Abstract
The mitochondrial genome of Chlamydomonas reinhardtii only encodes three expressed tRNA genes, thus most mitochondrial tRNAs are likely imported. The sharing of tRNAs between chloroplasts and mitochondria has been speculated in this organism. We first demonstrate that no plastidial tRNA is present in mitochondria and that the mitochondrial translation mainly relies on the import of nucleus-encoded tRNA species. Then, using northern analysis, we show that the extent of mitochondrial localization for the 49 tRNA isoacceptor families encoded by the C. reinhardtii nuclear genome is highly variable. Until now the reasons for such variability were unknown. By comparing cytosolic and mitochondrial codon usage with the sub-cellular distribution of tRNAs, we provide unprecedented evidence that the steady-state level of a mitochondrial tRNA is linked not only to the frequency of the cognate codon in mitochondria but also to its frequency in the cytosol, then allowing optimal mitochondrial translation.
INTRODUCTION
To compensate the evolutionary loss of mitochondrial tRNA genes, import of nucleus-encoded tRNAs into mitochondria occurs in all eukaryotic kingdoms (1). An intriguing aspect of tRNA import concerns the proportion of nucleus-encoded tRNA found in mitochondria. It has long been believed that only a few percent of a cytosolic tRNA is present in mitochondria (2). However, some evidence supports the idea that a differential distribution of nucleus-encoded tRNAs between the cytosol and mitochondria does exist. Indeed, in Leishmania tarentolae, some tRNAs can be classified as mainly cytosolic and others as mainly mitochondrial (3,4). In wheat mitochondria tRNALeuUAA was shown to be in higher amount in mitochondria than in the cytosol (5). So far, such a difference in the cytosolic and mitochondrial localization of tRNAs has not been carefully studied, as well as the basis for that.
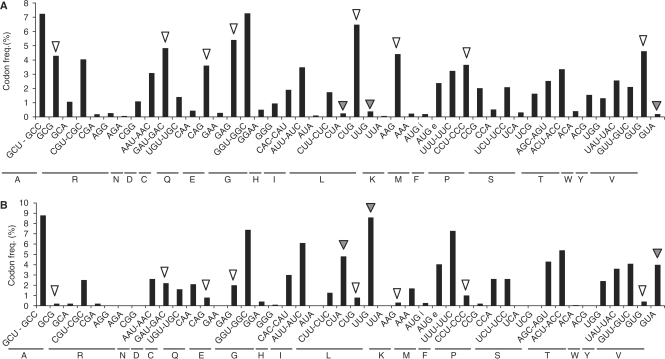
The mitochondrial genome of Chlamydomonas reinhardtii only encodes three tRNAs (6). The idea that, in this organism, plastids can provide tRNAs to mitochondria has been put forward (7,8). Here, we first show that no plastidial tRNA is imported into Chlamydomonas mitochondria and that most of the mitochondrial tRNAs are nucleus-encoded and imported from the cytosol. As both cytosolic and mitochondrial codon usages are highly and differentially biased (6,9) (Figure 1), we thought that this alga was the appropriate model to answer the following question: is the efficiency of the nucleus-encoded tRNA import into mitochondria adapted to the need of the mitochondrial translation machinery? Here the sub-localization of the tRNAs representing the 49 isoacceptor families encoded by the C. reinhardtii nuclear genome (9) and their extent of import into mitochondria were analyzed. We propose that the mitochondrial localization of tRNA isoacceptors is tightly linked to both the nuclear and the mitochondrial codon usages.
Figure 1.
Histograms for codon frequency in C. reinhardtii. (A) Cytosolic codon usage. (B) Mitochondrial codon usage. For (A) and (B) codons have been grouped according to classical wobble rules and aminoacids are represented with the one-letter code. White (high frequency in the cytosol but weak in the mitochondria) and grey (low frequency in the cytosol but high in the mitochondria) arrow heads indicate some codons differentially used in the cytosol and mitochondria.
MATERIALS AND METHODS
Purification of mitochondria and chloroplasts
Crude mitochondrial fraction was isolated from the cell-wall-less mutant cw15 mt+ by digitonin treatment according to (10). The mitochondrial fraction was then loaded on a discontinuous Percoll gradient (13%/21%/45%). Purified mitochondria were recovered at the 45/21 interface and washed two times in MET buffer (Mannitol 280 mM, Tris–HCl pH 7 10 mM, EDTA 0.5 mM, 0.1% BSA) by 10 min centrifugation at 11 000 g. Chloroplast fraction was recovered according to ref. (11).
Total, mitochondrial and plastidial RNA isolation
Total tRNA extract was prepared from whole cells as described in ref. (12). For mitochondrial and plastidial RNA preparation, the same method was used, but because of the low complexity of the RNA population, the LiCl precipitation step was omitted.
Northern analysis and quantitation of tRNA import
Hybridization of oligonucleotides (Supplementary data, Table S1) to northern blots of total, mitochondrial or plastidial RNAs from C. reinhardtii were performed as in ref. (9). Signals obtained on a phosphorimager were quantified using the MacBas software. For quantitation, RNA fractions were analyzed by northern blots such that hybridization signal obtained with each specific probe was linear with respect to the amount of RNA loaded on the gel. To determine the percentage of a given tRNA localized in mitochondria, we first determined the fraction of the three mitochondria-encoded tRNAs present in total (i.e. whole cell) tRNA fraction. Hybridization of 3 µg of total or mitochondrial tRNA fractions with probes directed against the three mitochondria-encoded tRNAs (Figure 2B) have shown that 4% of the whole-cell fraction is mitochondrial. In other words, 3 µg of mitochondrial tRNAs are extracted from 25 times more cells than 3 µg of total tRNAs. This value allowed us to normalize the signal for a given tRNA in total and mitochondrial tRNA fractions with respect to the same number of cells. Then, the extent of mitochondrial localization was estimated by calculating the percentage of a given tRNA in the mitochondrial fraction relative to the abundance of that RNA in the whole cell fraction as follow: 100× (Phosphorimager units in mitochondrial fraction)/(Phosphorimager units in total fraction × 25) = 4× (Phosphorimager units in mitochondrial fraction)/(Phosphorimager units in total fraction). For tRNAAla isoacceptors hybridizing to the same oligonucleotides (Table 1), RT–PCR followed by sequence analysis of clones allowed us to calculate the percentage of each isoacceptor among the whole-cell and mitochondrial tRNA populations.
Figure 2.
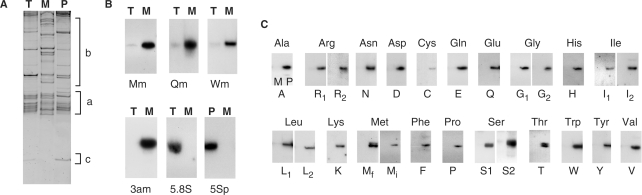
Mitochondria of C. reinhardtii do not import plastidial tRNAs. (A) Whole-cell (T), mitochondrial (M) and plastidial (P) RNA fractions separated on polyacrylamide gel and stained with ethidium bromide. Regions corresponding to tRNAs (a), rRNAs (b) and plastidial 3S rRNA (c) are indicated. (B) Hybridization of probes specific to mitochondria-encoded tRNAMet (Mm), tRNAGln (Qm) and tRNATrp (Wm) and to mitochondrial 3a rRNA (3am), cytosolic 5.8S rRNA (5.8S), plastidial 5S rRNA (5Sp) to northern blots of whole-cell (T), mitochondrial (M) or plastidial (P) RNAs. (C) Hybridization of probes specific to plastidial tRNA genes to northern blots of mitochondrial (M) and plastidial (P) RNAs. Name of probes is given below each northern experiment (see Supplementary data Table S1 online).
Table 1.
Steady-state levels of imported tRNAs in mitochondria relative to their abundance in the cytosol in Chlamydomonas reinhardtii
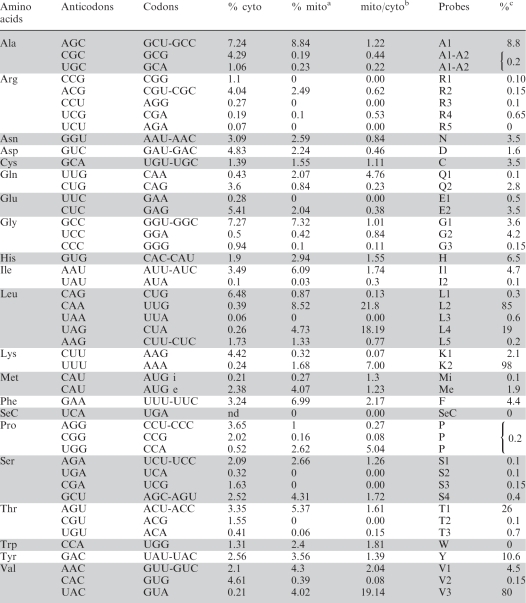 |
Nuclear tRNA gene content (identified by anticodons), minimal potential codon recognition pattern (codons), cytosolic codon usage (% cyto) and name of probes are given as in (9), (see also supplementary data, Table S1 online).
aMitochondrial codon usage (% mito) is according to (6).
bRatio codon frequency (mito/cyto).
cThe steady-state levels of imported tRNAs relative to their abundance in the cytosol are given in percentage.
Mean values are given (see legend of Figure 3 for detailed explanations).
Cloning and sequencing of RT–PCR products
Transfer RNAs purified on 15% denaturing polyacrylamide gel (12) were used as substrate for RT–PCR amplification and cloned into PCR-2-TOPO (Invitrogen).
Misceallenous
All available sequence data for C. reinhardtii used in this paper were previously described (9,13,14). This information was used to design oligonucleotide probes specific for the corresponding rRNAs and tRNAs (Supplementary data, Table S1).
RESULTS
The three tRNA genes encoded by the mitochondrial genome are expressed
As a first step towards the analysis of the C. reinhardtii mitochondrial tRNA set, mitochondrial RNA was extracted. This RNA fraction (Figure 2A) mainly contains a dozen of RNA fragments corresponding to ribosomal RNA (rRNA) pieces (13) and tRNAs. The mitochondrial tRNA profile was different to the profile observed with whole cell or plastidial tRNA fractions. Northern blots (Figure 2B) indicate that the three mitochondrial tRNA genes are expressed. Whereas a probe directed against the mitochondrial L3a rRNA [one of the 10 rRNA fragments constituting the large rRNA; (13)] gave a strong signal, almost no cytosolic 5.8S rRNA or plastidial 5S rRNA (<0.15%) was detected in the mitochondrial fraction. Thus this fraction is not significantly contaminated either by cytosolic or by plastidial RNAs and can be considered as a true mitochondrial fraction.
Plastidial tRNAs are not present in C. reinhardtii mitochondria
With only three mitochondria-encoded tRNAs, tRNA import is a prerequisite for the Chlamydomonas mitochondrial protein synthesis. While the import of nucleus-encoded tRNAs into mitochondria is a wide spread phenomenon (1), previous genetic studies suggest that, in this alga, chloroplasts could provide mitochondria with tRNAs (7). To address the question of the nuclear and/or plastidial origin of the imported tRNAs, we first performed northern experiments with probes specific to each plastidial tRNA (Supplementary data, Table S1). For each probe, a signal was detected in the plastidial RNA fraction whereas no signal was obtained in the mitochondrial one (Figure 2C), thus demonstrating that the mitochondrial translation machinery does not rely on the import of plastidial tRNAs.
The steady-state levels of imported tRNAs found in mitochondria are highly variable
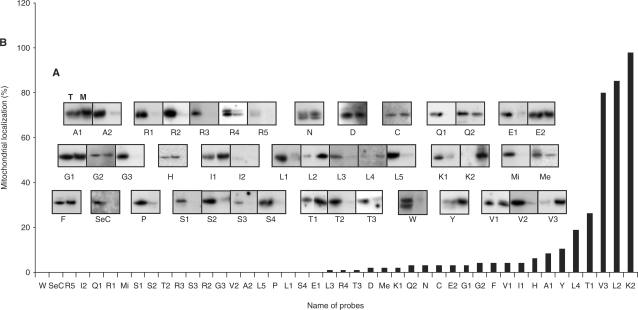
The above data suggest that the mitochondrial tRNA population mainly consists of nucleus-encoded tRNA species. Northern blots containing known amounts of the whole-cell and mitochondrial tRNA fractions hybridized with oligonucleotide probes specific for each cytosolic tRNA (Supplementary data, Table S1) allowed us to determine their sub-cellular distribution (Figure 3A). For each tRNA, we calculated the ratio between its steady-state level in mitochondria and in the whole-cell fraction (Figure 3B and Table 1). The most striking aspect of this analysis is the high variation of the partitioning among the tRNAs (from 0% to 98%). The following conclusions can be drawn.
If we assume that a percentage less than or equal to 0.15% (the maximal level of 5.8S rRNA cytosolic marker estimated in the mitochondrial fraction) is most likely the result of cytosolic contamination, then 15 tRNAs can be considered as cytosol-specific.
For 31 tRNAs, the extent of mitochondrial localization ranges from 0.2% to 26%. This reflects the important fluctuation of the level of nucleus-encoded tRNAs present in mitochondria.
For three tRNAs, the percentage is equal or above 80%. Up to now, in all organisms in which tRNA import has been studied, no nucleus-encoded tRNA has been localized exclusively in mitochondria. With an import level of 98%, tRNALysUUU can be considered as the first example of such a tRNA.
Comparing tRNA isoacceptors, the extent of mitochondrial localization is highly variable. For example, tRNALeuCAG and tRNAValCAC are mainly cytosol-specific whereas tRNALeuCAA and tRNAValUAC are mainly localized in mitochondria. No correlation between the amount of a given tRNA in the cytosol and the extent of its mitochondrial localization was found.
Figure 3.
Expression and sub-localization of the nucleus-encoded tRNAs of C. reinhardtii. (A) Whole-cell RNA fraction (T) and mitochondrial RNA fraction (M) separated on polyacrylamide gel and hybridized against probes specific to cytosolic tRNA genes (Table 1 and Supplementary data, Table S1). Name of probes is given below each northern experiment. (B) Extent of mitochondrial import for each cytosolic tRNA in percentage. For each tRNA, two to four independent northern blot experiments were performed. Three and four independent total and mitochondrial RNA preparations were used for the whole set of northern experiments. For each probe, a typical experiment is presented in (A) but the percentages of extent of mitochondrial import given in (B) are the mean values obtained with two to four independent northern blot experiments. Error bars are not presented on the histogram because, at this scale, they will not be visible for most tRNAs. The standard deviations observed are usually less than 5%. Only for two probes, Y and Me, a higher deviation of around 10% was observed.
A substantial variation of the distribution of cytosolic tRNAs in mitochondria was described in wheat (5) or Leishmania (3,4), but such a detailed analysis on the important variability of the mitochondrial localization of nucleus-encoded tRNAs was never reported. We then addressed the question of the reasons for such a differential import.
Biological significance for the differential import of cytosolic tRNAs into mitochondria
The three mitochondria-encoded tRNATrp, tRNAGlnUUG and tRNAMet genes are expressed in Chlamydomonas mitochondria. As expected, cytosolic tRNATrp and tRNAGlnUUG are not in the organelle. In contrast, the second cytosolic tRNAGln isoacceptor, tRNAGlnCUG, is imported into mitochondria. The mitochondria-encoded tRNAMet gene encodes an elongator tRNAMet-e. An initiator tRNAMet-i and an elongator tRNAMet-e are expressed from C. reinhardtii nuclear DNA (9). Surprisingly, cytosolic tRNAMet-i is not brought into mitochondria whereas cytosolic tRNAMet-e is.
In Chlamydomonas mitochondria, the codon distribution is highly biased: seven codons occur very infrequently and nine codons are not used (Figure 1) (6). In the cytosol of Chlamydomonas, selenocysteine is inserted into selenoproteins by a tRNASec that recognizes the UGA stop codon (15). In Chlamydomonas mitochondria, there is no UGA stop codon and no selenoproteins, and, as expected, tRNASec remains in the cytosol. The other unused codons are recognized by tRNALeuUAA, tRNASerUGA, tRNASerCGA, tRNAThrCGU, tRNAGluUUC, tRNAArgCCG, tRNAArgCCU and tRNAArgUCU. These tRNAs, a priori not required by the mitochondrial translation machinery, are either cytosol-specific or very inefficiently addressed to the organelles.
In the mitochondrial genome, 77 Arg codons out of 80 are CGU and CGC. These codons must be read, according to wobble rules, by the cytosolic tRNAArgACG, assuming that the A is post-transcriptionaly modified in inosine. Surprisingly, the tRNAArgACG is mostly exclusively in the cytosol. In mammalian mitochondria, tRNAArgUCG is sufficient for reading all four CGN codons (16). Whether the tRNAArgUCG present in Chlamydomonas mitochondria is sufficient for reading all arginine codons will need to be addressed. Similarly, two tRNA isoacceptor families (tRNAPro and tRNASer) gave unexpected data: the amount of imported tRNA was very low whatever the isoacceptor tRNA considered. In order to assess this result, probes covering other regions of tRNAPro and tRNASer isoacceptors were used but gave similar results (data not shown).
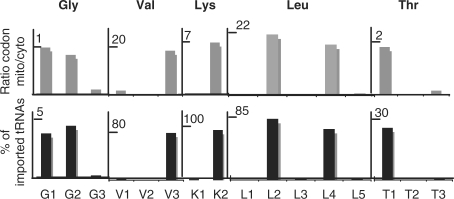
Among the other tRNA isoacceptor families, the extent of mitochondrial localization is highly variable and there is no correlation between the abundance of a given tRNA in the cytosol and the extent of its mitochondrial localization. For some of them, it was just the opposite: although a tRNA was present in large amount in the cytosol (e.g. tRNALeuCAG or tRNAValCAC), the level of imported tRNA in mitochondria was very low whereas tRNAs present at low concentration in the cytosol (e.g. tRNALeuCAA or tRNAValUAC) could be highly found in mitochondria. For some tRNA isoacceptors, the preferential mitochondrial localization is clearly linked to the high frequency of the corresponding codon in the organelle (e.g. tRNALeuCAA). However, the level of different imported tRNAs relative to their abundance in the whole-cell fraction can be within the same range of magnitude although the frequency of the mitochondrial codons they can recognize greatly differs (e.g. tRNAGlyGCC versus tRNAGlyUCC). Looking for other reasons to explain that, the ratio mitochondrial codon frequency/cytosolic codon frequency was calculated for codons read by tRNAGly, tRNALeu, tRNALys, tRNAThr and tRNAVal isoacceptors (Figure 4A). A strong link between the extent of their mitochondrial localization and the ratio was observed. When this ratio is high, meaning that the corresponding codons are frequently used in mitochondria as compared to the cytosol, then the percentage of imported tRNAs is high. In contrast, this percentage is low when the corresponding codons are rare in mitochondria but often used in the cytosol. Thus, the level of imported tRNAs is linked not only to the mitochondrial codon usage, but also to the cytosolic one.
Figure 4.
Link between codon usages and the steady-state levels of imported tRNAs. Histograms (in grey) showing the ratios mitochondrial codon frequency/cytosolic codon frequency (mito/cyto) for five aminoacids (Gly, Val, Lys, Leu, Thr) and histograms (in black) showing the percentages of the steady-state levels of imported tRNA isoacceptors into mitochondria relative to their abundance in the whole-cell fraction. Name of tRNAs is as in Figure 3.
DISCUSSION
The Chlamydomonas mitochondrial translation machinery does not rely on the import of plastidial tRNAs and depends on the import of nucleus-encoded tRNAs. Out of 49, 31 cytosolic tRNA isoacceptors (representing all amino acids but one, tryptophan) were found in mitochondria. This process is quite specific and a few nucleus-encoded tRNAs not required by the mitochondrial translation machinery are not or not significantly detected in Chlamydomonas mitochondria. These data are in agreement with what was observed so far: only essential nucleus-encoded tRNAs are present within mitochondria (1).
However, a few interesting discrepancies are worth to be discussed. In higher plant mitochondria, a single tRNAGln, with a UUG anticodon, is sufficient to decode the two CAA and CAG glutamine codons (16). The presence of two isoaccepting tRNAGln (with the CUG and UUG anticodons) in Chlamydomonas mitochondria thus raises the question of their apparent redundancy in term of codon recognition. The most likely explanation is that the mitochondria-encoded tRNAGlnUUG contains a modified U in the ‘wobble’ position so as to prohibit the recognition of the CAG codon.
Concerning tRNAMet, there is, on one hand, an apparent redundancy of two potential tRNAMet-e and, on the other hand, an apparent lack of a tRNAMet-i in Chlamydomonas mitochondria. Bacterial and organellar tRNAMet-i are only functional when carrying a formylated methionine. The formyl group is added to the methionine of the charged tRNAMet-i by a methionyl–tRNAMet formyltransferase. Such an enzyme is likely present in Chlamydomonas mitochondria (Supplementary data, Figure S1) suggesting that one out of the two tRNAMet-e is formylated within the organelle. A similar situation was found in Trypanosoma brucei where no mitochondrial tRNAMet genes exist. In this organism, whereas the eukaryotic tRNAMet-i remains exclusively in the cytosol, a fraction of the cytosolic tRNAMet-e becomes formylated after import into mitochondria and is then used as initiator (17).
According to the mitochondrial codon usage, the CCA proline codon and the UCY and AGY serine codons are frequently used but the corresponding nucleus-encoded tRNAs were not found in the organelle. Obviously, Chlamydomonas mitochondrial translation requires a minimal set of one tRNAPro and two tRNASer but these tRNAs remain to be identified. Four possibilities may be hypothesized to explain these results. First, we cannot exclude that our northern experiments failed to detect these tRNAs. However, the use of several independent probes for the same tRNA isoacceptor makes this hypothesis unlikely to occur. Second, we could hypothesize that the same set of tRNAs is not always present in the organelle because Chlamydomonas mitochondrial genes would not be expressed at the same time or would not use the same codon usage. This hypothesis is not correct because, with the exception of the reverse-transcriptase like gene, the seven other mitochondrial genes encode proteins of respiratory complexes which must be expressed at the same time. Furthermore, the codon usage pattern of the eight mitochondrial genes is mostly identical (6). Third, a single copy gene can be sufficient to produce a tRNA mostly localized in mitochondria [e.g. tRNAVal UAC, tRNALysAAA; (9)]. Thus, we cannot exclude that some nuclear tRNA genes, giving raise to tRNAs mostly present in mitochondria, may have escaped detection. In this context, it is worth to note that very recent findings such as the existence of permuted tRNA genes, split tRNA genes or tRNA genes with multiple introns demonstrate that tRNA genes may easily escape detection (18). Finally, as shown in Leishmania (19) editing events modifying the anticodon of imported tRNAs also represent a possibility to explain this discrepancy, although editing has not yet been demonstrated in Chlamydomonas mitochondria. To clarify the situation, the precise characterization of the tRNASer and tRNAPro species used during mitochondrial translation in Chlamydomonas will need to be assessed. In this respect, fractionation on 2D-gel of a large amount of Chlamydomonas mitochondrial tRNA extract will provide material to identify by aminoacylation tests the tRNAs of interest.
Up to now, only tRNAs of eukaryotic origin were shown to be imported into mitochondria. The presence of nucleus-encoded tRNAs mainly localized in mitochondria raises the question of their origin. Do they correspond to prokaryotic tRNA genes that were once inserted into the nuclear genome and are now expressed and addressed back to the organelle? Phylogenetic trees show that these tRNAs are not related to prokaryotic tRNAs but to eukaryotic ones (9). Thus, in course of evolution, a very few nuclear tRNA genes of eukaryotic origin have been recruited exclusively to provide mitochondria with essential tRNAs.
In Chlamydomonas, the cytosolic tRNA abundance is adjusted to the codon usage (9). Here, in most cases, we found a good correlation between the set of nucleus-encoded tRNAs present in mitochondria and the mitochondrial codon usage. This reveals an unprecedented degree of adjustment of the tRNA populations not only within the cytosol but also within the mitochondria of this green alga. In the trypanosomatid L. tarentolae preferential import of some tRNAs has been observed (3,4) but whether the abundance of the imported tRNAs is correlated to the frequency of the cognate codons in this organism has not been studied yet. In another trypanosomatid, T. brucei, the extent of mitochondrial localization varies from 1 to 7.5% for the 15 nucleus-encoded tRNAs studied but, contrary to what we observed in Chlamydomonas, no correlation between the differential import efficiencies and the mitochondrial codon usage was found (2). Another piece of evidence suggesting that the tRNA mitochondrial import pathway is regulated comes from studies on the S. cerevisiae mitochondrial tRNALys. Import of this tRNA is essential under stress conditions (20). It is probably too early to draw conclusions but we can wonder whether the level of regulation of tRNA mitochondrial import depends on the mechanism involved and on the organism considered. In Chlamydomonas, with regards to the mitochondrial localization of tRNA, the system seems to be able to adapt relatively easily not only by acquiring the possibility to bring new cytosolic tRNA species into mitochondria depending on the set of mitochondrial tRNA genes, but also by regulating the abundance of each tRNA so as to optimize mitochondrial translation. The steady-state level of a nucleus-encoded tRNA in mitochondria is mainly determined by two parameters: its import efficiency and its rate of degradation in the mitochondria. The question of mitochondrial tRNA stability has not often been addressed. Whereas cytosolic tRNAGly isoacceptors are differentially localized in vivo in dicotyledonous plant mitochondria, no difference in their stability has been seen in vitro in potato mitochondria (21). In T. brucei, no significant differences in the kinetics of degradation of several tRNAs were observed (2). Thus, it seems that the abundance of a nucleus-encoded tRNA in mitochondria depends on its import efficiency rather than on its degradation rate once inside the mitochondrial matrix. A strong correlation between the abundance of the nucleus-encoded tRNAs and codon usage is observed in Chlamydomonas mitochondria, and we now need to understand how such a correlation can be established. Two major hypotheses, which are not exclusive, can be proposed. First, this correlation can be a result of co-evolution between the mitochondrial genome and tRNA import processes. As previously noted, in Chlamydomonas, during evolution, the number of tRNA genes has been adapted so as to adjust the cytosolic tRNA abundance to the codon usage (9). It is also worth to mention that, in higher plants, the set of nucleus-encoded tRNA species found in mitochondria is complementary to the set of the mitochondria-encoded tRNA species and both the mitochondria-encoded and nucleus-encoded tRNA populations vary in parallel in different species and maintain the complementarity between the two tRNA sets (22). These variations are likely to be the result of selective pressure during evolution. In the same line, we can speculate that the steady-state levels of imported tRNAs in Chlamydomonas mitochondria result from evolution. A second equally attractive explanation which also should be explored, is that the mitochondrial genome codes for factors that regulate tRNA import. In the latter case, fine-tuning of tRNA import would be necessary to rapidly and efficiently adapt the tRNA population to the needs of Chlamydomonas mitochondria. Such regulation requires feed back control from the mitochondria to the cytosol by unknown factors and has not been observed so far. Whether tRNAs themselves or other factors play an important role, and if so how, is a crucial question to be solved. The availability of efficient mitochondrial transformation in Chlamydomonas (23) will provide an invaluable tool for manipulating the mitochondrial tRNA gene content or the codon usage in this organelle. Looking for the modification of the balance of nucleus-encoded tRNAs between the cytosol and the mitochondria will then allow us to determine if the information residing in the mitochondrial genome can regulate tRNA import efficiency and/or if it is the result of a co-evolution between the mitochondrial genome and tRNA import process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the French Centre National de la Recherche Scientifique, the Belgian Fonds de la Recherche Fondamentale et Collective and Fonds National de la Recherche Scientifique (grants 2.4582.05 and 1.5.255.08 to C.R.), a joint France-Wallonie Tournesol Grant (to L.M.D. and C.R.), a short-term EMBO fellowship (to E.V.) and a long-term Marie Curie fellowship (to T.S.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Salinas T, Duchêne AM, Maréchal-Drouard L. Recent advances in tRNA mitochondrial import. Trends. Biochem. Sci. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Tan TH, Pach R, Crausaz A, Ivens A, Schneider A. tRNAs in Trypanosoma brucei: genomic organization, expression, and mitochondrial import. Mol. Cell Biol. 2002;22:3707–3717. doi: 10.1128/MCB.22.11.3707-3716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X, Chen DTC, Suyama Y. A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear-encoded tRNAs between the cytosol and mitochondria. Mol. Biochem. Parasitol. 1994;65:23–37. doi: 10.1016/0166-6851(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 4.Kapushoc ST, Alfonzo JD, Simpson L. Differential localization of nuclear-encoded tRNAs between the cytosol and mitochondrion in Leishmania tarentolae. RNA. 2002;8:57–68. doi: 10.1017/s1355838202012281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glover KE, Spencer DF, Gray MW. Identification and structural characterization of nucleus-encoded transfer RNAs imported into wheat mitochondria. J. Biol. Chem. 2001;276:639–648. doi: 10.1074/jbc.M007708200. [DOI] [PubMed] [Google Scholar]
- 6.Michaelis G, Vahrenholtz C, Pratje E. Mitochondrial DNA of Chlamydomonas reinhardtii: the gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol. Gen. Genet. 1990;223:211–216. doi: 10.1007/BF00265056. [DOI] [PubMed] [Google Scholar]
- 7.Bennoun P, Delosme M. Chloroplast suppressors that act on a mitochondrial mutation in Chlamydomonas reinhardtii. Mol. Gen. Genet. 1999;262:85–89. doi: 10.1007/s004380051062. [DOI] [PubMed] [Google Scholar]
- 8.Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Cognat V, Deragon JM, Vinogradova E, Salinas T, Remacle C, Marechal-Drouard L. On the evolution and expression of Chlamydomonas reinhardtii nucleus-encoded transfer RNA genes. Genetics. 2008;179:113–123. doi: 10.1534/genetics.107.085688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardol P, Matagne RF, Remacle C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J. Mol. Biol. 2002;319:1211–1221. doi: 10.1016/S0022-2836(02)00407-2. [DOI] [PubMed] [Google Scholar]
- 11.Klein U, Chen C, Gibbs M, Platt-Aloia KA. Cellular Fractionation of Chlamydomonas reinhardii with Emphasis on the Isolation of the Chloroplast. Plant Physiol. 1983;72:481–487. doi: 10.1104/pp.72.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maréchal-Drouard L, Small I, Weil J-H, Dietrich A. Transfer RNA import into plant mitochondria. In: Attardi GM, Chomym A, editors. Mitochondrial Biogenesis and Genetics. Vol. 260. Inc, York: Spectrum Publisher Services; 1995. [Google Scholar]
- 13.Boer PH, Gray MW. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell. 1988;55:399–411. doi: 10.1016/0092-8674(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 14.Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002;14:2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao M, Carlson BA, Novoselov SV, Weeks DP, Gladyshev VN, Hatfield DL. Chlamydomonas reinhardtii selenocysteine tRNA[Ser]Sec. RNA. 2003;9:923–930. doi: 10.1261/rna.5510503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maréchal-Drouard L, Dietrich A, Weil JH. Adaptation of tRNA population to codon usage in cellular organelles. In: Hatfield D, Lee BJ, Pirtle RM, editors. Transfer RNA in Protein Synthesis. Boca Raton: CRC Press; 1992. pp. 125–140. [Google Scholar]
- 17.Tan THP, Bochud-Allemann N, Schneider A. Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc. Natl Acad. Sci. USA. 2002;99:1152–1157. doi: 10.1073/pnas.022522299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randau L, Soll D. Transfer RNA genes in pieces. EMBO Rep. 2008;9:623–628. doi: 10.1038/embor.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alfonzo JD, Blanc V, Estevez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol. Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Salinas T, Schaeffer C, Maréchal-Drouard L, Duchene AM. Sequence dependence of tRNA(Gly) import into tobacco mitochondria. Biochimie. 2005;87:863–872. doi: 10.1016/j.biochi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R, Maréchal-Drouard L, Akama K, Small I. Striking differences in mitochondrial tRNA import between plant species. Mol. Gen. Genet. 1996;252:404–411. doi: 10.1007/BF02173005. [DOI] [PubMed] [Google Scholar]
- 23.Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl Acad. Sci. USA. 2006;103:4771–4776. doi: 10.1073/pnas.0509501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.