Abstract
Missense point mutations in the TP53 gene are frequent genetic alterations in human tumor tissue and cell lines derived thereof. Mutant p53 (mutp53) proteins have lost sequence-specific DNA binding, but have retained the ability to interact in a structure-selective manner with non-B DNA and to act as regulators of transcription. To identify functional binding sites of mutp53, we established a small library of genomic sequences bound by p53R273H in U251 human glioblastoma cells using chromatin immunoprecipitation (ChIP). Mutp53 binding to isolated DNA fragments confirmed the specificity of the ChIP. The mutp53 bound DNA sequences are rich in repetitive DNA elements, which are dispersed over non-coding DNA regions. Stable down-regulation of mutp53 expression strongly suggested that mutp53 binding to genomic DNA is functional. We identified the PPARGC1A and FRMD5 genes as p53R273H targets regulated by binding to intronic and intra-genic sequences. We propose a model that attributes the oncogenic functions of mutp53 to its ability to interact with intronic and intergenic non-B DNA sequences and modulate gene transcription via re-organization of chromatin.
INTRODUCTION
Mutations in the TP53 gene represent the most frequent genetic alterations in a single gene in human cancer, ranging between different types of tumors from 10% to 70% (1). Unlike other tumor suppressor genes, which are functionally eliminated by deletions, truncations, nonsense mutations, or gene silencing, ∼80% of all TP53 alterations in human tumors are missense point mutations resulting in the expression of a functionally altered mutant p53 (mutp53) protein, with an exchange of a single amino acid. Most missense point mutations (∼80%) are localized in the central DNA-binding core domain (2,3). P53 binds as tetramer to symmetrical DNA target sequences (p53 response elements) in a sequence- and geometry-specific fashion (4). Experimental work (5) and molecular dynamics simulations (4) provided evidence that mutations in the core domain dramatically reduce the ability of mutp53 proteins for sequence-specific DNA binding; however geometry-specific DNA binding is retained and detectable under in vitro conditions (6). In tumor cells, mutp53 is able to regulate the expression of a large number of tumor-associated genes (7). Although several recent reports demonstrated that mutp53 can physically interact with promoter regions of target genes identified by microarray analyses (8), the molecular mechanism of the transcriptional activity exerted by mutp53 is still far from being understood. Two different, but not mutually exclusive possibilities are currently considered: (i) mutp53 interacts specifically with other transcription factors, such as Sp1 (9), Ets1 (10) and NF-Y (11), and modulates their activity; (ii) mutp53 has retained a residual transcriptional activity from wild-type (wt) p53 (12) and modulates transcription by interacting with components of the transcription machinery. This view is supported by the finding that the transcriptional competence of mutp53 requires intact N-terminal (13) and C-terminal (14) domains. These domains are rarely mutated in human tumor cells (2,3) and are crucial for interactions with proteins of transcription complexes (15,16). In both cases it is assumed that mutp53 acts within the frame of a standard mechanism encompassing reactions and interactions that take place on regulatory sequences surrounding a transcription start site (TSS). However, the highly complex transcriptional program of a cell is not only directed by binding of regulatory proteins to promoters. Additionally, multiple long-range interactions within the chromatin provide a platform for higher-order levels of gene regulation that complement the promoter-dependent mechanism by the possibility of integrating a single gene into a gene regulatory network (17). Locus control regions (LCR), enhancers, silencers and insulators (18–21) are examples for functional, promoter-distant sequence elements which are embedded into variably sized non-coding, intra- and inter-genic DNA regions. These regions also contain the heterogeneous family of conformation-flexible DNA sequences that represent S/MARs (Scaffold/Matrix Associated Regions) (22), characterized by their strong binding to a nuclear, salt and detergent-insoluble proteinaceous structure (nuclear matrix or scaffold) (23). Through co-operative interaction with nuclear matrix proteins and a wealth of regulatory proteins, S/MARs constitute nuclear matrix anchorage sites for topologically separate, DNase I-sensitive chromatin loops, and form organizing centers for transcription-, replication-, recombination- and repair-factories (24). S/MAR sequences exhibit a high content of repetitive DNA elements and adopt non-B DNA conformations under favoring conditions, for example under superhelical tension (25).
On the basis of the ability of mutp53 to bind in vitro DNA sequences capable to adopt non-B DNA conformations (6), and to interact with repetitive DNA sequences in vivo (26), we put forward the concept of mutp53 as a nuclear component involved in transcriptional events. This concept is strongly supported by the fact that mutp53 behaves like a S/MAR-binding protein (27). The murine p53D168G/M234I protein (MethA) was found to interact with DNA fragments from the murine immunoglobulin heavy chain gene enhancer locus and the interferon β gene locus, which exhibit properties of S/MARs, and the binding characteristics of MethA mutp53 to these DNA fragments were similar to that of S/MAR–DNA-binding proteins (28). Mutp53 proteins (MethA, and human p53R175H, p53R273H, p53R273P) bind to single-strand/double-strand DNA junctions and, by promoting further DNA-unwinding, generate single-stranded regions (29,30). Such a mechanism may contribute to the conformational dynamics of S/MAR sequences that lead to activation or repression of transcription.
So far, however, no genome-wide correlation between in vivo DNA-binding of mutp53 and its transcriptional activity has been shown. A prerequisite for establishing such a correlation is the identification of functional genomic mutp53-binding sites, in conjunction with gene expression analysis in the presence of mutp53 and after shutoff of mutp53 expression, e.g. by using an RNAi approach. In the present work we combined chromatin immunoprecipitation (ChIP) (31) with DNA cloning and sequencing (ChIP cloning) (32) to identify functional binding sites of mutp53 and provide data supporting the proposed model of mutp53 activity. As proof of principle we established a small library of genomic sequences bound by p53R273H in human U251 glioblastoma cells and confirmed the specific binding of mutp53 to isolated DNA fragments by electrophoretic mobility shift assay (EMSA). ChIP sequences, like S/MARs, were found to be rich in repetitive DNA sequences and dispersed over non-coding DNA regions. Supporting specificity of mutp53 binding, despite the small library size we found a tendency for a local enrichment of ChIP sequences at distinct chromosomal positions, specifically, close to genes whose expression was affected after manipulation of the mutp53 level. Thereby, our approach led to the identification of the PPARGC1A and FRMD5 genes as targets of mutp53. We propose that in U251 cells p53R273H exerts a regulatory effect on gene transcription by acting as DNA/chromatin associated factor that participates in the organization of chromatin into functional domains.
MATERIALS AND METHODS
Cell lines
The human glioblastoma cell line U251 was a kind gift from Dr Kazuo Washiyama (Brain Research Institute, Niigata University, Japan). Mutation (R273H) in the TP53 gene was confirmed by sequencing of PCR-amplified p53 cDNA (see Supplementary Data). Human non-small lung carcinoma cell line H1299 (NCI-H1299) was kindly provided by Dr Moshe Oren (The Weizmann Institute, Israel). The cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen GmbH, Karlsruhe, Germany) supplemented with 10% fetal calf serum (FCS; PAA Laboratories GmbH, Linz, Austria) in a humid atmosphere containing 5% CO2.
Stable transfection of shRNA expressing vectors
To obtain U251 derived clones with a stable reduction of p53 expression, 106 U251 cells were transfected either with 1.8 µg pSuper-p53/0.2 µg pCI-neo (Oligoengine), or 2 µg pENTR/H1-p53 using the NHDF Neo kit, program U20 (Amaxa AG, Cologne, Germany) and plated into three 10 cm dishes. As control for off-target effects of the shRNA, U251 cells were transfected with pENTR/H1-scr for the expression of scrambled shRNA. To generate the pENTR/H1-p53 and pENTR/H1-scr constructs the respective expression cassette [see (33) and Table S8 for sequences] were cloned into the vector backbone of pENTR/H1/TO (Invitrogen GmbH) according to the manufacturer's protocol. Forty-eight hours after transfection the cell culture medium was supplemented with 0.4 mg/ml G418 sulphate (PAA Laboratories GmbH) or 100 µg/ml Zeocin (Invitrogen GmbH). G418- and Zeocin-resistant clones were picked using cloning rings and propagated for further characterization. Integration of vectors into U251 genome was checked by PCR from genomic DNA using vector specific primers (see Table S8). Reduction of p53 expression in the established clones was measured by real-time qRT–PCR, immunoblotting and immunofluorescence staining. Expression of scrambled shRNA was verified by stem-loop RT–PCR (34) using specific primer set (see Table S8).
ChIP
Cells were cultured in three 15 cm diameter tissue culture dishes. At 80% confluence, cells were treated with DMEM supplemented with 1% formaldehyde (Sigma-Aldrich Chemie Gmbh, Munich, Germany) for 10 min at 37°C. Cells were then washed with warm PBS (pH 7.4) buffer, and crosslinking was stopped by adding 125 mM glycin in PBS. Treated cells were washed twice with cold PBS, once with KM buffer (10 mM HEPES, pH 7.5, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 5 mM dithiothreitol, 10% glycerol), and lysed for 30 min on ice in KM buffer supplemented with 1% Nonidet P-40 (NP-40) and protease inhibitors (5 µg/ml leupeptin (Sigma-Aldrich), 1 µg/ml pepstatin (Sigma-Aldrich), 1% Trasylol (Bayer), 0.1 mg/ml Pefabloc SC (Biomol GmbH, Hamburg, Germany)). After centrifugation at 900× g at 4°C, the supernatant was discarded and the pellet was slowly shaken with KM buffer adjusted to 2 M NaCl overnight at 4°C. Finally, the residual cell structures were harvested in TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA) and centrifuged at 15 000× g for 5 min. The cell pellet was resuspended in 300 µl TE buffer and sonicated for 10 min in a Bioruptor UCD-200 (Diagenode sa, Liege, Belgium) according to the manufacturer's instructions. Debris was eliminated by centrifugation for 20 min at 20 000× g at 4°C. Protein concentration was quantified by Bradford Assay (Bio-Rad Laboratories GmbH, Munich, Germany). DNA size was estimated after reverse crosslinking and DNA purification by agarose gel electrophoresis. Optional precipitation with 1% N-cetyl-N,N,N,-trimethylammonium bromide was performed (Cetavlon; Merck KGaA, Darmstadt, Germany) to remove non-crosslinked proteins from samples as previously described (32). For immunoprecipitation (IP), goat polyclonal (sc-6243; Santa Cruz Biotechnology, Heidelberg, Germany) or monoclonal (DO1 and Pab421) anti-p53 antibodies, goat polyclonal anti-lamin B antibody (sc-6217; Santa Cruz Biotechnology), and protein G-sepharose (GE Healthcare Europe GmbH, Munich, Germany) were used. IP was done in 500 µl reaction volume as described elsewhere (32). Sau3A (Fermentas GmbH, St Leon-Rot, Germany) digestion of chromatin was conducted during the capture of immunocomplexes on sepharose beads. Protein–DNA complexes were eluted with 250 µl elution buffer (0.5% SDS, 20 mM Tris–HCl, pH 8.0, 2 mM CaCl2) for 15 min at 65°C. Supernatants were collected and elution was repeated once more. To reverse protein–DNA crosslinks, samples (500 µl) were supplemented with 500 µg/ml Proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) and incubated at 50°C for 1 h, followed by 4 h at 65°C. DNA was extracted twice with phenol–chloroform–isoamylalcohol (25:24:1; Biomol GmbH), precipitated with ethanol in the presence of 10 µg glycogen (Roche Diagnostics GmbH), washed with 70% ethanol, air-dried and dissolved in water. Double-stranded Sau3A-adapters were prepared by annealing of 5′phosphorylated 24-mer and non-phosphorylated 20-mer oligonucleotides (see Table S8). Adapters were ligated to Sau3A-cleaved immunoprecipitated DNA using T4 DNA ligase (New England Biolabs GmbH, Frankfurt am Main, Germany). Adaptor-bearing DNA molecules were amplified by PCR using Sau3A-primer (see Table S8) under the following conditions: 2 min at 94°C, 29 cycles (94°C for 60 s, 55°C for 30 s, 72°C for 60 s) and 10 min at 72°C. PCR products were analyzed on a 1.5% agarose gel, purified, and either ligated into the cloning vector pCRII applying the TA cloning kit (Invitrogen GmbH) or cleaved with Sau3A and ligated into the BamHI site of the pBluescript II SK vector (Stratagene). Ligated DNA was transformed into the competent E. coli TOP10 (Invitrogen GmbH) or SURE (Stratagene) strain. Recombinant plasmid DNAs were purified using NucleoSpin®Plasmid (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany) and their inserts were sequenced by Eurofins MWG Operon (Ebersberg, Germany).
Sequence analysis
ChIP sequences were aligned with the human genome using the BLAT algorithm (35) implemented in the Genome browser (http://genome.ucsc.edu/index.html). The coordinates for the highest-scoring alignments were considered for further analysis. Randomization of chromosomal coordinates was done using random package within the R environment (http://www.r-project.org). The length of each single chromosome (NCBI Build 36.1; hg18) was used as a range for random numbers generation. Nucleotide sequences around random positions ±1000 nt were retrieved from the Genome browser database. Nucleotide sequences of promoters (2000 nt upstream of TSS) for randomly selected gene accession numbers were retrieved from the PromoSer database (http://cagt.bu.edu/page/Promoser_submit) (36). The content of repetitive sequences was analyzed using the RepeatMasker server (A.F.A. Smit, R. Hubley and P. Green; http://www.repeatmasker.org/). The coordinates of ChIP and random sequences were mapped onto the human chromosomes using the ChromoMapper program (http://mcluster.tu-graz.ac.at/clustercontrol/modules/ChromoMapper/) (37). S/MAR-specific and short oligomeric motifs were inspected using MAR-Wiz (http://www.futuresoft.org/MAR-Wiz) (38) and RSAT web-tools (http://rsat.ulb.ac.be/rsat/index.html) (39), respectively. P53RE was searched using PatSearch program (http://bighost.area.ba.cnr.it/BIG/PatSearch) (40).
Electrophoretic mobility shift assay (EMSA)
Recombinant human p53R273H was expressed in Escherichia coli strain C41 (DE3), derived from BL21 (DE3) using a two-step induction at 18°C to limit protein aggregation. p53R273H was purified according to a protocol described previously (41) with some minor modifications. In this study tetrameric form of p53 was used, obtained by a final purification step by gel filtration chromatography on Superdex HR200. Plasmid DNA was propagated in E. coli TOP10 and purified using the NucleoBond® PC EF kit (MACHEREY-NAGEL GmbH & Co. KG). Optionally, plasmid DNA was treated with the restriction enzyme PvuII (New England Biolabs GmbH) and purified. To perform EMSA, samples containing 200 ng of supercoiled plasmid DNA or PvuII-digested plasmid DNA were incubated with p53R273H protein at the indicated p53 tetramer/DNA molar ratios in binding buffer (5 mM Tris–HCl, pH 7.6, 0.01% Triton X-100, 0.5 mM EDTA, 0.5 mM DTT, 150 mM KCl) for 30 min on ice to reach equilibrium. After binding, samples were supplemented with gel loading buffer (5×: 40 mM Tris–HCl, pH 7.6, 10% glycerol, 0.025% bromophenol blue) and separated by electrophoresis on a 1% agarose gel (Bio-Rad) in 0.33× Tris/Borate/EDTA buffer for about 5 h at 4°C in an electric field of 5 V/cm. DNA was stained with ethidium bromide and visualized under UV light.
Immunofluorescence staining
Cells were plated in a six-well culture plate on glass coverslips at a density of 5 × 104 cells/well, fixed with 4% paraformaldehyde (PFA) in PBS and permeabilized with 1% Triton X-100 in PBS, pH 7.4. Optionally, cells were extracted with 1% Triton X-100 in PBS and then fixed with 4% PFA in PBS. After washing and blocking in 1% BSA/PBS, cells were stained with goat polyclonal anti-p53 (sc-6243; FL-393) antibodies alone or in combination with rabbit polyclonal anti-SATB2 (kindly provided by Dr Victor Tarabykin, MPI for Experimental Medicine, Göttingen, Germany), anti-Sp1 (07-645; Millipore GmbH, Schwalbach/Ts., Germany), anti-YY1 (sc-1703; Santa-Cruz), or anti-RNA polymerase II (sc-899; Santa-Cruz) antibodies. Donkey anti-goat and anti-rabbit fluorochrome-coupled antibodies were obtained from Invitrogen. The images were captured as Z-stacks using a Plan-Apochromat 63×/1.4 NA oil DIC objective and an Axiovert 200 microscope equipped with a LSM 510 META confocal scanner (Carl Zeiss MicroImaging GmbH, Jena, Germany). To acquire images of samples probed with two fluorophores and sequential scanning with DIC/transmitted light, multi-tracking settings were used. Following image acquisition, raw data was exported to the Huygens Essential software (version 2.7.2p0, Scientific Volume Imaging B.V., Hilversum, The Netherlands) and digital deconvolution was performed using the Maximum Likelihood Estimation (MLE) algorithm. The restored image data set was visualized and processed with the Imaris software package (version 4.1.3, Bitplane AG, Zürich, Switzerland). Co-localization was analyzed with the ImarisColoc module integrated into the Imaris software package, and a map of the co-localized voxels was saved as separate channel.
Immunoblotting
Proteins were extracted by washing the cells once with warm PBS and lysed directly in 200 µl (per six wells) of Laemmli buffer (62.5 mM Tris–HCl, pH 6.8, 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.1% bromphenol blue). Proteins were separated on SDS–polyacrylamide gels and electroblotted onto nitrocellulose membranes (GE Healthcare). To reverse protein–DNA crosslinks, samples were boiled for 30 min in Laemmli buffer prior electrophoresis. P53, lamin B, HSC70 and actin were detected with goat polyclonal anti-p53 (sc-6243), anti-lamin B (sc-6217), anti-HSC70 (sc-1059) and anti-actin (sc-1616) antibodies. HRP-conjugated donkey anti-goat antibody was obtained from Santa Cruz. Proteins were visualized using luminol-based ECL western blotting reagent (GE Healthcare).
Expression analysis
Total RNA was isolated applying the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality and integrity of the total RNA was evaluated with the 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany) and the concentration was measured using a NanoDrop spectrophotometer (Peqlab Biotechnologie GmBH, Erlangen, Germany). For microarray profiling on ABI human genome survey microarray version 1 chip, biological replicates (cells at different passages) were prepared. The ABI human genome survey microarray chip contained 32 878 60-mer oligonucleotide probes representing 29 098 known genes. Synthesis of cRNA, hybridization and image data processing was completed by the Core Facility at the Molecular Diagnostic Center of the University of Muenster according to the ABI standard protocol. Quantile normalization was done using the limma package within the R environment. A gene was considered to be differentially expressed between groups, if a Student's t-test statistic had an associated P ≤ 0.05 and a fold change (FC) ≥ 2. The microarray data sets discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession numbers: GSM351‱481, GSM351‱486, GSM351‱488 and GSM351‱490.
For qRT–PCR analysis, RNA was purified using the Trizol reagent (Invitrogen), and reverse transcribed with the High Capacity RT kit (Applied Biosystems) according to the manufacturer's protocol. PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) in a standard program (10 min 95°C; 15 s 95°C, 1 min 60°C; 40 cycles) running in an ABI 7500 Fast thermal cycler (Applied Biosystems). PCR reactions for each sample were repeated in triplicates. The integrity of the amplified products was confirmed by melting-curve analysis. PCR primers were selected from Primer Bank (http://pga.mgh.harvard.edu/primerbank/index.html). PCR efficiency was measured for each primer pair using serial dilution of cDNA. The housekeeping gene, HPRT1, was used as endogenous control. Relative quantitation of transcript levels with respect to the calibrator (U251 stably transfected with pENTR/H1-scr vector) was done based on 2−ΔΔCT algorithm (42).
RESULTS
Binding of mutp53 to chromatin/DNA in U251 cells
U251 cells express large amounts of p53R273H which mainly localizes to the nucleus (Figure 1A; shown in green). There it is excluded from nucleoli and partially co-localizes with the nuclear matrix protein SATB2 (43) (Figure 1A; shown in red; co-localization is shown in yellow representing 37% of red-green pixels). Extraction of non-fixed cells with the non-ionic detergent Triton X-100, which removes most membrane lipids and soluble proteins (44), significantly reduced the amount of nuclear p53R273H. However, a small fraction of mutp53 resisted extraction and remained bound to the detergent-insoluble structures composed of chromatin fibers and a nuclear proteinaceous scaffold, represented by the SATB2 protein (Figure 1B; 28% co-localization). The observed nuclear punctuate distribution of detergent-resistant mutp53 and its anchorage to the nuclear scaffold is reminiscent to that of many transcription factors and of RNA polymerases (45–47). Therefore, we tested for co-localization of p53R273H with the nuclear matrix-associated transcription factors Sp1 (48) and YY1 (49), both known as binding partners for wtp53 (50,51). Before and after Triton X-100 extraction a fraction of mutp53 co-localized with Sp1 and YY1 (Figure 1C–F; C—19%, D—6.5%, E—5.7%, F—2.1% co-localization). Also immunostaining with RNA polymerase II-specific antibodies revealed co-localization with the fraction of mutp53 in punctuate structures that resisted extraction with Triton X-100 (Figure 1G–H; G—7%, H—2.9% co-localization). We concluded that in U251 cells a fraction of p53R273H is bound to components of detergent-insoluble nuclear structures and thus could interact with genomic DNA.
Figure 1.
p53R273H is targeted to the nucleus in U251 glioblastoma cells. (A–H) Confocal images of U251 cells stained with anti-p53 (green) and anti-SATB2 (A, B), anti-Sp1 (C, D), anti-YY1 (E, F), and anti-RNA polymerase II (G, H) antibodies (red) according to a standard protocol (A, C, E, G), or extracted with Triton X-100 prior to PFA-fixation (B, D, F, H). Z-stack sections were deconvoluted using the Huygens software and the images were processed with the Imaris software. The co-localization channel (yellow) was generated using the ImarisColoc module. Scale bar—5 µm.
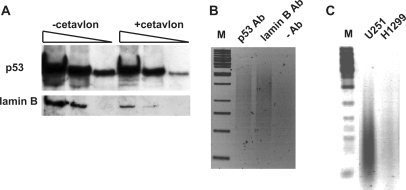
To test this conclusion, we performed ChIP after formaldehyde crosslinking. The membrane soluble crosslinking reagent formaldehyde produces covalent adducts of interacting proteins and nucleic acids (31) by forming reversible methylene bridges. In standard ChIP protocols, the samples are submitted to the IP reaction directly after crosslinking. However, the fraction of non-covalently bound, i.e. non-crosslinked chromatin proteins will interfere with the further procedure and lead to misinterpretation of the crosslinking efficiency. Therefore, we applied the cationic detergent Cetavlon, which specifically precipitates nucleic acids and covalently bound proteins, and has a strong dissociating effect on non-covalent protein–nucleic-acid complexes (see Materials and Methods section for details) (32). As shown in Figure 2A, mutp53 as well as the chromatin-associated protein lamin B (as a control for crosslinking) were detected in the Cetavlon-treated DNA–protein complexes, providing unequivocal evidence for the binding of mutp53 to chromatin in U251 cells. For the nucleotide sequence analysis of mutp53-crosslinked DNA, we optimized the ChIP protocol to specifically purify covalent mutp53-chromatin complexes and thereby maximize the specificity for LM–PCR mediated amplification of mutp53-bound DNA (Figure 2B). ChIP for lamin B, which has similar DNA-binding properties as mutp53 (52,53), served as further control for the suitability of the method. After LM–PCR of mutp53 bound DNA fragments, followed by agarose gel electrophoresis, a prominent broad band representing amplified ChIP fragments could be detected only in ChIP experiments performed with the corresponding antibodies. Three different p53 antibodies were used in our study. The specificity of the monoclonal antibodies DO1 and Pab421 has been already documented (54,55). To verify the specificity of the polyclonal goat anti-p53 antibody (FL-393; Santa Cruz), a sample from p53-null H1299 cells was processed together with a sample from U251 cells. No visible PCR-products were detected in the sample from H1299 cells (Figure 2C), thereby supporting the specificity of this p53 antibody.
Figure 2.
Formaldehyde-mediated crosslinking of p53R273H with genomic DNA. (A) Detection of mutp53 and lamin B by immunoblotting in protein–DNA complexes purified from formaldehyde-treated U251 cells. DNA-bound proteins were separated by SDS–PAGE either directly or after treatment with Cetavlon and blotted onto a nitrocellulose membrane. (B) Detection of DNA crosslinked with mutp53 and lamin B by formaldehyde via ChIP and LM–PCR. Amplified DNA was separated by electrophoresis in an agarose gel and stained with ethidium bromide. In the control experiment antibodies were omitted. (C) Demonstration of the specificity of the anti-p53 antibody (FL-393; Santa-Cruz). P53-deficient H1299 cells and U251 cells were treated with formaldehyde. Protein–DNA complexes were subjected to ChIP followed by LM–PCR and agarose gel electrophoresis. M—DNA size marker.
Analysis of genomic DNA fragments bound by mutp53 in vivo
Genomic DNA fragments co-precipitated with mutp53 (ChIP sequences) were amplified by LM–PCR, cloned and 135 randomly selected E. coli clones were picked for plasmid purification and sequencing. The total number of ChIP sequences submitted to bioinformatic analysis was 165 (see Tables S1 and S2), because 30 recombinant plasmids contained two genomic DNA fragments. The ChIP sequences were aligned with the human genome (hg18, 2006) using the Blat program (35); their coordinates are listed in Table S2. 143 ChIP sequences were matched to unique positions in the human genome. Two ChIP sequences, AA23 and UC24B, were mapped to duplicated genomic regions. The genomic position of 20 ChIP sequences could not be ascertained because they originated from highly repetitive sequence families. Of these, ChIP sequences UC15a and UC5 are derived from an intact copy of long interspersed nuclear element (LINE) and satellite II repeat DNA, respectively. Eighteen non-identical ChIP sequences (see Figure S1, Table S1) represented beta-satellite repeat-rich chromosomal regions, indicating a preferential binding of mutp53 to these moderately repetitive sequences. The beta-satellite sequences are composed of ∼69 bp monomeric units, repeated in long arrays up to 1 Mb in length and located on the short arm of acrocentric chromosomes (56) and on some other chromosomes (57). All ChIP sequences matching to unique positions were inspected for the presence of repetitive sequences using the RepeatMasker web-program. As the mutp53 cross-linked chromatin has been trimmed by the four-cutter restriction enzyme Sau3A, the flanking sequences were added to the ChIP sequences to increase the size of the inspected genomic region to 2000 nucleotides (see Table S3). The content of repetitive sequences was also determined in a set of random genomic fragments selected as described in Methods section (2000 nucleotides centered on randomly selected positions; see Table S4), and randomly selected promoter regions (2 kb upstream of the TSS; see Table S5) from the PromoSer database (36). As shown in Figure 3A, no significant differences were found between ChIP and random DNA datasets, whereas LINE1 sequences were significantly underrepresented in the promoter regions, and to a lesser extent also MaLRs, ERVLs and ERV-class I sequences.
Figure 3.
Content of repetitive DNA and S/MAR motifs in ChIP and random datasets. (A–B) Nucleotide sequences from the ChIP library, random genomic fragments (2000 nucleotides centered on random positions) and random promoter regions (2-kb upstream of TSS) were analyzed for the presence of repetitive elements and S/MAR motifs using RepeatMasker and Mar-Wiz web-tools, respectively. Although S/MARs are rather defined by functional tests than by sequence motifs, some sequence motifs can help to roughly identify the presence of S/MARs (38). The graphs show the content of each class of repetitive sequences (A) and S/MAR-specific motifs (B) in all groups. Names of the repetitive sequences are marked under the column graph. The content of the major classes of dispersed repetitive sequences is presented: Alu- and MIR-SINE, short interspersed nucleotide element; LINE1, LINE2 and L3/CR1, long interspersed nucleotide element; MaLR, mammalian LTR retrovirus; ERVL, ERV_classI and ERV_classII, endogenous retrovirus repeat family; MER1_type and MER2_type, medium reiteration frequency; simple repeats. The MAR-Wiz tool was operated with its default settings (Ori rule, TG richness rule, curved-DNA rule, kinked-DNA rule, Topo II recognition rule, AT richness).
Considering that a high content of repetitive DNA is a hallmark of S/MAR regions, it is likely that some mutp53-binding sites are S/MARs. As expected, both ChIP and random sequences are distinguished from the promoter dataset by a higher content of AT-rich motifs (AT-richness and Ori rule) (Figure 3B). Curved-DNA and kinked-DNA patterns show a tendency for a slight enrichment within the ChIP sequences, which would support the in vitro specificity of mutp53 for non-B DNA conformation (6). The ChIP and random datasets were also screened for the presence of shared (conservative) motifs (10–20 nucleotides) and the occurrence frequency of short oligomeric sequences (6–8 nucleotides) using regulatory sequence analysis tools (RSAT) (39). No significant enrichment for any motif or oligomer was detected in both datasets (data not shown). Computational search for the p53 Response Element (p53RE [two tandem repeated decamers 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ separated by 0–13 bp]) using the PatSearch program (40) and PatSearch syntax for the p53RE (58) also revealed no significant differences in the occurrence of p53RE in the random and ChIP datasets (data not shown). These results therefore fit into our concept that the genome contains numerous mutp53-binding sites, which are defined by structural properties rather than by primary sequence and are differentially occupied by mutp53 in the functional context of chromatin.
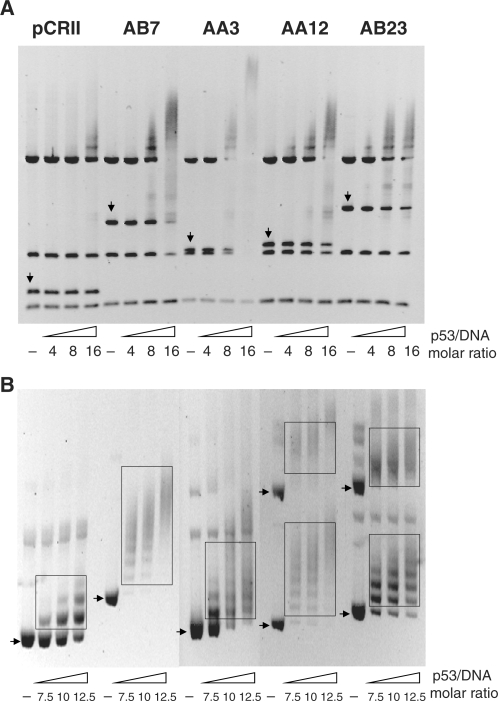
Mutp53 might either compete or cooperate with other proteins for binding to a particular DNA site and, therefore, might not necessarily bind directly to DNA. To provide evidence for direct binding of mutp53 to DNA, we analyzed a small set of ChIP sequences for in vitro binding by recombinant bacterially expressed p53R273H protein applying EMSA. As the interaction of mutp53 with DNA is structure-selective (6), it should be strongly influenced by alterations in DNA conformation. We therefore compared binding of p53R273H to ChIP sequences prepared either as linear DNA or as inserts in supercoiled pCRII vector DNA. Figure 4A shows the mobility-shift profiles of EMSA performed with a mixture of PvuII restriction fragments from control or insert-containing pCRII vector DNA and recombinant p53R273H at various molar ratios. The profiles demonstrate that in comparison to restriction fragments of vector DNA, ChIP sequences showed superior binding to mutp53, although their affinity varied greatly. The strongest interaction with p53R273H was observed for ChIP sequence AA3. This ChIP DNA fragment contains a 72-bp long polypurine/polypyrimidine-rich region which can adopt an intramolecular triple-helix H-DNA conformation (59). In control experiments we noted that wtp53, p53R273H and another ‘hot spot’ DNA-binding domain mutant protein, p53R249S, binds to linearized AA3 and AB23 sequences with comparable affinity, whereas the conformational mutant protein p53R175H binds very weakly (see Figure S2). The band-shift profiles in Figure 4B demonstrate that p53R273H binds to pCRII vector DNA in a stoichiometric fashion, forming three slowly migrating complexes at the highest tested protein/DNA ratio. In contrast, plasmids containing ChIP sequences interact much stronger with mutp53, forming multiple, slowly migrating complexes already at a 7.5 M excess of protein. Wtp53 (41,60,61) and mutp53 (60) bind strongly to supercoiled DNA via an interaction of the C-terminal domains of p53 tetramers with strand crossings, thereby forming complexes with electrophoretic properties reminiscent to the mobility of DNA topoisomers in agarose gel. Altogether, the in vitro binding data strongly support the notion that the ChIP sequences reflect genomic DNA regions, to which p53R273H binds directly in U251 cells.
Figure 4.
Recombinant p53R273H interacts preferentially with ChIP DNA sequences in vitro. (A–B) Four selected ChIP DNA sequences were tested for their interaction with p53R273H using EMSA. For the binding reaction, either PvuII-restriction fragments of empty or insert containing pCRII vector (A), or supercoiled forms of plasmid DNA (B) were used. Increasing amounts of p53R273H protein (marked by p53/DNA molar ratio) were incubated with 200 ng of PvuII-restriction fragments (A) or supercoiled DNA (B) in binding buffer and then separated in an agarose gel. After staining with ethidium bromide, gels were photographed. Restriction DNA fragments containing the multi-cloning site and inserted ChIP sequences are marked by vertical arrows. Positions of supercoiled forms (monomer and dimer) of plasmid DNA after electrophoretic separation are marked by horizontal arrows. Slowly migrating complexes of p53R273H protein and supercoiled DNA are marked by boxes.
Functionality of mutp53-binding sites
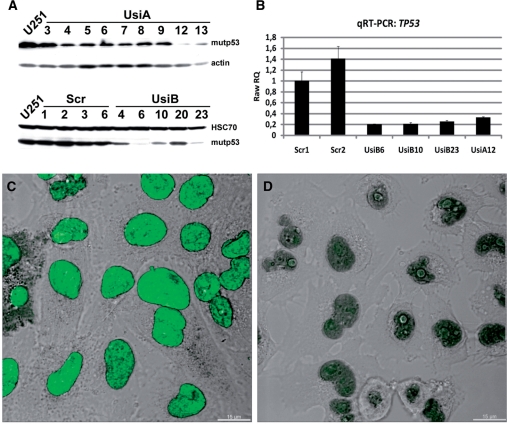
To analyze the postulated effects of mutant p53 on the regulation of transcription, we applied shRNA-mediated knock-down of the p53 protein level, which allows for a comparison between parental cells and cells with reduced p53 expression. U251 cells were transfected with H1-promoter-based vectors encoding either a well-established anti-p53 shRNA (33) or, as a control for off-target effects, scrambled shRNA. After antibiotic selection, stable clones, named UsiA (transfected with vectors pSuper-p53 and pCI-neo), UsiB (transfected with vector pENTR/H1-p53), and Scr (transfected with vector pENTR/H1-src) were analyzed for mutp53 expression by western blotting (Figure 5A). While the level of mutp53 in all clones expressing scrambled shRNA remained unchanged, mutp53 expression was strongly reduced in several clones selected after transfection with anti-p53 shRNA-expressing constructs (Figure 5A). As shown in Figure 5B, the amount of p53 transcripts was significantly reduced in the clones UsiA12, UsiB6, UsiB10 and UsiB23 in comparison to Scr1 and Scr2 clones expressing scrambled shRNA. In line with this result, mutp53 nuclear staining typically observed for the parental cells (Figure 5C) was dramatically reduced in anti-p53 shRNA-expressing clones (UsiA12 clone shown as an example in Figure 5D).
Figure 5.
Stable reduction of p53R273H expression using shRNA. (A) U251 cells were transfected with vectors encoding either anti-p53 or scrambled shRNA, and after antibiotic selection, single clones (named as UsiA, UsiB and Scr) were tested by immunoblotting for the expression of mutp53. Actin or HSC70 were used as a loading control. (B–D) Reduction of mutp53 expression in UsiA12 cells was validated by qRT–PCR (B) and immunofluorescence microscopy (C–D). (B) For relative quantitation of mutp53 transcription by qRT–PCR, total RNA from U251 clones expressing either scrambled (Scr1 and Scr2) or p53-specific shRNA (UsiA12, UsiB6, UsiB10, UsiB23) was isolated three times. qRT–PCR was done in triplicates. RQ (relative quantitation) values were calculated by normalizing to the transcription of the HPRT1 gene and selecting Scr1 sample as calibrator. (C–D) Confocal images of U251 (C) and UsiA12 cells (D), immunostained with anti-p53 antibody (green). Z-stack sections were deconvoluted using the Huygens software and the images were processed with the Imaris software. 3D reconstructed confocal images were merged with a differential interference contrast (DIC) micrograph to show the cellular outlines and nuclei. Scale bar—15 µm.
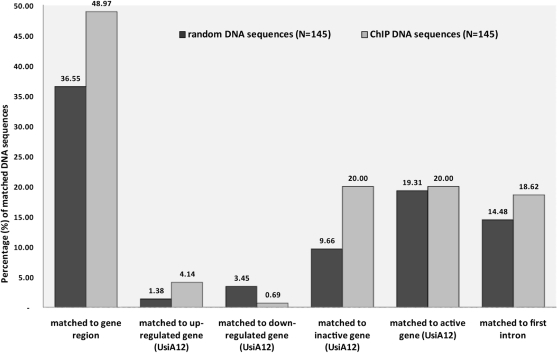
To study the consequences of mutp53 reduction on gene expression, total RNA from UsiA12 and parental U251 cells was prepared from two independent cell culture experiments (biological replicates) and processed for microarray-based profiling of gene expression. A detailed analysis of the microarray data will be presented elsewhere (manuscript in preparation). Using the t-test as criterion for statistical significance (P ≤ 0.05), we identified 805 genes whose mRNA levels were either increased (379 genes) or decreased (426 genes) >1.8-fold in both UsiA12 replicates compared to parental U251 cells (see Table S6). Assuming that some of the ChIP sequences originate from differently regulated genes, we correlated the ChIP results with the expression microarray data by mapping the ChIP sequences to the genome. ChIP and random sequences were inspected for their chromosomal positions and proximity to known genes (UCSC and Refseq gene tracks in Genome Browser, hg18). In total, ∼49% of the analyzed ChIP sequences (71 out of 145) were mapped to genes, mostly to introns and never close to TSS (Figure 6; Table S2), whereas ∼37% of the randomly selected genomic sequences (53 out of 145) are derived from intra-genic regions (Figure 6; Table S7), thereby indicating a tendency for a preferred location of mutp53BS in intergenic regions. Inspection of the microarray expression profiles of the groups of genes to which ChIP (Table S2) and random (Table S7) sequences were mapped yielded two important observations: First, binding of p53R273H was mainly associated with up-regulation of the respective genes in UsiA12 cells (Figure 6). Since mutp53 is down-regulated in UsiA12 cells, activation of these genes in UsiA12 cells implies that these genes are directly or indirectly repressed by mutp53 in parental U251 cells. The mRNA amount of six out of seven regulated genes, FRMD5, PPARGC1A, NRG1, C3ORF26, TMEM108 and JAK2, was increased 1.8–4.4-fold in UsiA12 cells (Table S2). Secondly, despite equal frequency of mapping to active, non-regulated (‘on’) genes, two times more ChIP sequences than random sequences mapped to inactive (‘off’) genes (Figure 6). cis-Regulatory sequences within the first intron are the target sites for transcription factors including p53 (62–64). We found that there is a tendency for binding of mutp53 to sequences within the first intron (Figure 6). The location of ChIP sequences within the first introns of four genes up-regulated in UsiA12 cells (i.e. repressed by mutp53 in U251 cells), FRMD5, NRG1, C3ORF26 and JAK2, supports the significance of this observation.
Figure 6.
Correlation of ChIP and expression microarray data. The graph shows the statistic evaluation of chromosomal positions of ChIP and random genomic sequences which were inspected with respect to the coordinates of known genes (UCSC and Refseq gene tracks in Genome Browser, hg18), location in the first intron, transcriptionally regulated genes (up- or down-regulated in UsiA12 cells), and active (‘on’) or inactive (‘off’) gene as judged from the expression microarray data (UsiA12 cells). Ordinate axis represents the percentage of sequences from both groups identified in each analysis.
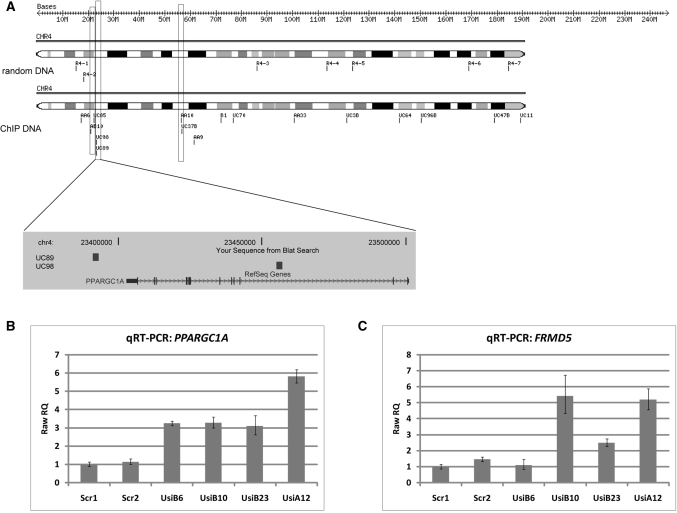
Besides location in the first intron, the proximity of ChIP sequences in the genome might be indicative of their functionality. Therefore we plotted the coordinates of ChIP and random sequences using the ChromoMapper program (37) and found a tendency for a local enrichment of ChIP sequences at distinct chromosomal positions. The distribution of ChIP sequences on chromosome 4 is shown in Figure 7A (see Figure S3 for all chromosomes), illustrating that a significant number of ChIP sequences (a total of 42) localizes as closely linked pairs (duplets) within a range of 2 Mb. One duplet (ChIP sequences UC89 and UC98; Table S1) maps inside and close to the PPARGC1A gene, up-regulated in UsiA12 cells, encoding a transcriptional co-activator involved in controlling energy metabolism (65); a second duplet (AB10 and UC85; Table S1) localizes in an adjacent window of ∼2 Mb (Figure 7A). qRT–PCR analysis strongly supports the assumption that the PPARGC1A gene is a transcriptional target of mutp53. Indeed, in comparison to clones expressing scrambled shRNA, the activity of the PPARGC1A gene was significantly affected by anti-p53 shRNA in all four clones, resulting in 3–6-fold higher transcript levels (Figure 7B). Next, by qRT–PCR we measured the expression of FRMD5, NRG1, C3ORF26 and JAK2 as potential target genes regulated by mutp53 via binding to cis-regulatory sequences within the first intron. For the FRMD5 gene, encoding a putative cytoskeletal protein, we detected an up to 6-fold up-regulation in three anti-p53 shRNA-expressing clones (Figure 7C). Thereby, we identified two genes, PPARGC1A and FRMD5, as p53R273H targets, whose transcription is down-regulated either by binding of mutp53 to multiple sites that are distant from the TSS (PPARGC1A) or to sequences within the first intron (FRMD5). However, transcription of the other tested genes was either regulated only in clone Usi12A or indistinguishable from the control clones (data not shown), indicating that mutp53-dependent regulation of potential target genes may differ in individual U251 derived clones.
Figure 7.
p53R273H binding correlates with the regulation of PPARGC1A gene activity. (A) Chromosomal coordinates of ChIP and random sequences on chromosome 4 were plotted using the ChromoMapper software. Closely linked positions of ChIP sequences in a window of 2 Mb are marked by boxes. Genome browser snapshot shows the location of ChIP sequences in PPARGC1A gene. (B–C) Relative quantitation of PPARGC1A (B) and FRMD5 (C) gene transcription by qRT–PCR. Total RNA from cell clones expressing scrambled (Scr1 and Scr2) or p53-specific shRNA (UsiA12, UsiB6, UsiB10, UsiB23) was isolated three times. qRT–PCR was done in triplicates. Raw RQ values were calculated by normalizing to the transcription of the HPRT1 gene and selecting Scr1 cell clone as calibrator.
DISCUSSION
In the present work we provide evidence that mutp53, specifically the R273H hot-spot mutant protein, is targeted to the nucleus of U251 cells where it interacts with DNA and participates in the regulation of gene transcription. The R273H mutation in p53 disrupts the DNA-binding surface and abolishes the specific interaction of the core domain with wtp53 target nucleotide sequences (66). However, it does not interfere with nuclear localization and endows the protein with an increased half-life, leading to its accumulation in the nucleus. In accordance with FRAP measurements demonstrating that both p53-GFP and mutp53-GFP (R273H, R175H) fusion proteins overexpressed in p53-null H1299 cells undergo constrained diffusion with similar kinetics within the nucleus (67), we detected p53R273H largely in the soluble nuclear fraction which contains freely diffusing proteins. Such a dynamic behavior seems to be a general phenomenon for transcription factors, which are predominantly located in the non-bound fraction under steady-state conditions, while only a small fraction is occupying DNA target sites (68). It seems that p53R273H shares this property with wtp53 and other transcription factors. The small fraction of p53R273H bound to the detergent-insoluble nuclear scaffold could represent proteins which at the time of extraction were recruited to chromatin sites to exert a regulatory function on gene transcription. The retention of mutp53 at the nuclear scaffold could be mediated by interaction either with proteins or DNA alone or by additive effect of both interactions. Indeed, our results point to a possibly direct interaction of p53R273H with components of the transcription machinery, including the RNA polymerase II complex and the regulatory proteins Sp1 and YY1, known as binding partners for wtp53 (50,51,69). On the other hand, EMSA studies using ChIP sequences support an association of p53R273H to chromatin by direct binding to DNA.
The bioinformatic evaluation of our ChIP sequencing data confirmed our assumption that the primary nucleotide sequence of a mutp53-binding site would not be much different from any randomly selected genomic fragment, since we presume that rather than its sequence, its location in the genome and its conformational features would determine its functionality. Consequently, the ChIP DNA fragments are rich in repetitive DNA elements distributed more or less randomly over the whole genome and are not enriched for any short nucleotide motif (e.g. p53 response elements). Two lines of evidence argue against the apparent random binding of p53R273H to genomic DNA: (i) over-representation of beta-satellite repeats in the collection of the sequenced ChIP DNA fragments, thereby qualifying p53R273H as a beta-satellite binding protein, and (ii) a significant enrichment of ChIP sequences over randomly selected sequences in inactive genes or genes whose activity was enhanced after stable, shRNA-mediated reduction of mutp53 expression. According to these data it is likely that mutp53-binding sites are largely located in transcriptionally inactive, constitutive and facultative heterochromatin regions, represented by beta-satellite-rich DNA and inactive genes, respectively. This would imply that the transcriptional activity of mutp53 mainly, but not exclusively, results in repressive effects mediated by mutp53-dependent damping of target gene transcription, or alternatively by gene silencing or heterochromatization of the whole chromatin domain. A likely prerequisite for such a role of mutp53 is its ability to interact with repetitive DNA sequences, whose role in gene regulation is well documented (70,71). Importantly, repetitive DNA sequences contribute to the fraction of non-B structured DNA (72,73), in this way providing multiple binding sites for regulatory proteins lacking sequence specificity, but endowed with the capacity to sense DNA conformation. p53R273H fulfills the criteria of a DNA-binding protein recognizing DNA conformation, as it binds to non-linear DNA substrates, but is virtually inactive toward linear DNA (6). In support, we detected a much stronger binding of p53R273H to non-linear DNA represented by a polypurine/polypyrimidine tract than to other ‘linear’ ChIP DNA fragments. Furthermore, p53R273H bound significantly better to ChIP sequences inserted into supercoiled DNA than to linearized plasmid DNA.
The mode of mutp53 DNA interaction in vitro (6) allows us to hypothesize that in vivo mutp53 can bind stably only to pre-existing non-B DNA structures, whose real number and location depends on chromatin conformation and composition, and cannot be predicted in silico solely based on nucleotide sequence. Taking this into consideration one would expect that mutp53 will bind with low preference to multiple genomic regions representing a fraction of potentially available sites. Indeed, quantifying by sequence-specific real-time PCR the binding events of mutp53 to ChIP sequences mapped to the first introns, we could not detect significant enrichment over control genomic DNA (data not shown). The low specificity would be compensated by the high abundance of mutp53 in the nucleus, and the tendency for co-operative binding and formation of higher-order oligomeric forms (6). The latter notion is indirectly supported by Triton X-100 extraction which results in multiple dot-like structures decorated with p53 antibodies. The preference of mutp53 for one or the other genomic site could also be determined by the local affinity to the actual DNA structure and the interaction with co-factors, for example, other DNA-binding proteins which could assist in recruitment of mutp53 to a particular site and co-operate in the transcriptional activity of mutp53.
The finding that potential mutp53-binding sites lack sequence specificity and are distributed over the whole genome argues for a promoter-binding independent, long-range mechanism for the regulation of gene transcription by mutp53. Long-range regulation is not a special attribute of mutp53, but reflects the notion that transcription factors may act as dual-purpose regulators, controlling gene expression either via direct targeting of a gene or via (re)organizing higher-order chromatin structure. In this regard, recent genome-wide studies of the binding of sequence-specific transcription factors (74), including wtp53 (55), to DNA demonstrated that they not only bind to regulatory sequences around TSS, but also to numerous sites distributed over the whole genome. Extending the term ‘epigenetics’ to regulatory reactions primarily based on nuclear (re)organization, including changes of the global chromatin conformation (75,76) and the formation of new transcription factories (46) in expense of existing ones, we want to propose that mutp53 modulates the gene expression program as an epigenetic factor. In cooperation with other regulatory proteins mutp53 might be involved in the establishment and support of a transcription program. By binding to promoter-distant, cis-regulatory sequences, for example within the first intron, mutp53 might influence chromatin organization which determines the transcriptional activity of a particular gene. Our ChIP data support this hypothesis, especially the identification of four ChIP sequences from non-coding regions inside and close to the PPARGC1A gene, whose transcription was increased in all tested clones with a reduced expression of mutp53. The connection between mutp53 and the transcriptional co-activator PPARGC1A is interesting insofar, as low expression of this protein in breast cancer patients was found associated with poor clinical prognosis (77).
Correlation analyses performed for several tumor entities demonstrated that expression of mutp53 in human tumors is associated with unfavorable prognosis (78). Many mutp53 proteins, in particular all ‘hot spot’ mutp53 proteins, exert oncogenic functions which contribute to tumor progression. These functions appear in experimental systems e.g. in enhanced growth rates and in the modulation of the expression of tumor relevant genes (79,80). Transcriptional regulation of such genes by mutp53 has been demonstrated previously, and can be achieved by an altered sequence-specificity of mutp53 proteins (12), and by its interaction with other transcription factors, like SP1, Ets1 and NF-Y (9–11). In this article we describe another, more global mechanism for mutp53 dependent gene regulation which should also contribute to its oncogenic functions. Although much more work has to be done to understand the molecular details of mutp53 activity, we believe that establishing an adequate experimental cell model and applying genome-wide technologies will help to expand our knowledge of transcription in tumor cells in general, and the impact of mutp53, in particular.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This study was supported by grants from the Deutsche Forschungsgemeinschaft (to W.D.), the Fond der Chemischen Industrie (to W.D.), EC FP6 funding (to W.D.), the German-Israeli Foundation (G.I.F. No. I- 927-187.13/2006 to W.D.), the Czech Science Foundation (204/06/P369 and 204/08/1560 to M.B.), by the Ministry of Education of the CR (1K04119 to M.B.), by the Academy of Sciences of the CR (AV0Z50040702) and by the European commission (QLGA-CT-2001-52001 and MERG-6-CT-2005-014875 to M.B.). The Heinrich-Pette-Institut is financially supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms Marion Kühl and Gabriele Warnecke and Mr Jakub Dvorak for competent technical assistance. We are indebted to Dr Burkhard Brandt and Dr Dirk Kemming (University Medical Center Hamburg-Eppendorf, Department of Tumor Biology, Hamburg, Germany) for performing microarray expression experiments and for their help with the bioinformatic evaluation of microarray data.
REFERENCES
- 1.Soussi T, Kato S, Levy PP, Ishioka C. Reassessment of the TP53 mutation database in human disease by data mining with a library of TP53 missense mutations. Hum. Mutat. 2005;25:6–17. doi: 10.1002/humu.20114. [DOI] [PubMed] [Google Scholar]
- 2.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 3.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 4.Ma B, Levine AJ. Probing potential binding modes of the p53 tetramer to DNA based on the symmetries encoded in p53 response elements. Nucleic Acids Res. 2007;35:7733–7747. doi: 10.1093/nar/gkm890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ang HC, Joerger AC, Mayer S, Fersht AR. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 6.Gohler T, Jager S, Warnecke G, Yasuda H, Kim E, Deppert W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005;33:1087–1100. doi: 10.1093/nar/gki252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Deppert W. Transcriptional activities of mutant p53: when mutations are more than a loss. J. Cell Biochem. 2004;93:878–886. doi: 10.1002/jcb.20271. [DOI] [PubMed] [Google Scholar]
- 8.Weisz L, Oren M, Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–2211. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- 9.Chicas A, Molina P, Bargonetti J. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem. Biophys. Res. Commun. 2000;279:383–390. doi: 10.1006/bbrc.2000.3965. [DOI] [PubMed] [Google Scholar]
- 10.Xu D, Wilson TJ, Chan D, De Luca E, Zhou J, Hertzog PJ, Kola I. Ets1 is required for p53 transcriptional activity in UV-induced apoptosis in embryonic stem cells. EMBO J. 2002;21:4081–4093. doi: 10.1093/emboj/cdf413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol. Cell Biol. 2006;26:2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matas D, Sigal A, Stambolsky P, Milyavsky M, Weisz L, Schwartz D, Goldfinger N, Rotter V. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 2001;20:4163–4172. doi: 10.1093/emboj/20.15.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol. Cell Biol. 1998;18:3735–3743. doi: 10.1128/mcb.18.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 16.Liu WL, Midgley C, Stephen C, Saville M, Lane DP. Biological significance of a small highly conserved region in the N terminus of the p53 tumour suppressor protein. J. Mol. Biol. 2001;313:711–731. doi: 10.1006/jmbi.2001.5082. [DOI] [PubMed] [Google Scholar]
- 17.Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc. Natl Acad. Sci. USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 19.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 22.Gluch A, Vidakovic M, Bode J. Scaffold/matrix attachment regions (S/MARs): relevance for disease and therapy. Handb. Exp. Pharmacol. 2008;186:67–103. doi: 10.1007/978-3-540-72843-6_4. [DOI] [PubMed] [Google Scholar]
- 23.Razin SV, Gromova II, Iarovaia OV. Specificity and functional significance of DNA interaction with the nuclear matrix: new approaches to clarify the old questions. Int. Rev. Cytol. 1995;162B:405–448. doi: 10.1016/s0074-7696(08)62623-6. [DOI] [PubMed] [Google Scholar]
- 24.Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- 25.Bode J, Winkelmann S, Gotze S, Spiker S, Tsutsui K, Bi C, A KP, Benham C. Correlations between scaffold/matrix attachment region (S/MAR) binding activity and DNA duplex destabilization energy. J. Mol. Biol. 2006;358:597–613. doi: 10.1016/j.jmb.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 26.Koga H, Deppert W. Identification of genomic DNA sequences bound by mutant p53 protein (Gly245–>Ser) in vivo. Oncogene. 2000;19:4178–4183. doi: 10.1038/sj.onc.1203745. [DOI] [PubMed] [Google Scholar]
- 27.Deppert W. Binding of MAR-DNA elements by mutant p53: possible implications for its oncogenic functions. J. Cell Biochem. 1996;62:172–180. doi: 10.1002/(sici)1097-4644(199608)62:2<172::aid-jcb5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 28.Weissker SN, Muller BF, Homfeld A, Deppert W. Specific and complex interactions of murine p53 with DNA. Oncogene. 1992;7:1921–1932. [PubMed] [Google Scholar]
- 29.Will K, Warnecke G, Albrechtsen N, Boulikas T, Deppert W. High affinity MAR-DNA binding is a common property of murine and human mutant p53. J. Cell Biochem. 1998;69:260–270. doi: 10.1002/(sici)1097-4644(19980601)69:3<260::aid-jcb4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 30.Will K, Warnecke G, Wiesmuller L, Deppert W. Specific interaction of mutant p53 with regions of matrix attachment region DNA elements (MARs) with a high potential for base-unpairing. Proc. Natl Acad. Sci. USA. 1998;95:13681–13686. doi: 10.1073/pnas.95.23.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 32.Tolstonog GV, Mothes E, Shoeman RL, Traub P. Isolation of SDS-stable complexes of the intermediate filament protein vimentin with repetitive, mobile, nuclear matrix attachment region, and mitochondrial DNA sequence elements from cultured mouse and human fibroblasts. DNA Cell Biol. 2001;20:531–554. doi: 10.1089/104454901317094954. [DOI] [PubMed] [Google Scholar]
- 33.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halees AS, Weng Z. PromoSer: improvements to the algorithm, visualization and accessibility. Nucleic Acids Res. 2004;32:W191–W194. doi: 10.1093/nar/gkh433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackl H, Burkard TR, Sturn A, Rubio R, Schleiffer A, Tian S, Quackenbush J, Eisenhaber F, Trajanoski Z. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 2005;6:R108. doi: 10.1186/gb-2005-6-13-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh GB, Kramer JA, Krawetz SA. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brazdova M, Palecek J, Cherny DI, Billova S, Fojta M, Pecinka P, Vojtesek B, Jovin TM, Palecek E. Role of tumor suppressor p53 domains in selective binding to supercoiled DNA. Nucleic Acids Res. 2002;30:4966–4974. doi: 10.1093/nar/gkf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 43.Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- 44.Nickerson JA, Krockmalnic G, Wan KM, Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc. Natl Acad. Sci. USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He S, Dunn KL, Espino PS, Drobic B, Li L, Yu J, Sun JM, Chen HY, Pritchard S, Davie JR. Chromatin organization and nuclear microenvironments in cancer cells. J. Cell Biochem. 2008;104:2004–2015. doi: 10.1002/jcb.21485. [DOI] [PubMed] [Google Scholar]
- 46.Carter DR, Eskiw C, Cook PR. Transcription factories. Biochem. Soc. Trans. 2008;36:585–589. doi: 10.1042/BST0360585. [DOI] [PubMed] [Google Scholar]
- 47.Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, Zaidi SK, Young D, Choi JY, Gutierrez S, et al. Nuclear microenvironments support assembly and organization of the transcriptional regulatory machinery for cell proliferation and differentiation. J. Cell Biochem. 2004;91:287–302. doi: 10.1002/jcb.10777. [DOI] [PubMed] [Google Scholar]
- 48.van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- 49.Guo B, Odgren PR, van Wijnen AJ, Last TJ, Nickerson J, Penman S, Lian JB, Stein JL, Stein GS. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc. Natl Acad. Sci. USA. 1995;92:10526–10530. doi: 10.1073/pnas.92.23.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koutsodontis G, Vasilaki E, Chou WC, Papakosta P, Kardassis D. Physical and functional interactions between members of the tumour suppressor p53 and the Sp families of transcription factors: importance for the regulation of genes involved in cell-cycle arrest and apoptosis. Biochem. J. 2005;389:443–455. doi: 10.1042/BJ20041980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 53.Luderus ME, de Graaf A, Mattia E, den Blaauwen JL, Grande MA, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992;70:949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- 54.Hearnes JM, Mays DJ, Schavolt KL, Tang L, Jiang X, Pietenpol JA. Chromatin immunoprecipitation-based screen to identify functional genomic binding sites for sequence-specific transactivators. Mol. Cell Biol. 2005;25:10148–10158. doi: 10.1128/MCB.25.22.10148-10158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 56.Greig GM, Willard HF. Beta satellite DNA: characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics. 1992;12:573–580. doi: 10.1016/0888-7543(92)90450-7. [DOI] [PubMed] [Google Scholar]
- 57.Eichler EE, Hoffman SM, Adamson AA, Gordon LA, McCready P, Lamerdin JE, Mohrenweiser HW. Complex beta-satellite repeat structures and the expansion of the zinc finger gene cluster in 19p12. Genome Res. 1998;8:791–808. doi: 10.1101/gr.8.8.791. [DOI] [PubMed] [Google Scholar]
- 58.Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D'Erchia AM, et al. p53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinformatics. 2007;8(Suppl 1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 60.Mazur SJ, Sakaguchi K, Appella E, Wang XW, Harris CC, Bohr VA. Preferential binding of tumor suppressor p53 to positively or negatively supercoiled DNA involves the C-terminal domain. J. Mol. Biol. 1999;292:241–249. doi: 10.1006/jmbi.1999.3064. [DOI] [PubMed] [Google Scholar]
- 61.Palecek E, Brazda V, Jagelska E, Pecinka P, Karlovska L, Brazdova M. Enhancement of p53 sequence-specific binding by DNA supercoiling. Oncogene. 2004;23:2119–2127. doi: 10.1038/sj.onc.1207324. [DOI] [PubMed] [Google Scholar]
- 62.Chen J, Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc. Natl Acad. Sci. USA. 2005;102:4813–4818. doi: 10.1073/pnas.0407069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reczek EE, Flores ER, Tsay AS, Attardi LD, Jacks T. Multiple response elements and differential p53 binding control Perp expression during apoptosis. Mol. Cancer Res. 2003;1:1048–1057. [PubMed] [Google Scholar]
- 64.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J. Biol. Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 67.Hinow P, Rogers CE, Barbieri CE, Pietenpol JA, Kenworthy AK, DiBenedetto E. The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys. J. 2006;91:330–342. doi: 10.1529/biophysj.105.078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier JL, Triezenberg SJ, Reinberg D, Flores O, Ingles CJ, et al. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 71.Zuckerkandl E, Cavalli G. Combinatorial epigenetics, ‘junk DNA’, and the evolution of complex organisms. Gene. 2007;390:232–242. doi: 10.1016/j.gene.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 72.Hanke JH, Hambor JE, Kavathas P. Repetitive Alu elements form a cruciform structure that regulates the function of the human CD8 alpha T cell-specific enhancer. J. Mol. Biol. 1995;246:63–73. doi: 10.1006/jmbi.1994.0066. [DOI] [PubMed] [Google Scholar]
- 73.Howell R, Usdin K. The ability to form intrastrand tetraplexes is an evolutionarily conserved feature of the 3′ end of L1 retrotransposons. Mol. Biol. Evol. 1997;14:144–155. doi: 10.1093/oxfordjournals.molbev.a025747. [DOI] [PubMed] [Google Scholar]
- 74.Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackson DA. The principles of nuclear structure. Chromosome Res. 2003;11:387–401. doi: 10.1023/a:1024954123092. [DOI] [PubMed] [Google Scholar]
- 76.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 77.Jiang WG, Douglas-Jones A, Mansel RE. Expression of peroxisome-proliferator activated receptor-gamma (PPARgamma) and the PPARgamma co-activator, PGC-1, in human breast cancer correlates with clinical outcomes. Int. J. Cancer. 2003;106:752–757. doi: 10.1002/ijc.11302. [DOI] [PubMed] [Google Scholar]
- 78.Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 79.Bossi G, Marampon F, Maor-Aloni R, Zani B, Rotter V, Oren M, Strano S, Blandino G, Sacchi A. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 2008;7:1870–1879. doi: 10.4161/cc.7.12.6161. [DOI] [PubMed] [Google Scholar]
- 80.Heinlein C, Krepulat F, Lohler J, Speidel D, Deppert W, Tolstonog GV. Mutant p53(R270H) gain of function phenotype in a mouse model for oncogene-induced mammary carcinogenesis. Int. J. Cancer. 2008;122:1701–1709. doi: 10.1002/ijc.23317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.