration calorimetry with synthetic phospho-peptides (Table 1).
Table 1.
ITC dissociation constants and stoichiometry
| Peptide Sequence | Kd (µM) | N |
|---|---|---|
| YAGSTDENTDSE | No binding | – |
| YAGSTDENTDSE | No binding | – |
| YAGSTDENTDSE | No binding | – |
| YAGSTDENTDSE | 10.58 ± 0.28 | 0.61 |
| YAGSTDENTDSE | 13.49 ± 1.296 | 0.88 |
| YAGSTDENTDSE | 0.76 ± 0.22 | 0.51 |
| YAGSTDENTDSE | 1.23 ± 0.04 | 0.51 |
| YAGSTDENTDSE | 0.19 ± 0.02 | 0.46 |
| YAGSTDENTDSE | 0.14 ± 0.01 | 0.50 |
| YAGSTDENTDSE | 24.18 ± 1.44 | 0.56 |
| YAGSTDENVDSE | 1.7 ± 0.06 | 0.91 |
No binding of PNK–FHA was detected to a non-phosphorylated XRCC1-derived peptide, or to peptides mono-phosphorylated on either Ser518 or Ser525. PNK–FHA did however bind weakly to a peptide phosphorylated on Thr523 with Kd = ∼13.5 µM and an estimated molar ratio (N = 0.88) consistent with a single binding site (Figure 2A). Slightly tighter binding was observed with a pT519 peptide (Kd = 10.6 µM), but at a molar ratio more consistent with two FHA molecules binding to a single peptide (N = 0.61). When Thr519 and Thr523 were phosphorylated in the same peptide, PNK–FHA bound ∼16-fold tighter than to either of the mono-pT peptides, with a Kd = 0.7 µM. Again, the estimated stoichiometry of the interaction (N = 0.51), suggesting that two copies of PNK–FHA bound simultaneously to the XRCC1 peptide.
Figure 2.
Isothermal titration calorimetry of PNK binding to XRCC1 phospho-peptides. (a) ITC-binding curves for injection of XRCC1 peptides phosphorylated on Thr523 (left), Thr519 (middle), and on both Thr519 and Thr523 (right), into a microcalorimeter cell containing PNK–FHA (see Methods section). The mono-phosphorylated peptides show weak binding, while the doubly phosphorylated peptide binds with sub-micromolar affinity. Both peptides containing pThr518 show binding stoichiometry, (corresponding to the inflexion point of the curve) consistent with a 2:1 PNK–FHA:XRCC1 peptide complex. (b) As (a) but for peptide phosphorylated on pSer518 and pThr519 (left), and on pSer518, pThr519 and pThr523 (right). Affinities are >8-fold and >50-fold tighter, respectively, than the mono-phosphorylated pThr519 peptide. Binding stoichiometry in both cases indicates a 2:1 PNK–FHA:XRCC1 peptide complex. (c) Binding curves for injection of a pSer518 and pThr519 peptide (left) or a pSer518, pThr519, pThr523 and pSer525 peptide (right), into a cell containing full length human PNK. Affinities compare with those for the isolated FHA domain, and the binding stoichiometry remains consistent with a 2:1 PNK:XRCC1 peptide complex. (d) Inverted experiment, where PNK–FHA protein is injected into a cell containing an XRCC1 pSer518, pThr519 peptide. In this orientation of the experiment the binding stoichiometry is again consistent with formation of a 2:1 PNK:XRCC1 peptide complex, confirming that the observation is not a result of the experimental set-up. (e) Binding curves for injection of an XRRC1 peptide, phosphorylated on Ser518 and on Thr519, but with Thr523 mutated to valine. Removal of the threonine residue ablates the second binding site, so that only a single FHA domain now binds.
While no binding was observed to a peptide only phosphorylated on Ser518, when this residue was phosphorylated in addition to Thr519 within the same peptide, PNK–FHA bound >8-fold tighter (Kd = 1.23 µM) than to the peptide only containing pThr519, again with 2:1 stoichiometry (N = 0.51). Significantly greater affinity (Kd = 0.19 µM), but again with 2:1 stoichiometry (N = 0.46), was observed with the tris-phosphorylated pSer518/pThr519/pThr523 peptide (Figure 2B). However adding the fourth putative CK2 phosphorylated residue, pSer525, offered no significant further increase in affinity.
The consistent observation of a 2:1 stoichiometry for PNK–FHA-binding to all the peptides tested, other than that phosphorylated on Thr523 only, was surprising. To determine if this was related to the use of the isolated FHA domain, we repeated the titrations with full-length human PNK, against the pSer518/pThr519, and pSer518/pThr519/pThr523/pSer525 peptides. The measured affinities for these peptides (Kd = 1.57 µM and 0.10 µM, respectively) were comparable to those obtained with the isolated FHA domains, and the stoichiometries obtained for the full-length proteins (N = 0.61 and 0.49) were similarly consistent with formation of 2:1 complexes with the peptide in both cases (Figure 2C). Finally we inverted the ITC experiment from the normal arrangement with the protein in the cell and the peptide injected. When PNK–FHA was injected into the cell containing the pSer518/pThr519 peptide, the measured affinity (Kd = 2.23 µM) was comparable to that obtained in the inverted experiment, with the measured stoichiometry (N = 1.76) inverted, but fully consistent with formation of a 2:1 protein–peptide complex (Figure 2D).
The binding data are suggestive of two possible PNK–FHA-binding sites within the 12 residues of the XRCC1 CK2 cluster—a primary site centred on pThr519 and a secondary site centred on pThr523. The 1:1 stoichiometry for binding to the pThr523-only peptide as compared to the 2:1 stoichiometry for all the peptides containing pThr519, is further suggestive of hierarchical co-operative binding, whereby an FHA bound to Thr519 can facilitate binding of a second FHA to Thr523 (most tightly when phosphorylated), but not the other way round. To test this, we made a further pSer518/pThr519 peptide in which the kernal of the putative secondary site, Thr523, was mutated to valine—a residue isosteric with threonine, but unable to make the polar interactions that are likely to contribute to peptide binding. PNK–FHA bound this with a comparable affinity (Kd = 1.70 µM) to the equivalent wild-type peptide (Kd = 1.23 µM), but with a stoichiometry (N = 0.91) indicative of a single site of binding, consistent with abrogation of the secondary binding site by the Thr523Val mutation (Figure 2E).
Structural basis for bis-phospho-peptide binding by PNK–FHA
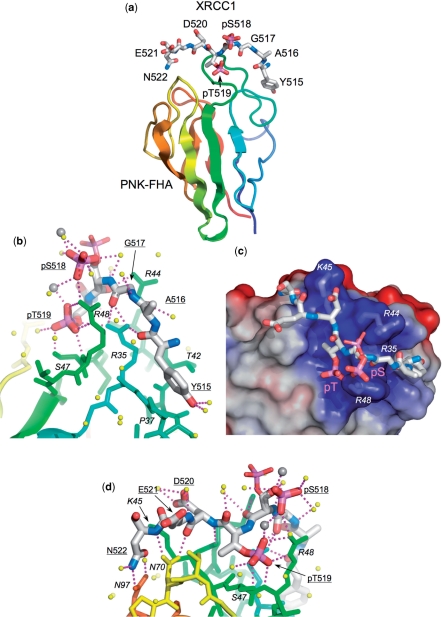
The strong additive interaction we observe for the PNK–FHA binding to the bis-phosphorylated pS518/pT519 motif is clearly different from the specificity previously described for FHA domains in general, but most importantly also differs from that ascribed to the PNK–FHA domain itself in previous studies (9). To investigate this further we sought to obtain co-crystals of human PNK–FHA/bound to XRCC1 phospho-peptides whose binding we had characterized. However initial attempts only yielded crystals of the unliganded FHA domain (PDB: 2BRF), which had an unusually low solvent content (∼38% v/v) and diffracted strongly to high resolution. Careful analysis of the crystal packing in this structure revealed a crowded environment around the expected peptide-binding site, incompatible with retention of the peptide ligand on formation of this very stable lattice. We therefore introduced a mutation into the protein (Leu99Glu) in a residue involved in a substantial hydrophobic crystal packing interface, but distant from the peptide-binding site, with the aim of preventing formation of this crystal form. Using the mutant FHA domain we obtained a new crystal form containing a complex of the FHA domain and the tris-phosphorylated XRCC1 peptide (pSer518/pThr519/pThr523).
As in the previously described murine PNK–FHA–XRCC4 peptide complex (10), the XRCC1-peptide binds across the tips of the loops connecting the β-strands that form either face of the FHA domain structure (Figure 3A). Clear electron density is visible for the bound peptide from XRCC1 residue Tyr515 to Asn522, including the phosphorylated residues pSer518 and pThr519. However the third phosphorylated residue pThr523 and the three residues following that, are not resolved despite our ‘crystal engineering’ attempts; the packing of the FHA domains within the new crystal lattice seemingly incompatible with a 2:1 protein; peptide-binding mode.
Figure 3.
Structure of PNK–FHA–XRCC1 phosphopeptide complex. (a) Overview of the binding of the XRCC1 peptide (stick model) to residues forming the inter-strand loops in the PNK–FHA domain (cartoon rainbow coloured N-terminal:blue → C-terminal:red). This and all other molecular graphics were made with MacPyMol (DeLano Scientific). (b) Interactions of the N-terminal end of the XRCC1 peptide with PNK–FHA (see text for details). The phosphate group on pSer518 shows dual conformations in the crystal structure, (shown as thick and thin sticks), due to interactions with a calcium ion involved in formation of the crystal lattice. Ordered water molecules are shown as yellow spheres, Ca2+ ions as larger grey spheres. (c) The central part of the XRCC1 peptide consisting of the negatively charged residues pSer518, pThr519 and Asp520 binds in a very basic recess on the FHA domain surface, generated by the side chains of arginine residues 35, 44 and 48, and Lys 45. The molecular surface of PNK–FHA is shown coloured by electrostatic potential – positive:blue → negative:red. (d) Interactions of the C-terminal end of the XRCC1 peptide (see text for details). The C-terminal Asn522 of the XRCC1 peptide is anchored by a bidentate amide-amide interaction with PNK residue Asn97.
The peptide makes extensive polar and hydrophobic interactions with the FHA domain (Figure 3B). At the N-terminus XRCC1-Tyr515 packs between the side chains of PNK Pro37 and Thr42—the conformation of Tyr515 suggests that the chain N-terminal to this would not contact the FHA domain. C-terminal to this, the main chain carbonyls of Tyr515 and the two consecutive residues Ala516 and Gly517, are all directed down towards the FHA domain, and make hydrogen bonding interactions with the guanidinium head group of Arg35. This residue also hydrogen-bonds to the Oγ of the phosphorylated pThr519. The interaction with Arg35 fixes the conformation of the XRCC1 peptide from residue 515 to residue 519, and locks it between the side chains of Arg44 and Arg48, which form a narrow hydrophobic but intensely charged clamp, around the main chain at Gly517 and the phosphorylated serine, pSer518 (Figure 3C). The Nε and NH moieties of Arg48 also provide additional hydrogen bonds to the phosphate group of pThr519.
The pendant phosphate group of pSer518 has two alternate conformations in the crystals, related by a ∼120° rotation around the Cα-Cβ bond. In either conformation, the phosphate group is in close proximity to the guanidinium head-groups of one or other of the side chains of Arg44 or Arg48, making direct or water bridged hydrogen-bonding interactions. In any case, the proximity of the negatively charged phosphate group to both these positively charge side-chains would provide a very favourable electrostatic interaction.
Whether both these favourable alternative conformations would persist in free solution is unclear, as the tip of the phosphate group is involved in interactions with a calcium ion required for crystal formation, and this likely perturbs the conformational energetics of the pSer518 side-chain conformation. In contrast, the pendant phosphate of pThr519 is locked down in a cage of hydrogen bonds provided by the side chain hydroxyl of PNK Ser47, and peptide and side-chain nitrogens of PNK Arg48 (Figure 3D).
Beyond pThr519, the conformation of the XRCC1 peptide is determined by main-chain to side-chain hydrogen bonding interactions with the side chain of PNK Asn70 and Lys45. The visible C-terminus of the XRCC1 peptide is locked down by a bidentate amide–amide interaction between the side chains of XRCC1 Asn522 and PNK Asn97.
DISCUSSION
The FHA domain is a widely utilized module for recognition of phosphorylated motifs, in a range of cellular systems (18,19). Recognition centres on the core pThr residue, which interacts with a Ser-Arg sequence (47 Ser-Arg 48 in hPNK) that is very highly conserved amongst FHA domains. However, the determinants of specificity outside of the core pThr vary considerably, with residues preceding and/or following the pThr making substantially different contributions in the different classes of FHA domain (20). Previous studies of the PNK–FHA domain and its interaction with XRCC4 (9,10) have highlighted the importance of residues −2, −3 and +1 relative to the pThr, and in particular the apparent absence of significant +3 interactions, unlike many other FHA domains, where this is an important feature.
Specificity of PNK–FHA
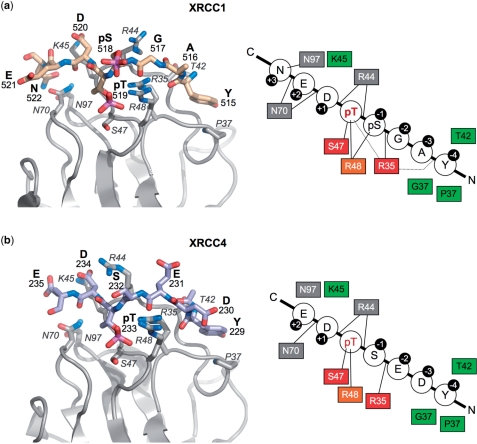
In the XRCC1 peptide complex described here however, PNK–FHA shows a clear and highly specific +3 interaction involving the side chains of XRCC1 Asn522 and PNK Asn97, indicating that, at least in this respect, PNK–FHA may not be as distinctive as originally suggested. The equivalent to XRCC1 Asn522 in XRCC4 is a glutamic acid (Glu236), which is disordered in the XRCC4 peptide complex. Interestingly, Asn (as in XRCC1) is indicated as the preferred +3 substituent in a peptide array analysis of PNK–FHA-binding preferences (9) (Figure 4C therein), whereas Glu (as in XRCC4) appears somewhat disfavoured.
Figure 4.
Comparison of PNK–FHA interactions with XRCC1 and XRCC4. PNK–FHA interacts in a similar way with the XRCC1 phosphopeptide (a), as with the XRCC4 peptide (b) apart from the additional interactions with pSer518 and Asn522 in the XRCC1 complex. That Ser518 was not phosphorylated in the XRCC4 complex is probably responsible for the different conformation observed for Arg44 in the two complexes. In both cases, the residues at −2 and −3 relative to the pThr, make no side-chain contacts with the protein that could mediate selectivity for these positions, and the specificity of FHA for the XRCC1 and XRCC4 sequences resides in the polar and electrostatic interactions with phosphorylated and acidic residues at 0, −1, +1 and +2, the hydrophobic interaction with Tyr at −4, and (for XRCC1) with the amide-side chain of the Asn at +3. In the schematics on the right, residues with a red (strictly) or orange (highly) background are conserved across FHA domains. Those with a green background are involved in hydrophobic contacts with the bound XRCC1/XRCC4 peptide, whilst those with a grey background are involved in making specific hydrogen bonds (as indicated by a line).
The unusual sensitivity to the nature of the −2 and −3 substituents observed in the peptide array analysis (9), is difficult to reconcile with the crystal structure of the PNK–FHA–XRCC1 peptide complex described here, and indeed with the previously described PNK–FHA–XRCC4 peptide complex (10). In both crystal structures the −2 and −3 residues of the peptide (XRCC1: G,A – XRCC4: E,D) are actually distinguished by having no side chain interactions with the FHA domain that could mediate specificity. Furthermore, the consensus derived from the peptide array data, would apparently preclude the −2 and −3 residues, Gly and Ala, that actually occur in XRCC1, so its biological relevance is open to question (Figure 4A and B).
PNK–FHA binds bis-phosphorylated motifs
Where PNK–FHA does genuinely appear distinctive, is in its preferential binding to a bis-phosphorylated motif. Thus while PNK–FHA binds the pThr519-XRCC1 peptide with an affinity ∼10 μM, its affinity for the pSer518/pThr519 peptide is nearly 10 times greater. The contribution of the phosphorylated −1 residue is purely additive to a necessary core pThr-dependent interaction, so that a pSer518 monophosphorylated peptide shows no significant binding. The pSer in the −1 position relative to the core pThr, is able to make direct hydrogen bonding interactions with residues in the FHA domain, but will also provide a strong favourable electrostatic interaction with the intense positive charge generated by the side chains of Arg 35, Arg 44 and Arg 48. Indeed, this, rather than any specific recognition process, may be the explanation for the apparent preference for acidic residues at the −2 and −3 positions in the XRCC4 peptide array study (9).
Mass spectrometry analysis of tryptic digests of XRCC1 extracted from HeLa cells (6) indicated that an isolated fragment (corresponding to amino acids 503–546) was a mixture of tri- and tetra-phosphorylated species. Additionally, it was noted that residues Ser518 and Thr523 are consensus CK2 phosphorylation sites, whilst those of Thr519 and Ser525 were considered to be atypical or hierarchical sites. Together, these data strongly suggest that pThr519 most likely only exists in cells in a bis-phosphorylated context (i.e. along with pSer518).
Previous studies of the interaction of PNK–FHA with XRCC4 did not discover the preferential binding to the adjacent bis-phosphorylated motif we describe here for XRCC1. However, both XRCC1 and XRCC4 scaffold proteins share the core Ser-Thr-Asp-Glu motif, in which the downstream acidic side chains directs CK2-phosphorylation of the serine and threonine residues. Thus, it is highly likely that this motif is bis-phosphorylated in both cases, and bound by PNK–FHA in the same way.
PNK is one of three mammalian DNA repair enzymes with distinctive and related N-terminal FHA domains that have so far been shown to bind to XRCC1 (6,21,22). Although only the structure of the PNK–FHA is currently available, the FHA sequences of PNK, APTX and APLF show strong conservation of the distinct sequence motifs that in PNK mediate interaction with the main specificity determinants in XRCC1. While yet to be formally demonstrated, the likelihood is therefore, that the FHA domains of APTX and APLF will also show preferential binding to the adjacent bis-phosphorylated pSer518-pThr519 motif. Indeed, Aprataxin has been shown to bind with a higher affinity to a tris-phosphorylated XRCC1 peptide (pSer518/pThr519/pThr523) than a mono-phosphorylated peptide (pT523) (22).
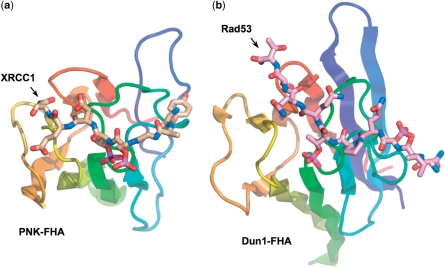
While this work was in progress, the FHA domain of the yeast checkpoint kinase Dun1p, was shown to bind preferentially to a di-phospho-threonine motif generated by ATM/ATR in the SQ/TQ cluster domain of the upstream kinase Rad53 (23). However, unlike the adjacent phosphorylated residues in the XRCC1 and XRCC4 motifs, the two pThr residues in Rad53 are separated by two residues. Furthermore, the mode of interaction, as indicated by an NMR structure of the Dun1-FHA bound to a bis-phosphorylated Rad53 decapeptide, is markedly different, with no equivalent to the Ser-Arg ‘cage’ interaction seen in PNK and other FHA domains (20), and with the N-terminal of the two pThr binding in a similar region on the FHA domain as the tyrosine in the XRCC1/XRCC4 motifs (Figure 5A and B).
Figure 5.
Different modes of bis-phosphopeptide-binding in PNK and Dun1. The binding mode to PNK–FHA of the XRCC1 bis-phosphopeptide (a) with adjacent pSer-pThr residues is markedly different to that recently observed for a yeast Rad53 phosphopeptide containing two non-adjacent pThr residues, to the FHA of the DNA damage checkpoint kinase Dun1 (PDB code 2JQL) (b). While the C-terminal phosphorylated residue is in a similar topological position on the FHA domain, the N-terminal pThr in Dun1 binds in the −4 subsite, which is occupied by a tyrosine in PNK–FHA complexes with XRCC1 and XRCC4 peptides.
Tandem binding sites in XRCC1
The sequence C-terminal to the common bis-phosphorylated binding motif is quite different between the two DNA repair scaffold proteins with which PNK interacts. XRCC1 possesses an additional threonine residue, again in the context of downstream acidic residues (523 Thr-Asp-Ser-Glu-Glu 527) that can also be phosphorylated by CK2 (6). Using an affinity purified polyclonal antibody specific to a tris-phosphorylated XRCC1 peptide (see Methods section) we found that Thr523, like the upstream pSer518 and pThr519 appears protected from dephosphorylation by PNK–FHA, and that PNK–FHA bound to a pThr523 mono-phosphorylated peptide with similar affinity as to a mono-phosphorylated pThr519 peptide. Taken together, these data indicate that both the threonine residues in this region of XRCC1 are capable of being phosphorylated by CK2, and when phosphorylated, of providing binding sites for PNK. Significantly, the ITC data shows that the two sites co-operate, and an XRCC1 peptide phosphorylated on both threonines binds two PNK–FHA molecules, with >15-fold higher affinity than for either site in isolation, and with >50-fold higher affinity when Ser518 is also phosphorylated (Figure 6).
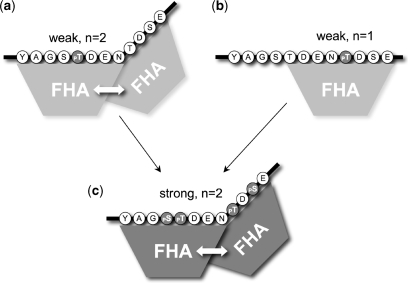
Figure 6.
Hierarchical co-operative binding of PNK–FHA domains. PNK–FHA binds with modest affinity (Kd ∼10 μM) to the XRCC1 FHA-binding region when phosphorylated on either Thr518 (a) or Thr523 (b). An FHA bound to the pThr518 site can stabilize the very weak interaction of a second FHA domain bound to the Thr523 site, even when not phosphorylated, probably by direct protein–protein contacts. However an FHA domain bound to pThr523 cannot stabilize the binding of a second FHA to the unphosphorylated Thr518 site. When both sites are fully phosphorylated (c), which is the likely result of the hierarchical phosphorylation of this region by CK2 in vivo, two FHA domains can bind with >50-fold higher affinity (Kd ∼0.1 μM), suggesting the presence of a far more stable complex than had been suggested by previous studies.
The co-operation between the two sites is however hierarchical, with binding to the pThr519 (more so with simultaneous pSer518) being able to facilitate binding to the Thr523 site, most tightly when phosphophorylated, but to some degree even when not. That the second binding site is indeed focussed on Thr523 is demonstrated unequivocally by the effect of the Thr523Val mutation, which shifts the binding stoichiometry to ∼1:1. The nature of the interactions between the FHA domains, that mediate co-operative binding to the pThr-sites remain to be determined.
CONCLUSION
Recognition of a bis-phosphorylated motif provides PNK, and probably the other FHA-domain DNA end-processing enzymes, APTX and APLF, with a unique mechanism for highly specific recognition of their binding sites in the XRCC1 and XRCC4 DNA repair scaffolds. XRCC4 is known to be dimeric (24) and might in principle therefore recruit two copies of PNK, APTX or APLF, potentially in homo- and hetero-combinations, although this is yet to be tested. XRCC1, although showing a degree of dimerization at higher concentrations in vitro, is primarily monomeric and interacts with nicked and gapped DNA substrates as a monomer (25). We show here that XRCC1 is also able to recruit two copies of PNK, and possibly two copies of APLF or APTX. However our preliminary data (not shown) suggests that it is not possible to obtain a PNK–XRCC1–APTX hetero-complex, and this is certainly consistent with the observation that these enzymes reside in distinct XRCC1 complexes in vivo (22). As PNK is a dual function enzyme, removing 3′-phosphates from one end of a ssDNA gap and adding 5′-phosphates at the other end, the presence of two binding sites on the XRCC1 scaffold may facilitate specific access of one or other of the two PNK active sites to their particular substrate ‘end’ of the gap (Figure 6B). Further work will be required to test this hypothesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by a Cancer Research UK Programme Grant (C302/A8265) (to L.H.P). R.M.J. is in receipt of a Cancer Research UK Studentship. A.A.E.A. was supported by an MRC Research Studentship. Funding for open access charge: Cancer Research UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Mark Roe for help with crystallographic data collection and processing, and Chris Prodromou for advice on ITC. We thank both Olaf-Georg Issinger (University of Southern Denmark, Odense, Denmark) for the original clone, and Cara Vaughan (ISMB, Birkbeck, UK) for the E. coli expression vector of maize CK2.
REFERENCES
- 1.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly(ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 3.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 4.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3'-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 5.Mani RS, Karimi-Busheri F, Cass CE, Weinfeld M. Physical properties of human polynucleotide kinase: hydrodynamic and spectroscopic studies. Biochemistry. 2001;40:12967–12973. doi: 10.1021/bi011383w. [DOI] [PubMed] [Google Scholar]
- 6.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 7.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 8.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 2002;21:2827–2832. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Prodromou C, Pearl LH. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 12.Leslie AGW. MRC Laboratory of Molecular Biology. U.K: Cambridge; 1995. [Google Scholar]
- 13.Collaborative Computational Project, N. The CCP4 Suite: programs for protein crystallography. Acta. Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 14.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta. Crystallogr. D. Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP's model-building algorithms. I. The main chain. Acta. Crystallogr. D Biol. Crystallogr. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallographica. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Durocher D, Smerdon SJ, Yaffe MB, Jackson SP. The FHA domain in DNA repair and checkpoint signaling. Cold Spring Harb. Symp. Quant. Biol. 2000;65:423–431. doi: 10.1101/sqb.2000.65.423. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Lee GI, Van Doren SR, Walker JC. The FHA domain mediates phosphoprotein interactions. J. Cell Sci. 2000;113(Pt 23):4143–4149. doi: 10.1242/jcs.113.23.4143. [DOI] [PubMed] [Google Scholar]
- 20.Liang X, Van Doren SR. Mechanistic insights into phosphoprotein-binding FHA domains. Acc. Chem. Res. 2008;41:991–999. doi: 10.1021/ar700148u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, Yuan C, Hammet A, Mahajan A, Chen ES, Wu MR, Su MI, Heierhorst J, Tsai MD. Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade. Mol. Cell. 2008;30:767–778. doi: 10.1016/j.molcel.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani RS, Karimi-Busheri F, Fanta M, Caldecott KW, Cass CE, Weinfeld M. Biophysical characterization of human XRCC1 and its binding to damaged and undamaged DNA. Biochemistry. 2004;43:16505–16514. doi: 10.1021/bi048615m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.