Abstract
Homing endonucleases (HEs) cut long DNA target sites with high specificity to initiate and target the lateral transfer of mobile introns or inteins. This high site specificity of HEs makes them attractive reagents for gene targeting to promote DNA modification or repair. We have generated several hundred catalytically active, monomerized versions of the well-characterized homodimeric I-CreI and I-MsoI LAGLIDADG family homing endonuclease (LHE) proteins. Representative monomerized I-CreI and I-MsoI proteins (collectively termed mCreIs or mMsoIs) were characterized in detail by using a combination of biochemical, biophysical and structural approaches. We also demonstrated that both mCreI and mMsoI proteins can promote cleavage-dependent recombination in human cells. The use of single chain LHEs should simplify gene modification and targeting by requiring the expression of a single small protein in cells, rather than the coordinate expression of two separate protein coding genes as is required when using engineered heterodimeric zinc finger or homing endonuclease proteins.
INTRODUCTION
Genome engineering requires the ability to direct biochemical activities (such as single- or double-strand cleavage, recombination or integration) to target sites with a high degree of site and/or sequence specificity. Several different types of genome engineering reagents are being developed to target specific genes. Among the best developed of these highly sequence-specific reagents are homing endonuclease (HE) proteins. The HEs consist of several families of nucleases that catalyze the lateral transfer (or ‘homing’) of parasitic DNA elements in all kingdoms of life (1). HEs are most often encoded as open reading frames (ORFs) in mobile introns or inteins. They catalyze and target the lateral transfer of their coding intron or intein by cleaving homologous alleles that lack the intron or intein sequence to initiate recombination dependent repair off the intron-/intein-containing allele (2). The DNA target or ‘homing’ sites for HEs consist of long (14–40 bp) sequences that are recognized and cleaved with high specificity in vitro and in vivo. Despite their high site specificity, many HEs can tolerate at least some target site base pair changes without the loss of site binding or cleavage specificity (3,4). The ability to maintain high site specificity while tolerating limited target site sequence divergence may represent an evolutionarily advantageous strategy for many HEs by maximizing the opportunities for continued mobility while minimizing off-target, cleavage-dependent toxicity.
We have focused on engineering novel variants of the largest and best characterized of the five HE families, the LAGLIDADG HEs (LHEs). These proteins were identified, as were other homing endonuclease families, on the basis of several consistent features: location within mobile or optional intervening sequences as ORFs; the presence of conserved short sequence motifs within their coding sequences; similar DNA target or homing site lengths of 18–24 bp; and similar mechanisms of DNA recognition and catalysis. LHEs all contain two copies of a conserved αββαββα core fold that includes a single ‘LAGLIDADG’ motif (1). This motif contributes several residues to the intersubunit or interdomain interface as well as a C-terminal acidic acid residue to the LHE active site (4–9). LHE active sites are located directly at the center of the protein domain interface, and are flanked by two separate DNA-binding surfaces (comprised of antiparallel β-sheets from each protein domain). LHE proteins containing a single LAGLIDADG motif per peptide chain, such as I-CreI and I-MsoI, function as homodimers that cleave pseudo-palindromic DNA target sites. In contrast, LHEs with two LAGLIDADG motifs per protein chain, such as I-SceI and I-AniI, are pseudo-symmetric monomers that are able to recognize asymmetric DNA target sites.
The modular architecture of LAGLIDADG proteins suggested that DNA cleavage should be tightly coupled to target site recognition, and that it should be possible to redesign site recognition specificity without disrupting catalytic efficiency. This hypothesis has been verified by experiments demonstrating the separation of site binding and cleavage activities of homodimeric I-CreI (10–15) and I-MsoI (16), as well as for the monomeric I-SceI (17) and PI-SceI (18) LHE proteins. Several novel chimeric LHEs have also been generated by taking advantage of the modular structure of LHEs: an I-CreI monomer has been fused with one domain of the thermophilic LHE I-DmoI (19) to generate H-DreI (11,20), and a portion of the DNA recognition region of monomeric PI-SceI has been successfully substituted with a DNA recognition module from the Candida tropicalis VMA1 LAGLIDADG intein (21).
Monomeric LHEs are the most appropriate starting point for creating gene targeting reagents because they are not constrained to recognize symmetric DNA target sites. However, the homodimeric LHE proteins have thus far proven more amenable to high resolution crystallographic analyses (1,4) and have been used for design and selection experiments as well as for in vivo targeted gene modification or correction (10–16). In vivo applications have most recently employed two different LHE variants that form obligate heterodimers when coexpressed in cells (22). However, this latter strategy involves the delivery and coordinated expression of two proteins, and thus may be difficult or problematic to implement.
In this article, we report the generation of hundreds of catalytically active, single-chain variants of the well-characterized homodimeric LHEs I-CreI and I-MsoI. We refer to these monomerized I-CreI and I-MsoI proteins below as mCreI and mMsoI variants. These new monomeric LHE proteins were generated by inserting a randomized polypeptide linker library between two divergent copies of the I-CreI or I-MsoI ORF, followed by in vivo selection in Escherichia coli for catalytic activity at 30°C. Furthermore, the characterization of several mCreI and mMsoI proteins demonstrated site-specific DNA-binding affinities and cleavage activities similar to their homodimeric precursors I-CreI and I-MsoI. A high-resolution co-crystal structure of one mMso variant with a 33 residue linker revealed a canonical LHE structural fold with partial ordering of the linker adjacent to the now-linked I-MsoI subunit C- and N-termini. We also demonstrated that both mCre and mMso proteins were catalytically active in human cells and able to promote cleavage-dependent recombination to generate green fluorescent protein-positive (GFP+) cells. These well-characterized single-chain LHEs should facilitate the engineering of mCre or mMso variants with the ability to target and cleave asymmetric DNA target site sequences in vivo.
MATERIALS AND METHODS
Plasmids, bacterial and chemicals
The bacterial selection plasmids pEndo and pCcdB were generous gifts from David Liu (Harvard University) (17). The bacterial protein expression plasmids pET15b and pET24d, and the E. coli host strain NovaXGF′ used for in vivo selection and protein expression were obtained from Novagen (Gibbstown, NJ). Other reagents, including restriction enzymes, Taq DNA polymerase and ligase, were obtained from New England Biolabs (Ipswich, MA) or from Sigma-Aldrich (St Louis, MO).
Construction of monomeric homing endonuclease libraries
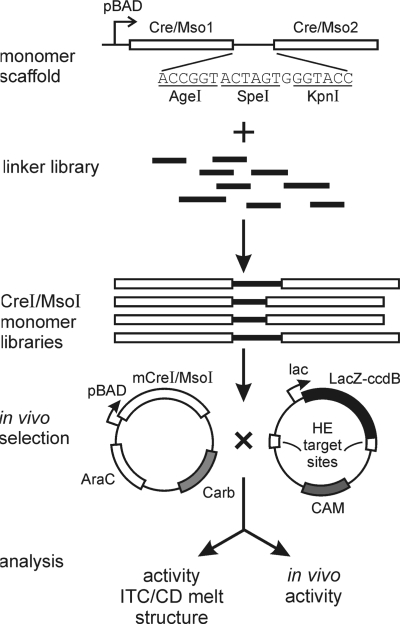
Pairs of ORF cassettes encoding native I-CreI or I-MsoI proteins were designed in which codon usage was optimized for expression in E. coli and between which nucleotide sequence divergence was maximized without changing coding properties. These divergent ORF pairs, synthesized by Blue Heron (Bothell, WA), were then cloned into pENDO separated by a short linker oligonucleotide containing AgeI, SpeI and KpnI cleavage sites to create linker recipient expression vectors for monomeric I-CreI or I-MsoI (pENDOmCre and pENDOmMso, respectively; Figure 1). A previously described random linker library, kindly provided by Michelle Scalley-Kim and David Baker (University of WA) (23), was PCR-amplified with two primers:
Figure 1.
Generation and characterization of catalytically active, monomeric versions of I-CreI and I-MsoI. Two divergent copies of the I-CreI or I-MsoI ORF were synthesized, and cloned into expression vectors together with an intervening multiple cloning site linker. A library of previously generated, randomized linkers was then PCR-amplified, cleaved and inserted at the multiple cloning site to generate libraries of monomeric mCre and mMso proteins with linker insertions. Active monomeric proteins were identified on the basis of an activity-dependent selection in E. coli, then characterized by a combination of biochemical, biophysical and functional assays. A high resolution co-crystal structure was also generated for one of the mMsoI proteins with a 33 residue linker (see Figure 5).
RLLf: 5′-ATCAGACCGGTAGCGGCTCAGGATC-3′ and
RLLr: 5′-CACAAGGTACCGCTTCCCGACCCAGATCC-3′.
The resulting linkers were cleaved with AgeI and KpnI, and then ligated into AgeI/KpnI-cleaved pENDOmCre and pENDOmMso to generate linker insert libraries of monomeric I-CreI and I-MsoI proteins for in vivo selection. The target site plasmids used for in vivo selection, pCcdB-Crewt2 and pCcdB-Msowt2, each contained single copies of the native I-CreI or I-MsoI recognition site inserted at the pCcdB Nhe I/Sac II and Afl III/Bgl II cloning sites (Figure 1).
In vivo selection of active monomeric homing endonucleases
Catalytically active mCreI and mMsoI homing endonucleases were selected from linker insert libraries by two rounds of positive selection in a CcdB in vivo selection system (17). In brief, pENDO coding plasmid libraries were transformed into bacterial containing either a pCcdBCrewt2 or a pCcdBMsowt2 target site plasmid, followed by selection for survival and colony formation on plates containing 0.02% arabinose and 0.2 mM IPTG. Colonies formed after growth for 3 days at 30°C were streaked onto plates containing carbenicillin, or carbenicillin and chloramphenicol in order to identify bacterial that retained the pENDO coding plasmid and either lost or retained the pCcdB selection plasmid. Plasmid DNA from clones surviving on carbenicillin alone was purified and retransformed into bacteria containing either pCcdBCrewt2 or pCcdBMsowt2 for a second round of selection in order to confirm that survival was pENDO plasmid-dependent.
Protein expression and purification
The mCreI and mMsoI ORFs from plasmids that had undergone two rounds of in vivo selection were sequenced and then subcloned into the NcoI/NotI sites of pET15b and pET24d (Novagen) prior to transformation into E. coli strain C2566 (New England Biolabs). Overnight cultures of single colonies were grown at 30°C, inoculated into fresh LB media containing carbenicillin or kanamycin and then grown to A600 = 0.8 at 37°C prior to inducing protein expression by the addition of 1 mM IPTG followed by growth for 3 h at 37°C. Proteins expressed from pET15b had N-terminal 6× His tags, and were purified using Ni-NTA agarose resin (Qiagen) according to the manufacturer's protocol. For crystallographic analyses, proteins expressed from pET24d were purified as previously described (4).
In vitro cleavage assays
pCcdB target vectors containing single copies of the native I-CreI or I-MsoI target site were linearized by AatII digestion for cleavage assays. Cleavage reactions contained 200 ng of linearized pCcdB plasmid DNA and different amounts of purified mCre or mMso protein in 20 μl of 20 mM Tris pH 8.0, 10 mM MgCl2 buffer. Reactions were incubated at 37°C for 1 h, stopped by the addition of loading buffer containing 0.1% SDS, and the digestion products were separated by agarose gel electrophoresis.
Circular dichroism (CD) analysis
Far UV CD spectra (250–190 nm) were recorded at 20°C on a Jasco J-815 chiro-optical spectrometer equipped with a PFD-425 Peltier thermoelectric temperature controller. Protein samples (5 μM) in PBS were analyzed in a 0.1 cm path length quartz cuvette (Hellma) to acquire continuous (1 nm band width) spectra with an 8 s response time and scan speed of 10 nm/min. Five scans were compiled to generate final spectra for each sample. Thermal denaturation curves were obtained at a protein concentration of 10 μM. Ellipticity at 222 nm was recorded at 1°C/min intervals over a temperature range of 10–95°C. The mean residue ellipticity [θ] was calculated from the equation: [θ] = θ° MRW/10 l c where [θ] is the mean residue ellipticity in degrees/cm2/decimole, θ° the measured ellipticity angle in millidegrees at wavelength λ, MRW is the mean residue weight, l is the optical path length of the cell in centimeters and c is the protein concentration in milligrams per milliliter (24). The percentage of α-helix in secondary structure was estimated using the equation: fraction of α-helix = (–[θ]222 nm + 3000)/39 000 (25).
Isothermal titration calorimetry (ITC)
ITC experiments were performed on a VP-ITC MicroCalorimeter (MicroCal LLC) following the manufacturer's protocol using protein samples that had been dialyzed overnight against 50 mM Tris pH 8.0, 75 mM NaCl and 10 mM CaCl2. Protein samples (3–5 μM) were placed in the ITC cell and target site DNA's (40–50 μM) in the auto-pipette injector. Individual experiments consisted of 25–35 injections of 5–9 μl of target site DNA (depending on sample concentration) at 30°C with a constant stirring speed of 300 r.p.m. Data were curve fit using Origin 7 SR2 ITP analytical software (OriginLab Corporation).
In vivo recombination assays
Mammalian expression vectors for mCreI and mMsoI were constructed by cloning Cre or Mso ORF's into the mammalian transient expression vector pCS2nlsMT where ORF expression is driven by a CMV promoter. Target plasmids for in vivo cleavage were modified from pDR-GFP (26) to replace the original I-SceI cleavage site with a single copy of the native I-CreI or I-MsoI cleavage site. Transient transfections were performed using 293T cells in 24-well plates. Cells were grown in Dulbecco-modified Eagle's medium (Cellgro) supplemented with 10% fetal bovine serum (Cellgro) and 1% penicillin/streptomycin (Gibco) at 37°C in a humidified 5% CO2 incubator. Cells were plated 24 h prior to transfection at 3 × 105 cells per well in 500 µl of complete growth medium, corresponding to 50–80% confluency at the time of transfection. A modified calcium phosphate protocol (27) was used to transfect cells with 1.5 µg per well of expression plus reporter DNA in a 3:1 molar ratio of expression to target plasmid DNA.
Transfected cells were analyzed for GFP fluorescence 48 h after transfection. Cells were tryspinized and washed in PBS, resuspended at ∼106 cells/ml and then stained in PBS containing 10 ng/µl propidium iodide prior to analysis on an Influx flow cytometer (Cytopeia). Typically 40 000 events were scored and gated for log side vs. linear forward scatter and for PI exclusion to identify viable single cells for GFP fluorescence analysis. The fraction of cells transfected in each experiment was estimated by determining the frequency of GFP+ cells that had been transfected with the positive control vector pEGFP-C1 (Clontech).
Crystallization, data collection and structural refinement
Co-crystals of mMsoI:DNA were grown using 4 mg/ml mMsoI protein and a 2-fold molar excess of I-MsoI target site DNA. The target site DNA (5′-GCAGAACGTCGTGAGACCGTTCCG-3′) was generated by annealing the oligonucleotide shown with a complementary 24mer at 18°C in 24–30% PEG 400, 100 mM Tris pH 8.0, 10 mM CaCl2, 50 mM NaCl as previously described (4). Data were collected in-house using an RAXIS-IV imaging plate area detector (Rigaku, USA), and a rotating anode X-ray generator with a copper source that provided X-rays at 1.54 Å wavelength. The resulting data were reduced using DENZO and SCALEPACK, and the structure was solved by molecular replacement using Phaser with the original I-MsoI:DNA structure as an initial search model. The resulting structure was modeled using Wincoot (http://www.ysbl.york.ac.uk/~lohkamp/coot/), and refined to 2.7 Å resolution using Refmac (http://mac.softpedia.com/get/Math-Scientific/refmac.shtml) with 5% of the data excluded for cross-validation. The crystal parameters and structural refinement statistics are summarized in Table 3.
Table 3.
Summary of data processing and refinement statistics for mMsoI co-crystal structure determination
| Parameter | mMsoI result/value |
|---|---|
| Space group | P1 |
| Cell parameters (Å) | a = 41.9 |
| b = 41.9 | |
| C = 71.0 | |
| α = 107.2 | |
| β = 95.4 | |
| γ = 109.4 | |
| Resolution range (Å) | 66.2–2.69 |
| Redundancy | 3.9 |
| Completeness (%)a | 96.6 (90.6) |
| Average I/σ (I)a | 20.9 (4.5) |
| Rsym (%)a | 5.2 (25.8) |
| Rwork (%) | 21.1 |
| Rfree (%) | 28.1 |
| RMSD bond length (Å) | 0.008 |
| RMSD angle (°) | 1.110 |
| Ramachandran plot (% of modeled residues) | |
| Most favored | 90.5 |
| Additionally allowed | 9.5 |
| Disallowed | 0.0 |
| Average B (Å2) (protein, DNA) | 59.2 |
aOuter resolution bin 2.76–2.69.
RESULTS
mCreI and mMsoI library construction
Our previous co-crystal structures of I-CreI and I-MsoI demonstrated that the C- and N-termini of adjacent subunits were separated by a substantial distance, ∼70 Å, that would need to be bridged with a polypeptide linker to generate monomeric versions of either protein (4). One previous effort to generate a monomeric version of I-CreI used an 11 amino-acid linker from the I-DmoI LAGLIDADG HE to connect intact and partially deleted I-CreI subunits. The resulting protein was soluble, though displayed reduced activity and thermal stability (11). In order to generate better-behaved, monomeric versions of I-CreI and I-MsoI we used longer linkers to join subunit C- and N-termini and to avoid truncating either protein subunit. This experimental strategy is shown in Figure 1.
We knew from previous structural analyses that long linkers were present in some LHE proteins (e.g. the 18 residue linker in I-AniI) (9). We were also inspired by experiments, which demonstrated that peptide linkers ranging from 30 to 120 amino-acid residues could be tolerated in the loop region of an SH2 domain without disrupting structure or binding in a phage surface display-based SH2 domain ligand-binding assay (23). These experiments demonstrated that linker insertions could be tolerated in a folded host protein without destabilizing local structure or function, and provided a library of partially randomized, experimentally verified linkers for our experiments (Figure 1).
Complex libraries of monomeric I-CreI and I-MsoI subunits were generated in two steps. First, we synthesized two different genes encoding the I-CreI or I-MsoI ORFs and cloned these into an expression vector separated by a short multiple cloning site to generate a ‘linker recipient’ expression vector for each protein. The ORFs were codon optimized for expression in E. coli, and designed to minimize nucleotide sequence identity between the tandem I-CreI or I-MsoI coding sequences without changing the encoded protein sequences. This was done to minimize deletions or other homology-mediated rearrangements between the directly repeated I-CreI or I-MsoI coding sequences. Libraries of polypeptide linkers (previously created in selection experiments to identify well-tolerated loop insertion elements) (23) were then inserted into each linker recipient expression vector to generate libraries of I-CreI or I-MsoI ORFs in which the two protein subunits were joined by polypeptide linkers of varying length and amino acid composition (Figure 1).
The complexities of the resulting mCreI or mMsoI libraries were both ∼1 × 106 (results not shown). DNA sequence analysis of randomly chosen clones from the unselected libraries identified linkers ranging from 13 to 93 residues. We also identified several ‘linkerless’ clones in each unselected library in which the two protein subunits were joined by six residues encoded by our multiple cloning site (Figure 1). The predominant linker lengths in both selected libraries were 13 and 33 residues, corresponding to the two most prevalent lengths present in our unselected libraries (Supplementary Figure 1). Thus, a majority of linkers in both unselected and selected libraries were ≥33 residues, or long enough to bridge the ∼70 Å distance between the N- and C-termini of I-CreI and I-MsoI.
All linker-containing clones in unselected libraries shared two common flanking polypeptides: TGSGSGS at linker N-termini and GSGSGSGT at linker C-termini. These Gly-Ser repeats were incorporated during construction of the original loop library to provide flexible regions flanking linkers in order to minimize disruption of well-ordered domains or folds following linker insertion (23). The N- and C-terminal Thr-Gly (AgeI) and Gly-Thr (KpnI) termini provided additional flexibility for further manipulation or linker transfer. The 20 residue periodicity observed in longer linkers (Supplementary Figure 1 and Table 1) reflected the use of randomized 18 residue blocks separated by a Gly-Ser pair encoded by a BamHI site to construct the original loop library (23).
The deduced amino-acid sequences of linkers from both libraries contained all 20 amino acids (Supplementary Figure 1). This was surprising as cysteine was originally excluded from linker sequences to limit entropic costs resulting from intra-loop disulfide bond formation. The presence of small numbers of cysteines likely reflects mutations generated during library construction, propagation or PCR to transfer the linker library into mCreI and mMsoI (23). Small and/or polar amino acid residues such as Thr, Gly, Ala, Ser and Asn were among the most common residues in linker sequences. Large aromatic residues such as Phe, Trp and Tyr, in contrast, were the least common.
In vivo selection of active, monomeric HEs
We used an in vivo, E. coli-based selection protocol to identify catalytically active single-chain HEs in both of our libraries. This method was a modification of the selection system originally developed by Liu and colleagues (17). In outline, a low copy number plasmid was used to express mCreI or mMsoI under control of an arabinose-regulatable pBAD promoter. A second, high copy-number target plasmid consisted of HE cleavage sites flanking a LacZα-ccdB gene fusion gene encoding the CcdB (controlled cell death protein B) E. coli DNA gyrase inhibitor protein that could be induced with IPTG (Figure 1). Cell viability in this system depends on HE cleavage and elimination of the pCcdB plasmid encoding the toxic LacZα-ccdB protein (17).
In minimal media at 30°C, expression of native homodimeric I-CreI and I-MsoI rescued almost 100% of cells harboring a pCcdB plasmid DNA carrying two copies of the I-CreI or I-MsoI target site (Table 1). In contrast, expression of catalytically inactive forms of either protein (D20N I-CreI or D22N I-MsoI) under the same selection conditions led to a ∼104-fold reduction in survival to fewer than 1 in 104 cells plated (Table 1). Similar low surviving fractions were observed when catalytically active I-CreI or I-MsoI were expressed in cells containing pCcdB plasmids with non-cognate HE target sites (Table 1). These results indicated that this activity-based selection was robust, activity-dependent and sufficiently sensitive to discriminate between the I-CreI and I-MsoI target sites that differed at only two base pair positions.
Table 1.
In vivo selection results for specific endonuclease–target site pairs
| Protein | Target site | Survivala (%) |
|---|---|---|
| I-CreI | I-CreI native | 97.8 ± 19.6 |
| I-CreI D20N | I-CreI native | (8.8 ± 1.4) × 10−3 |
| I-MsoI | I-CreI native | (1.1 ± 0.1) × 10−3 |
| I-MsoI | I-MsoI native | 98.6 ± 4.3 |
| I-MsoI D22N | I-MsoI native | (7.9 ± 1.4) × 10−3 |
| I-CreI | I-MsoI native | (1.7 ± 0.4) × 10−3 |
aSurvival rates were calculated by the number of colonies grown on selective media over the total number of cells plated. Survival for each endonuclease–target site pair were calculated from determinations performed in triplicate.
Two rounds of positive selection of our mCreI and mMsoI libraries yielded several hundred linker insert-positive plasmids from each library (results not shown). We randomly selected and sequenced the mCre ORF from 141 randomly selected mCreI or mMsoI clones (Supplementary Table 1), and further characterized the 19 different mCreI and 13 different mMsoI variants. Sequencing revealed single ORFs in selected variants in which I-CreI or I-MsoI subunits were separated by linkers ranging from 13 to 53 residues (mMsoI) or from 13 to 73 residues (mCreI; Supplementary Table 1). The most common linker length in each library was 33 residues (Supplementary Table 1 and Supplementary Figure 1). The amino acid composition of linkers in mCreI and mMsoI proteins selected on the basis of activity resembled the residue distribution observed in unselected libraries with two exceptions: a ∼2-fold drop in the abundance of glycine residues, and variable increases in residues with bulky (phenylalanine, histidine, proline, tryptophan and tyrosine) or negatively charged (arginine, lysine) side chains (Supplementary Figure 1). No additional sequence conservation was observed when linker sequences were aligned for either family of monomerized proteins (Supplementary Table 1; additional results not shown).
Biochemical characterization of mCreI and mMsoI
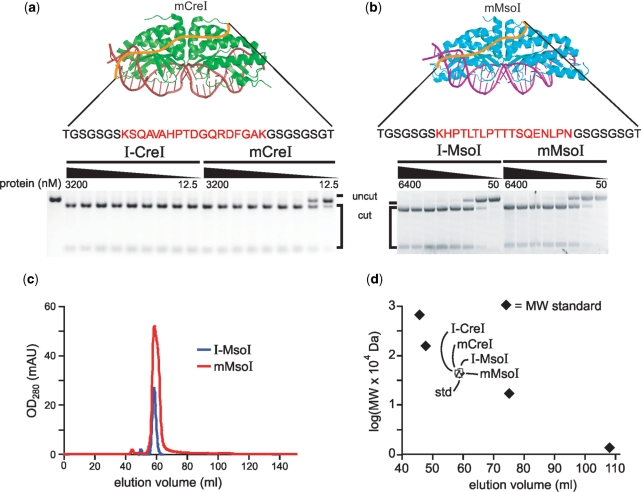
The biochemical characterizations of sequence-verified mCreI and mMsoI proteins were begun by examining in vitro cleavage of a linearized plasmid DNA substrate containing a single copy of the native I-CreI or the I-MsoI target site. All 19 mCreI proteins and all 13 mMsoI proteins demonstrated in vitro cleavage activity in these assays. We characterized the activity of 13 mCreI proteins and 8 mMsoI proteins in more detail as shown in Figure 2 and Supplementary Table 1. All 21 purified proteins showed in vitro cleavage activities comparable to their homodimeric counterparts (I-CreI or I-MsoI), with mCreI slightly less active than homodimeric I-CreI (Figure 2a). mMsoI, in contrast, appeared as active as homodimeric I-MsoI (Figure 2b). There was no detectable difference in cleavage activity as a function of protein linker length for mCreI variants with 13–73 residue linkers or for mMsoI variants with 13–53 residue linkers of variable composition. These results indicate that many different linker lengths and compositions are compatible with in vitro cleavage activity. We focused our subsequent analyses on the most straightforward and potentially useful of the monomeric mCreI and mMsoI proteins having linker lengths of 33 residues.
Figure 2.
In vitro cleavage and purification of monomeric and native, homodimeric versions of I-CreI and I-MsoI. (a, b) In vitro cleavage analyses using a linearized plasmid DNA substrate revealed comparable cleavage activities of monomeric and homodimeric versions of I-CreI and I-MsoI. Nineteen additional monomeric variants were characterized in similar fashion (Supplementary Table 1). Protein concentrations were 12.5–3200 mM for the I-CreI/mCreI proteins and 50–6400 nM for I-MsoI/mMsoI proteins. (c) Elution profiles of I-MsoI (blue) and mMsoI (red) were superimposable in size exclusion chromatography analyses on a HiLoad 16/60 Superdex 75 column (GE healthcare, Piscataway, NJ). Gel filtration analysis of all four proteins revealed predicted molecular weights when compared with an equivalent size gel filtration standard (MW std = 44 kDa chicken ovalbumin; Biorad, Hercules, CA).
One concern in both selection and cleavage assays was the potential of mCreI and mMsoI proteins to form active endonucleases as dimeric or higher order oligomers. We used physical and activity-based assays to address this issue. First, size exclusion chromatography demonstrated that both mCreI and mMsoI eluted at volumes similar to homodimeric native I-CreI or I-MsoI (Figure 2c). Both monomeric proteins and their homodimeric precursors when expressed in E. coli also had elution volumes that were similar to a 44 kDa chicken ovalbumin protein size standard (Figure 2d), in good agreement with the predicted molecular weights for I-CreI (37.4 kDa), mCreI (40.4 kDa), I-MsoI (38.9 kDa) and mMsoI (43 kDa). Corresponding western blot data indicated that all four proteins were of predicted molecular weight after transient expression in human cells (see below).
We also constructed and determined the catalytic activities of mCreI and mMsoI variants with single inactivating substitutions in one half of their active sites (D20N mCreI and D22N mMsoI). Prior structural and biochemical analyses of I-CreI (4,28) and I-MsoI (28) predicted that these single active site mCreI or mMsoI proteins should be catalytically inactive unless mCreI or mMsoI dimers or higher order oligomers were the active form of either endonuclease. D22N mMsoI did retain detectable strand nicking activity. However, neither D20N mCreI nor D22N mMsoI protein had detectable DNA double strand cleavage activity at protein concentrations up to 1.6 μM (data not shown).
Biophysical characterization of mCreI and mMsoI
In order to determine whether linker insertion affected the stability or binding affinity of either mCreI or mMsoI, we expressed and purified to homogeneity mCreI and mMsoI proteins with 33 residue linkers as previously described (4), and then analyzed each protein by far-UV CD analysis and ITC.
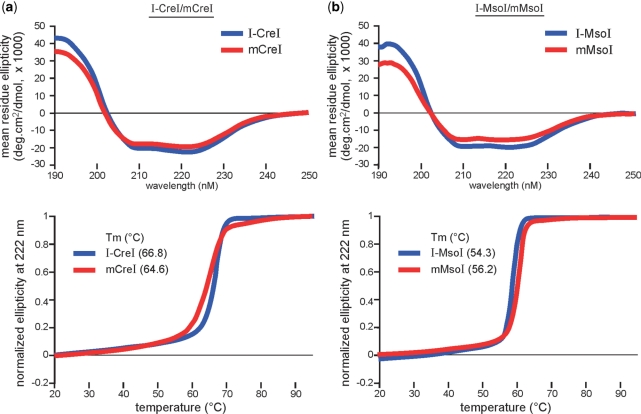
The far-UV CD analysis was performed to assess overall structural features of mCreI and mMsoI as compared with their homodimeric counterparts I-CreI and I-MsoI (Figure 3, top panels). The far-UV CD spectra of mCreI and mMsoI were very similar to their homodimeric counterparts and characteristic of proteins with mixed α/β topology and estimated α-helical contents of 58% and 46%, respectively. Homodimeric I-CreI and I-MsoI had CD spectra characteristic of proteins with mixed α/β topology, and had estimated α-helical contents of 67% and 58%, respectively. The values for monomeric and homodimeric protein pairs were virtually identical when differences in polypeptide chain lengths were normalized. This indicates that most of the linker regions in mCreI and mMsoI lack defined secondary structure. Thermal denaturation profiles were remarkably similar as were midpoint transition temperatures for homodimeric and monomeric I-CreI (Tm values of 66.8°C and 64.6°C, respectively), and for I-MsoI and mMsoI (Tm values of 54.3°C and 56.2°C, respectively; Figure 3, bottom panels). These results indicate that linker insertion did not disrupt the structure or stability of either monomeric protein, or promote the formation of potentially confounding dimer or higher order oligomers.
Figure 3.
Far-ultraviolet CD and thermal denaturation profiles of homodimeric and monomeric I-CreI (a) and I-MsoI (b). All four curves in both CD and thermal denaturation analyses were generated by best fit analyses for a two-state transition, and had 50% transition temperatures of 66.8 (I-CreI), 64.6 (mCreI), 54.3 (I-MsoI) and 56.2°C (mMsoI), respectively.
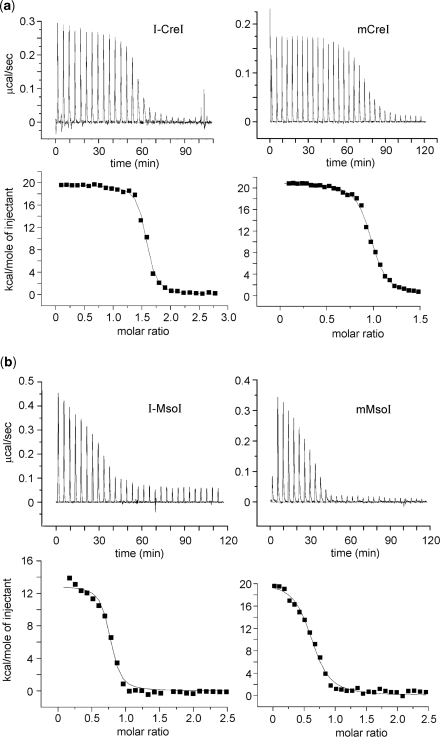
The binding of I-CreI, mCreI, I-MsoI and mMsoI to their cognate DNA target sites was analyzed by ITC. The thermodynamic parameters and profiles for each HE:DNA target site pair are shown in Figure 4, and summarized in Table 2. Homodimeric and monomeric protein pairs showed similar two-phase endothermic binding isotherms. The measured dissociation constants (KD) for I-CreI and I-MsoI were, as predicted, close to half the values determined for mCreI and mMsoI (13.3 nM/22.3 nM for I-CreI/mCreI, and 21.0 nM/40.2 nM for I-MsoI/mMsoI). This difference reflects the molar stoichiometry of protein–DNA interactions that were ∼2:1 for I-CreI and I-MsoI, and ∼1:1 for mCreI and mMsoI. These results indicate that mCreI and mMsoI have native DNA target site binding affinities that are similar to their homodimeric counterparts. This is also reflected in calculated free energies (ΔGbinding), enthalpic changes (ΔH) and entropic changes (–TΔS) for protein–DNA interactions that were found, again, to be virtually identical for each homodimer/monomer protein pair (Table 2).
Figure 4.
DNA binding and thermodynamic profiles for homodimeric and monomeric I-CreI and I-MsoI as determined by ITC. Heat absorption upon the injection of native target site DNAs can be seen in upper panels in (a) for I-CreI/mCreI and in (b) for I-MsoI and mMsoI. The molar ratio of injected target site DNA to protein is shown in bottom panels. Results for all proteins could be fitted to standard saturation curves, and all four proteins displayed endothermic binding profiles. A summary of thermodynamic parameters for all four proteins is given in Table 3. Panel (b) I-MsoI data were adapted from ref. 32.
Table 2.
Affinities and thermodynamic values for homing endonuclease–DNA binding
| Protein | KD (nM) | ΔH (kcal/mol) | ΔS (cal/mol/deg) | –TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| I-CreI | 13.3 ± 2.6 | 19.6 ± 0.20 | 101 ± 1.2 | –30.6 | –11.0 |
| MCreI | 22.3 ± 1.7 | 21.0 ± 0.12 | 104 ± 1.0 | –31.5 | –10.5 |
| I-MsoIa | 21.0 ± 5.0 | 12.7 ± 0.27 | 77 ± 1.1 | –23.3 | –10.6 |
| MMsoI | 40.2 ± 9.6 | 14.7 ± 0.4 | 82.4 ± 1.0 | –25.0 | –10.3 |
aAdapted from ref. (32).
Co-crystal structure of mMsoI
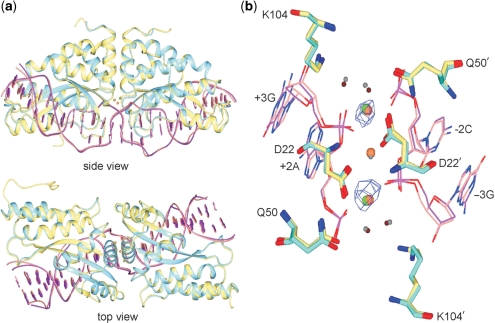
We determined the structure of mMsoI bound to its native DNA target site to provide additional insight into the structure of a monomerized LHE protein. This mMsoI:DNA substrate complex was crystallized in the presence of calcium ions under conditions previously described (4), and the structure was determined at 2.7 Å resolution (Figure 5 and Table 3). The mMsoI:DNA complex contained, as predicted, an intact 24 bp DNA target site that crystallized in space group P1 with one complex per unit cell, i.e. the same as previously observed for a homodimeric I-MsoI:DNA complex (4). The overall structures of mMsoI:DNA and I-MsoI:DNA were highly super-imposable, with an overall RMSD of 0.47 Å except for the linker region (Figure 5a). Substrate DNA bending was also very similar in the I-MsoI:DNA and mMsoI:DNA structures, including a significant distortion of the central ±1 bp region of target site DNA that is thought to facilitate cleavage (Supplementary Figure 2).
Figure 5.
Superposition of the I-MsoI:DNA and newly determined mMsoI:DNA co-crystal structures. (a) side and top views of the native homodimeric I-MsoI and newly determined monomeric mCreI co-crystal structures reveals backbone superposition with a 0.47 Å RMSD for protein and DNA. I-MsoI and mMsoI structures, and their DNA molecules, are shown respectively in cyan, yellow, purple and pink. Three calcium ions in I-MsoI:DNA structure and two calcium ions in mMsoI:DNA structure are shown, respectively, in green and coral. (b) Superposition of the I-MsoI and mMsoI active sites. Three active site residues—D22, Q50 and K104—with two calcium ions and two nucleotides flanking the scissile phosphodiester bond are shown. Four water molecules from I-MsoI:DNA and mMsoI:DNA structures are shown as small spheres in grey or tan, respectively. Anomalous difference mapping analysis revealed two calcium ions in the mMsoI:DNA active site. Figures were prepared using the CCP4 Molecular Graphics software package (33).
We were particularly interested in determining what portion of the mMsoI linker could be visualized in this structure, and whether this led to any significant distortion of the I-MsoI core fold adjacent to the linker insertion points. In the original cocrystal structure of the I-MsoI bound to substrate DNA, the first five N-terminal and last four C-terminal residues in each domain were not visible due to structural flexibility (4). In the mMsoI:DNA structure, composite omit mapping analyses allowed us to model 12 additional amino-acid residues: eight linker residues at the C-terminus of the Mso1 domain, and two linker residues at the N-terminus of the Mso2 domain. The remainder of the linker region in mMsoI is disordered and could not be visualized. These results are in good agreement with our biophysical characterization of mMsoI.
The active sites in both complexes were superimposable, containing two bound calcium ions as determined by anomalous difference mapping (Figure 5b). The discrepancy in number of calcium ions between the previously reported I-MsoI:DNA structure (4) and our mMsoI:DNA structures reflects the use of different methods to identify different metal ions during structure refinement. During the original I-MsoI structure determination, the binding of calcium was modeled based on a 2Fo – Fc difference Fourier analysis (4) which is not as definitive as the anomalous difference mapping we used to model calcium binding in the mMsoI structure. We can only visualize calcium and manganese (not magnesium) with anomalous differences. The use of this method indicated two bound calciums in uncleaved complexes with that metal, and three bound manganese ions in cleaved complexes formed with manganese. With magnesium in any of these structures, the best we can do is 2Fo – Fc difference mapping that indicates three bound metals. Thus, the most recent structural and biochemical data are consistent with the active forms of both I-MsoI/mMsoI and I-CreI/mCreI having three magnesium ions bound coordinately between two active sites, and sharing a common catalytic mechanism.
In vivo activity in human cells
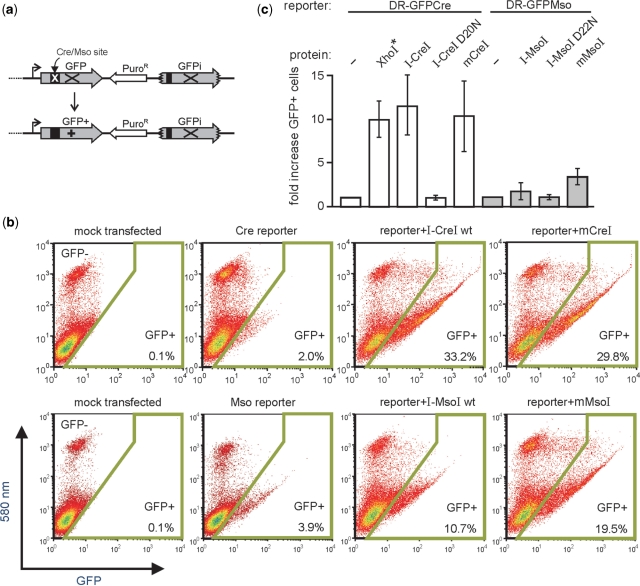
In order to determine whether monomeric I-CreI and I-MsoI were catalytically active in human cells, we co-transfected two plasmids into cells that encoding an endonuclease protein and a corresponding endonuclease-specific direct repeat recombination reporter plasmid. Site-specific cleavage of the reporter plasmid by the HE protein in vivo promotes homology-dependent gene conversion of the reporter plasmid to generate GFP+ cells that can be detected and quantified by flow cytometry (Figure 6a). Human 293T cells were used for these experiments as they are recombination-proficient and can be transiently transfected at very high (95–100%) efficiency. This assay can be performed rapidly with different combinations and amounts of input DNA, and thus allowed the rapid, quantitative functional characterization of mCreI, mMsoI and different versions of homodimeric I-CreI or I-MsoI in human cells.
Figure 6.
mCreI and mMsoI induce catalytic activity-dependent recombination in human cells. (a) In vivo recombination activity was assayed by co-transfecting coding plasmids for I-CreI or I-MsoI proteins together with a recombination reporter plasmid that contains a direct repeat of two genetically inactive copies of GFP. In vivo cleavage of the I-CreI or I-MsoI target site initiates gene conversion and repair of the cleaved copy to generate GFP+ cells that can be detected and quantified by flow cytometry (Materials and methods section). (b) Flow histograms of cells mock-transfected, transfected with reporter plasmid alone, or co-transfected with reporter: endonuclease plasmid pairs. The gate for GFP+ cells is shown by the boxed area, and the GFP+ frequency is given in the lower right of each histogram. (c) Frequency and fold increase in GFP+ cells for different reporter/coding plasmid combination. The frequency of GFP+ cells generated by in vitro XhoI-linearized DR-GFPCre reporter DNA (*) indicates that a substantial fraction of reporter molecules are likely cleaved in vivo by I-CreI and mCreI. D20N I-CreI and D22N I-MsoI are catalytically inactive mutants of I-CreI and I-MsoI which fail to induce GFP+ cells when cotransfected with a Cre or Mso-specific reporter plasmid. Error bars are means ± SDs.
Cells that were mock-transfected, or transfected with circular DR-GFPCre or DR-GFPMso recombination reporter plasmids had low autofluorescence or background recombination frequencies (0.1% autofluorescence vs. 2.0–3.9% of reporter-only transfected cells; Figure 6b). I-CreI and I-MsoI coding plasmids strongly induced the GFP+ cell fraction when transfected with a cognate reporter plasmid (GFP+ frequencies were 33.2% for I-CreI, and 10.7% for I-MsoI in experiments shown in Figure 6b). Comparable frequencies of GFP+ cells were seen following the co-transfection of a DR-GFPCre reporter plasmid with either an mCreI-coding or an I-CreI-coding plasmid (29.8% vs. 33.2%; Figure 6b). In contrast, mMsoI was more efficient at inducing GFP+ cells than was I-MsoI (19.5% vs. 10.7%; Figure 6b).
GFP+ frequencies in these assays provide a reliable estimate of the in vivo activity of specific homing endonuclease proteins. This is indicated by the high transfection efficiency (≥95%) in all experiments, the ability of in vitro XhoI-linearized DR-GFP reporter plasmids to consistently generate ∼25–30% GFP+ cells and the inability of catalytically inactive D20N I-CreI or D22N I-MsoI when co-transfected with reporter plasmid to induce GFP+ cells above background (Figure 6c). Western blot analyses indicated that all endonuclease proteins were expressed at comparable levels in these experiments, and were of predicted molecular weight for homodimeric or monomeric Cre or Mso variants (results not shown). Thus, these experiments demonstrate that mCreI and mMsoI proteins are catalytically active in vivo, and can efficiently promote site-specific, cleavage-dependent recombination to generate GFP+ cells.
DISCUSSION
We generated monomeric versions of the well-characterized and experimentally tractable homodimeric LHE proteins I-CreI and I-MsoI to further facilitate the engineering of LHEs to cleave asymmetric DNA target sites in living cells. Targeted gene modification or repair has relied thus far on engineered zinc finger nucleases (ZFNs) or homodimeric LHEs that are formed from pairs of co-expressed proteins. The in vivo use of either ZFNs or homodimeric LHEs obligatorily requires the coordinate expression of pairs of proteins in target cells. This strategy, as initially employed led to the formation of mixtures ZFN or LHE homo- and heterodimers that had variable degrees of in vivo site-specific activity or activity-dependent toxicity. More recent efforts have focused on the design of ZFN or LHE protein pairs that preferentially form heterodimers in solution as a way to ameliorate these problems (22,29).
The identification of native, monomeric LHEs having asymmetric DNA target sites provided us with the motivation and rationale to generate new, monomeric LHEs from pre-existing, homodimeric LHEs. We focused on two well-characterized, biochemically tractable LHEs, I-CreI and I-MsoI. These LHEs share partially degenerate palindromic sequences that differ by 1 base pair in each DNA half-site (4). Our previously determined a high-resolution co-crystal structures for each protein revealed nearly superimposable conserved αββαββα core folds, though substantially different β-sheet DNA–protein interfaces and contact maps that might facilitate engineering toward specific asymmetric DNA target sites.
We successfully generated libraries of several hundred active monomerized I-CreI and I-MsoI proteins (mCreIs and mMsoIs) by inserting random polypeptide linkers to join two LHE subunits into single ORFs. Characterization of selected examples from the mCreI and mMsoI protein libraries showed that both types of protein were of full length when expressed in E. coli or in human cells, were well-ordered and biochemically as well as biophysically well-behaved in vitro and catalytically active in human cells. Our experimental approach may also be useful for generating other novel proteins from pre-existing domains or subunits, especially if there is a facile selection or screen available to identify the desired novel fusion protein product(s).
The experimental approach we used may reflect the evolutionary history of LHEs. One model for LHE evolution begins with a small, ancestral monomeric protein precursor that formed first a functional homodimer that after gene duplication, fusion and domain divergence gave rise to present day monomeric LHEs (30). The ability to efficiently recognize asymmetric DNA target sites would be greatly enhanced by subunit fusion, and would provide a substantially broader range of potential DNA target sites and hosts to ensure evolutionary persistence. Target site diversification after fusion may have been facilitated in this model by pre-existing target site asymmetry in the ancestral homodimer fusion partners. This type of asymmetry exists, as shown by our recent structural analyses of the I-CeuI LHE. This structurally symmetric, homodimeric LHE was able to recognize asymmetric DNA target site base pairs in each half site by using different side chain rotamer torsion angles and bridging water networks (31).
The availability of well-characterized mCreI and mMsoI LHEs should further facilitate the engineering of protein variants to cleave asymmetric DNA target sites in vivo. The ability to develop gene- or genomic region-specific LHE variants may be further facilitated by having starting pairs of well-characterized monomeric proteins that use substantially different sets of DNA–protein contacts to recognize and bind closely related DNA target sites. The use of engineered versions of mCreI or mMsoI would be simpler than for either ZFNs or homodimeric LHEs as expression of a single protein, rather than co-ordinate expression of two different proteins, would be required in target cells. A final advantage of using LHE-derived genome engineering reagents is the inherently tight coupling of site binding to cleavage among LHE proteins. This desirable property ensures high site specificity and activity, while minimizing off-target cleavage events. Thus engineered target site-specific versions of mCreI, mMsoI or other well-documented LHEs should aid in vivo analyses of gene structure and function, as well as targeted, gene-specific modification or repair for purposes of disease therapy or prevention.
COORDINATE SUBMISSION
The structure and X-ray structure factor amplitudes of mMsoI bound to its DNA target site in the presence of calcium has been submitted to the RCSB database (PDB ID code 3FD2).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health U54 Interdisciplinary Research Consortium award (1RL1 CA133831 to R.J.M., Jr) National Institutes of Health Awards (R01 GM49857 and RL1 CA133833 to B.L.S.); Gates Foundation/FNIH ‘Grand Challenges in Global Health’ (subcontract awards to R.J.M., Jr and B.L.S). Funding for open access charge: 1RL1 CA133833.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Stoddard, Baker and Scharenberg Labs for continued help and advice throughout this work; David Liu and members of his lab for help with the in vivo selection system; Lei Zhao and Betty Shen for help with mMsoI biochemical and structural characterizations; and Bin Xu and Carina Xu in the lab of Dr Roland Strong at the Fred Hutchinson Cancer Research Center (Seattle, WA) for help with ITC analyses.
REFERENCES
- 1.Stoddard BL. Homing endonuclease structure and function. Quart. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M, Perlman PS. Mechanisms of intron mobility. J. Biol. Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 3.Argast GM, Stephens KM, Emond MJ, Monnat RJ., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- 4.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 5.Silva GH, Dalgaard JZ, Belfort M, Van Roey P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 6.Ichiyanagi K, Ishino Y, Ariyoshi M, Komori K, Morikawa K. Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol. 2000;300:889–901. doi: 10.1006/jmbi.2000.3873. [DOI] [PubMed] [Google Scholar]
- 7.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 8.Moure CM, Gimble FS, Quiocho FA. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat. Struct. Biol. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- 9.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Jr, Stoddard BL, Seligman LM. Isolation and characterization of new homing endonuclease specificities at individual target site positions. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 15.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 18.Gimble FS, Moure CM, Posey KL. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J. Mol. Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Silva GH, Belfort M. Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI. Nucleic Acids Res. 2004;32:3156–3168. doi: 10.1093/nar/gkh618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 21.Steuer S, Pingoud V, Pingoud A, Wende W. Chimeras of the homing endonuclease PI-SceI and the homologous Candida tropicalis intein: a study to explore the possibility of exchanging DNA-binding modules to obtain highly specific endonucleases with altered specificity. Chembiochem. 2004;5:206–213. doi: 10.1002/cbic.200300718. [DOI] [PubMed] [Google Scholar]
- 22.Fajardo-Sanchez E, Stricher F, Paques F, Isalan M, Serrano L. Computer design of obligate heterodimer meganucleases allows efficient cutting of custom DNA sequences. Nucleic Acids Res. 2008;36:2163–2173. doi: 10.1093/nar/gkn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scalley-Kim M, Minard P, Baker D. Low free energy cost of very long loop insertions in proteins. Protein Sci. 2003;12:197–206. doi: 10.1110/ps.0232003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelton JT, McLean LR. Spectroscopic methods for analysis of protein secondary structure. Anal. Biochem. 2000;277:167–176. doi: 10.1006/abio.1999.4320. [DOI] [PubMed] [Google Scholar]
- 25.Sparks DL, Lund-Katz S, Phillips MC. The charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. J. Biol. Chem. 1992;267:25839–25847. [PubMed] [Google Scholar]
- 26.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seligman LM, Stephens KM, Savage JH, Monnat RJ., Jr Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 30.Gimble FS. Broken symmetry in homing endonucleases. Structure. 2006;14:804–806. doi: 10.1016/j.str.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Eastberg JH, McConnell Smith A, Zhao L, Ashworth J, Shen BW, Stoddard BL. Thermodynamics of DNA target site recognition by homing endonucleases. Nucleic Acids Res. 2007;35:7209–7221. doi: 10.1093/nar/gkm867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 34.Ahmad S, Kono H, Arauzo-Bravo MJ, Sarai A. ReadOut: structure-based calculation of direct and indirect readout energies and specificities for protein–DNA recognition. Nucleic Acids Res. 2006;34:W124–W127. doi: 10.1093/nar/gkl104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.