Abstract
Structural properties of single λ DNA treated with anti-cancer drug cisplatin were studied with magnetic tweezers and AFM. Under the effect of low-concentration cisplatin, the DNA became more flexible, with the persistence length decreased significantly from ∼52 to 15 nm. At a high drug concentration, a DNA condensation phenomenon was observed. Based on experimental results from both single-molecule and AFM studies, we propose a model to explain this kind of DNA condensation by cisplatin: first, di-adducts induce local distortions of DNA. Next, micro-loops of ∼20 nm appear through distant crosslinks. Then, large aggregates are formed through further crosslinks. Finally, DNA is condensed into a compact globule. Experiments with Pt(dach)Cl2 indicate that oxaliplatin may modify the DNA structures in the same way as cisplatin. The observed loop structure formation of DNA may be an important feature of the effect of platinum anti-cancer drugs that are analogous to cisplatin in structure.
INTRODUCTION
Cisplatin [Pt(NH3)2Cl2] is one of the most widely used anti-cancer drug especially to treat testicular, head, neck, non-small-cell lung and cervical cancers (1). It is generally believed that the cytotoxicity of cisplatin derives from its adduct with DNA. In aqueous solution, the two chloride ions dissociate from the central platinum, and two water molecules or hydroxide ions locate there, transforming cisplatin to an active state (2,3). When the aqueous cisplatin reacts with DNA, its water molecules are easily substituted by the N7 atoms of guanine or adenine to form both mono-adduct and di-adduct (4,5). The biological effects of cisplatin on DNA are quite complicated. According to previous studies, cisplatin–DNA adducts may interact with cellular proteins in several modes, for instance, hijacking transcription factors to block DNA transcription, blocking off DNA polymerase or RNA polymerase bypassing (6,7).
The cisplatin–DNA adducts mainly contain intrastrand crosslinks: 65% 1,2-d (GpG), 25% 1,2-d (ApG), 5–10% 1,3-d (GpNpG), as well as a small portion of interstrand crosslinks (8). X-ray structure of a 12-bp double-strand DNA containing a single 1,2-d(GpG) adduct reveals that the adduct bends the double helix 50° towards the major groove. In addition, there is a 30° dihedral between the two adjacent guanines (9,10). The NMR solution structure of the same platinated DNA was also resolved, with a bending angle of 78°, and a 25° unwinding of the double helix (11). In brief, cisplatin molecules are able to bind to DNA target, make intrastrand and interstrand crosslinks, and distort severely the double helix towards the major groove as well as induce a partial unwinding near the adduct sites (12).
Over the past decade, single-molecule techniques have been developed as helpful methods to measure the properties of DNA–ligand-binding and DNA–protein interaction. For instance, Hatch et al. (13) have studied the salt-dependent stabilization of partially open λ-phage DNA by Escherichia coli SSB protein using magnetic tweezers. Williams et al. have done a lot of work on the interaction between DNA and proteins (i.e. HIV-1 nucleocapsid protein, T4 gene 32 protein, HMGB) by single-molecule stretching experiments using optical tweezers (14–16). Effects of DNA-binding proteins such as HU, IHF and H-NS on DNA mechanical properties have also been measured with magnetic tweezers (17–19). Besides, in recent years, some significant findings about ionic effects and ligand binding to DNA have been made with these single-molecule techniques. Different DNA-binding modes of small molecules have been observed by the single-molecule force spectroscopy (20). Through the measurement of WLC (worm-like chain) behavior of λ DNA, it is revealed that multivalent ions such as Co(NH3)63 + reduce the persistence length of DNA to 25–30 nm, obviously much below the normal value of 45–50 nm in monovalent salt (21). Also, DNA intercalators such as EtBr and YOYO-1 lead to a similarly decreased DNA persistence length (22). Furthermore, step-wise DNA condensation under the interaction of multivalent ions was revealed in single-molecule experiments (23). In addition to optical and magnetic tweezers, AFM is another widely used technique for DNA investigation due to its high-quality image resolution, and it has also been applied to drug discovery (24).
Previously, there were several studies exploring the effects of cisplatin on the mechanical properties of DNA. Gaub et al. (25) have used single-molecule force spectroscopy to measure the structural changes of DNA induced by cisplatin. Their results revealed that the B-S transition was very sensitive to the binding of cisplatin. Besides, the separation of the double helix was inhibited as a result of interstrand crosslinks. In addition to B-S transition, the elasticity of the DNA molecules has also been studied by single-molecule experiment qualitatively (26). The force versus extension curve was observed to be altered. However, the authors did not give out the persistence length and contour length. Besides, there were no results about the effect of cisplatin concentration, or about the change with time.
In the current work, we employed magnetic tweezers technique and AFM to systemically study the effect of cisplatin on the mechanical and structural properties of λ DNA. Single-molecule experiments using a magnetic tweezers showed that cisplatin may reduce the persistence length of DNA from ∼52 nm to less than 20 nm. AFM imaging revealed that the much reduced persistence length resulted from the formation of many local kinks on DNA. Besides, WLC fitting of force versus extension curve and AFM imaging both revealed that DNA contour length was basically not affected. At high cisplatin concentrations, we observed with the magnetic tweezers that the DNA extension gradually shortened under low force (∼1 pN) and this shortening was irreversible even at 20 pN stretching force. AFM imaging identified the mechanism for this kind of DNA condensation: first, micro-loops by long distance crosslinks were formed due to DNA thermal fluctuation, then large aggregates were formed, and finally the DNA molecule was condensed into a compact globule. We think the formation of this kind of loops by distant crosslinks is an important property of DNA–cisplatin interaction. At last, Pt(dach)Cl2 was also studied for comparison. It was found that it interacted with DNA in a similar manner as cisplatin. We believe the current study will be helpful for understanding more deeply the effects of cisplatin on mechanical and structural properties of DNA.
MATERIALS AND METHODS
Chemical reagents
Cisplatin was purchased from Sigma-Aldrich (USA), Pt(dach)Cl2 was obtained from the Institute of Precious Metal in Kunming of China. λ DNA for magnetic tweezers experiment was from New England Biolabs. It was isolated from λ phage c1857 S7, 48502 bp in length, with 12-nt single-strand overhanging at both ends. The catalog number was N3011. The 12-nt oligomers with modifications (see below) and λ DNA for AFM imaging were from the Sino-America Biotechnology Company in Shanghai of China. For the magnetic tweezers experiments, we used streptavidin-coated magnetic polystyrene microspheres with a diameter of 2.8 μm from Dynal Biotech (Norway). Solutions were made with 18.2 MΩ deionized water purified through the Milli-Q Water Purification System (Millipore Corporation, France). AgNO3 and all the other chemicals are all reagent grade.
In vitro platination
Cisplatin was converted to diaqueous derivative by reacting with two equivalents AgNO3 in solution at room temperature for 24 h in the dark. Then the mixture was centrifuged at 13 000 rpm for 10 min twice to remove the AgCl precipitation thoroughly (27). Reactive Pt(dach)Cl2 solution was prepared in the same way. DNA was incubated with platinum solution of various final concentrations at 37°C in the dark.
DNA construction for single-molecule study
The two 12-nt single-stranded ends of λ DNA molecules were annealed respectively with two 12-nt chemically labeled single-stranded oligomers, 3′-biotin-cccgccgctgga and 3′-digoxygenin-tccagcggcggg, according to the standard procedures (28). Afterwards, streptavidin-coated paramagnetic polystyrene microspheres with a diameter of 2.8 μm were bound to DNA through interaction with biotin (29). DNA molecules carrying a microsphere at one end and digoxygenin at the other end were then ready for use.
Magnetic tweezers setup
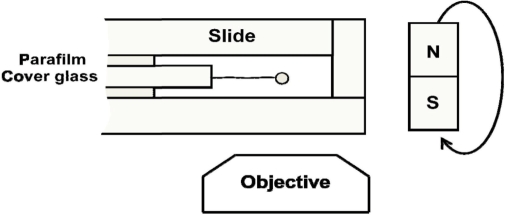
A home-built transverse magnetic-tweezers system was employed in our single-molecule experiments (Figure 1). A single DNA molecule with a paramagnetic microsphere at one end, as constructed above, was ligated to the polished edge surface, covered by anti-digoxygenin, of the sandwiched cover glass in the flow cell. With single biochemical links at its two extremities, the DNA molecule was torsionally unconstrained. DNA molecule was pulled by permanent magnets about 5 mm away at the right side of the flow cell. Applied force can only be altered by changing the distance between the magnets and microsphere through moving the magnets. Besides, rotation of the microsphere could be achieved by rotation of the magnets. The flow cell was placed in an inverted microscope (IX71, Olympus, Japan) which was employed to observe the movement of the microsphere relative to the polished edge surface of the cover glass in the flow cell. A CCD camera (DV885, Andor Technology) was used to record the position of the microsphere in real-time. DNA extension was determined as the distance between the microsphere and the polished edge surface of the cover glass by a homemade program. The applied force was calculated according to the position fluctuations of the microsphere in the direction perpendicular to the DNA extension (30).The buffer used in all the single-molecule experiments was 10 mM Tris–HCl, pH 7.5, with 5 mM NaCl.
Figure 1.
Experimental setup for transverse magnetic tweezers. The flow cell is constructed by sandwiching a 0.17 mm thick cover glass between two slides. The applied force is controlled by changing the distance between the magnetic microsphere and the magnets. DNA can also be rotated by rotating the magnets.
Single-molecule measurement
We carried out the single-molecule experiments with the magnetic tweezers in the following steps.
(i) Ligated the microsphere-bound DNA molecules to the polished edge surface of the cover glass in flow cell. Then rinse the cell with buffer to clean out the free particles.
(ii)Grabbed a ligated particle and confirm that it was connected to the surface through a single DNA molecule. First, the extension of the λ DNA should be close to 16.5 μm under a high tension (20 pN). Next, we set the applied force to about 0.2 pN, and rotated the particle for 400 turns. If no shortening was observed, there should be only one DNA molecule ligated. Then, for further confirmation, a force versus extension measurement was employed. By fitting the data with the WLC model, a persistence length at 52 ± 2 nm and a contour length at 16.5 ± 0.5 μm should be obtained (31). In each experiment, a molecule grabbed was checked to be single in the above way.
The persistence length and contour length of DNA were obtained by fitting the force versus DNA extension curves with the WLC model (32),
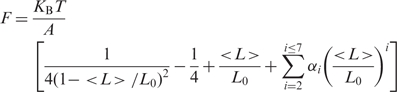 |
1 |
where F is the stretching force, L is the DNA extension, A and L0 are the persistence length and the contour length of the DNA molecule, respectively. The coefficients used are α2 = −0.5164228, α3 = −2.737418, α4 = 16.07497, α5 = −38.87607, α6 = 39.49944 and α7 = −14.17718.
(iii) Carried out the WLC behavior measurement. First, flow cisplatin at a given concentration to the flow cell. During this process, DNA was kept stretched under 20 pN force. Next, adjust the applied force. Then record the position of the microsphere in real-time. After the DNA extension becoming stabilized, perform force versus extension measurement (33).
For measuring the change of the DNA persistence length with time in the presence of cisplatin, after the DNA was tethered successfully to the surface, and confirmed to be a single molecule, 77 μM cisplatin was slowly injected to the flow cell. Then we repeated the force versus extension measurement at different times during a total time interval of more than 12 h. DNA was kept stretched under high tension between two consecutive measurements.
(iv) Carried out DNA shortening measurement. After the DNA ligation, and confirmed to be single molecule, 770 μM cisplatin was injected to the flow cell while 20 pN force was used to keep DNA under high tension. Afterwards, we adjusted the magnetic force to a lower value so that the DNA molecule could exist with a high flexibility. DNA extension was recorded in real-time. In control experiment, DNA was always stretched by a force of 20 pN. By comparing the different results at low and high forces, the influence of DNA thermal fluctuation on its interaction with cisplatin may be seen.
AFM sample preparation and imaging
Free unmodified λ DNA was incubated with cisplatin following the standard procedure described above. All manipulations were carried out in 10 mM Tris–HCl, pH 7.5. The concentration of DNA was 50 ng/μl.
To study the change of DNA configuration with time by AFM imaging, a 10 μl solution of 50 ng/μl DNA and 77 μM cisplatin (the cisplatin: nucleotide ratio is about 0.5) or 770 μM cisplatin (the cisplatin:nucleotide ratio is about 5) was incubated at 37°C in the dark. An aliquot of 1 μl was taken out at the indicated times and diluted to a DNA concentration of 1 ng/μl for AFM scanning. In the study of Pt(dach)Cl2, the experiments were performed in the same way as above.
We have employed two different techniques to adsorb DNA molecules onto mica surface.
(i) To stretch DNA on mica to an extended morphology for AFM imaging, we used one kind of molecular combing method: one freshly cleaved mica was immobilized on an inclined plane, then a 10 μl droplet with 1 ng/μl DNA and 1 mM Ni2+ was deposited on the top of the surface (34). After the droplet flowed from the surface top to bottom, the extra solution was absorbed away tenderly with a micropipette.
(ii) To adsorb DNA on mica surface with their natural configuration (35), samples were prepared by depositing a 10 µl droplet of 1 ng/μl DNA in 10 mM Tris–HCl (pH 7.5) and 5 mM Mg2+ onto freshly cleaved mica. After 5 min, mica surface was washed with 200 µl Milli-Q filtered water for several times and blown dry in a gentle stream of nitrogen gas.
The imaging was performed in air with a multi-mode AFM with nanoscope IIIa controller (Digital Instruments, Santa Barbara, CA, USA) in the tapping-mode. Silicon probe RTESP14 from Veeco (America) was employed, with a resonance frequency of 315 kHz. ‘E’ scanner was used. The scan frequency was 1 Hz per line, and the scan size was from 1 to 5 μm. DNA tracing and measurement were done semi-automatically using Image J software.
RESULTS
WLC elastic behavior
The WLC model describes the elastic behavior of duplex DNA under tension, where DNA is treated as a semi-flexible chain, and its flexibility is reflected by the persistence length (32). The contour length is another parameter, corresponding to the maximum DNA length when it is inextensible under high tension. For a single λ DNA molecule, previous experiments revealed a persistence length of 45–50 nm (21) and a contour length of about 16.5 μm. Both static bending and thermally induced dynamic bending may result in the change of persistence length, and Yan et al. (36) have systemically reported the theoretical modeling results of DNA persistence length reduction by DNA-binding proteins as well as intercalators with various binding modes.
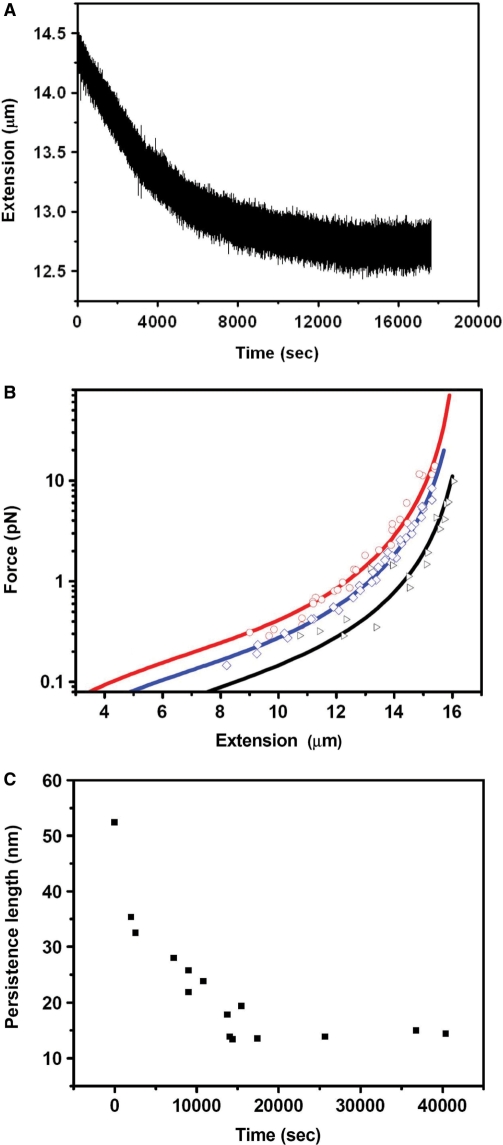
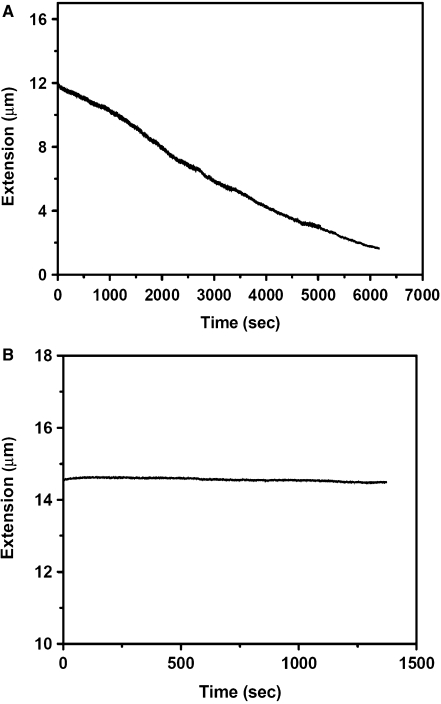
In our experiment, we first recorded the extension change of DNA with time after cisplatin was added. Figure 2A shows that at 77 μM cisplatin, the DNA extension gradually shortens with time, and after about 4 h, it reaches a steady state. Then we performed the force versus extension measurements. Figure 2B shows typical results for DNA alone (as a control), DNA interacting with 77 μM cisplatin and with 15.4 μM cisplatin. From the WLC fitting according to Equation 1, the obtained persistence lengths are 52.4, 15.0 and 25.0 nm, respectively. Their contour lengths obtained from the fitting are all close to 16.5 µm (16.4, 16.6 and 16.6 μm, respectively). In addition, a force versus extension measurement was also performed for DNA with 46.2 μM cisplatin. The final persistence length is 20.6 nm, between that for 77 and 15.4 μM cisplatin. It is clear that when DNA was treated with cisplatin for a plenty of time, its persistence length was decreased, while the contour length showed little variation.
Figure 2.
Single-molecule experiments under low concentration of cisplatin. (A) After 77 μM cisplatin was added to the flow cell, the force exerted on DNA was adjusted to 1.3 pN, and the end to end distance was recorded in real-time. After about 4 h, DNA extension reaches a steady state. (B) The force versus extension curves for bare DNA (black), DNA treated by cisplatin of 77 μM (red) and 15.4 μM (blue) after the extension reached the steady states. The lines are the best fits of the data to Equation 1. (C) The change of persistence length with time when 77 μM cisplatin was added to DNA in the flow cell. Persistence length was obtained from fitting of the force versus extension curves to Equation 1 measured at the indicated times. After about 4 h, the persistence length reaches a steady state.
To further study the decrease of DNA persistence length with time, we introduced 77 μM cisplatin to DNA in flow cell, and repeated measuring the force versus extension curves at different times. From fitting of the curves with Equation 1, we obtained contour lengths that did not change much with time (data not shown). The persistence length, however, decreased rapidly during the first 4 h and then became stabilized (Figure 2C), with the final value (∼14.9 nm) being in agreement with the result of direct measurement after DNA extension becoming stabilized (Figure 2B). This behavior of persistence change is not difficult to understand: at the beginning, there are quite a lot of binding sites on DNA for cisplatin, thus the number of bound cisplatin molecules increases with time, resulting in continuous variation of the DNA structure. After the binding sites are becoming saturated with cisplatin, cisplatin-binding and thus DNA structure change are slowed down and finally stabilized.
As to why cisplatin-binding changes the structure and persistence length of DNA, we need to consider the binding mode of cisplatin to DNA. We know cisplatin makes intrastrand crosslinks between 1,2-d (GpG), 1,2-d (ApG), 1,3-d (GpNpG) of DNA, as well as a few interstrand crosslinks, and distorts DNA to the major groove about 50° (9,10). According to Yan et al.'s theory (36), this kind of local bending will induce a decreased persistence length. In fact, there are also other DNA-binding ligands capable of altering DNA persistence length with various binding modes. For instance, single molecule experiments have revealed that the minor groove binder distamycin-A, the major groove binder α-helical peptide, and the DNA intercalator EtBr reduce the DNA persistence length to 26.7, 29.4 and 20.7 nm, respectively (22). Our present work is the first quantitative report of the effect of cisplatin on DNA persistence length measured with single molecule method.
To observe more directly the effect of cisplatin on DNA structure, we then used AFM scanning to image DNA in the presence of 77 µM cisplatin.
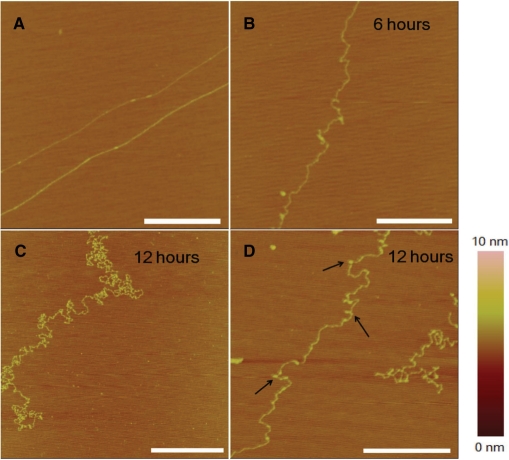
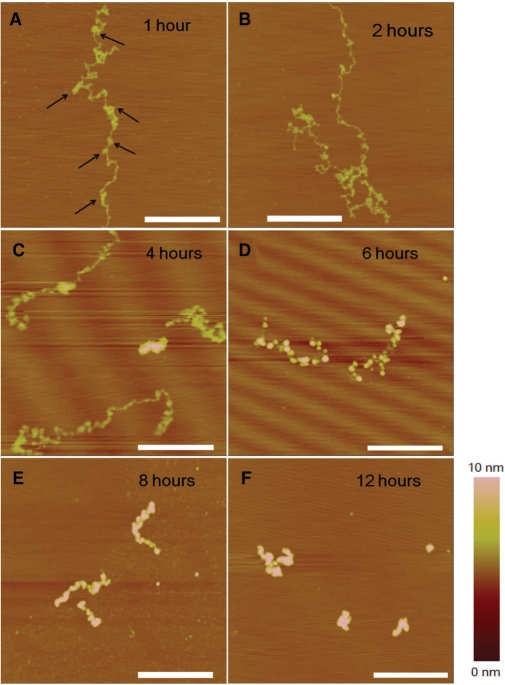
By using the method as described in the Materials and methods section, λ DNA molecules could be stretched linearly on the mica surface as shown in Figure 3A. This indicates that the force exerted by the withdrawing interface between mica and the liquid is strong enough. In the presence of cisplatin, after an incubation of 6 h, some local kinks appeared (Figure 3B), corresponding to the formation of adducts which bend DNA. This kind of DNA morphology is in agreement with the single-molecule observation which revealed a reduced persistence length. After 12 h, DNA coiling became more serious (Figure 3C and D). When observing carefully, we could see many tiny globules with a typical size of ∼20 nm and an average height of ∼1 nm (note that a single dsDNA strand on mica surface is about 0.5 nm in height). We assume that this kind of structures are micro-loops, formed in a mode different from the crosslinks of adjacent GpG, ApG or GpNpG bases. The identification of this kind of structures and mechanism of their formation will be discussed later.
Figure 3.
AFM images of λ DNA alone or incubated with 77 μM cisplatin at different times. (A) Unmodified DNA in the absence of cisplatin. The DNA was stretched on mica surface (see Materials and methods section). (B) In the presence of cisplatin, 6 h incubation. (C) and (D) 12 h incubation. DNA coiling became more serious, accompanied by formation of tiny structures (micro-loops, as indicated by arrows). All scale bars are 500 nm.
Actually, we have also scanned λ DNA molecules incubated with cisplatin of lower concentrations (1 and 15.4 µM) for 12 h. We observed that DNA was comparatively extended with only several local kinks under the condition of 1 µM cisplatin, while the DNA took a shape like that in Figure 3C with 15.4 µM cisplatin (data not shown).
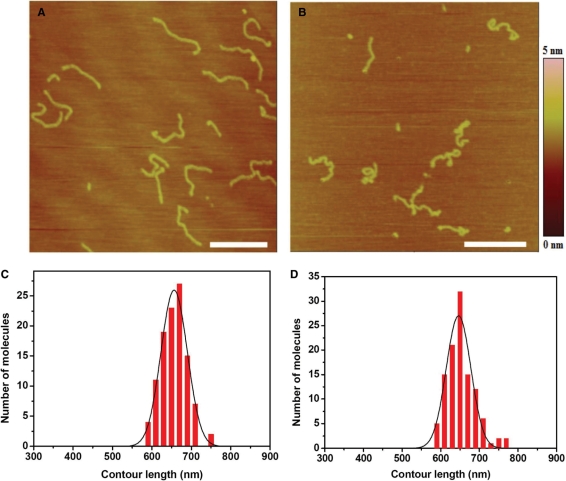
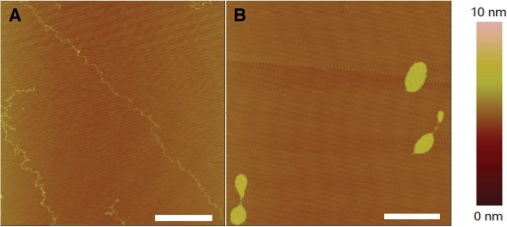
Furthermore, to study the change of DNA contour length after interaction with cisplatin by AFM, we carried out experiments with 2 kb DNA of which the contour was easy to trace (37). These 2 kb double-strand fragments were produced by PCR using λ DNA as a template and 5′-tggtcgttcagggttgtcgga-3′ and 5′-cgccttgccctcgtctatgta-3′ as primer sequences, then purified by gel extraction. Only linear DNA without loops on mica surface were measured. As the theoretical contour length of 2 kb DNA is ∼680 nm, we regarded DNA molecules much less than 680 nm as incomplete ones that were not included in the length statistics. The average contour lengths of the free and cisplatin-treated DNA on mica surface were 656.8 and 653.1 nm, respectively (Figure 4). It is clear that cisplatin just bends DNA backbone, and does not affect the contour length much, in agreement with the results for λ DNA obtained with magnetic tweezers.
Figure 4.
(A) AFM image of free 2 kb DNA adsorbed onto mica surface. (B) AFM image of 2 kb DNA treated by 15.4 µM cisplatin for 1 h. All scale bars are 500 nm. To avoid the influence of molecular combing on DNA length, samples were made by adsorption with Mg2+ and without combing according to the second technique for preparing AFM samples as indicated in Materials and methods section. (C, D) Distribution of lengths measured for free DNA molecules and molecules treated by 15.4 µM cisplatin. The mean contour lengths are 656.8 ± 31.5 nm (average of 108 molecules) and 653.1 ± 41.0 nm (average of 111 molecules), respectively, for the two cases of free and cisplatin-treated DNA.
DNA shortening
DNA-shortening effect has been previously reported in literatures studying cisplatin–DNA interaction. In one case, molecules of 260-bp DNA incubated with cisplatin in the dark at 37°C for 24 h was scanned by AFM and shown to have a 30% decrease in contour length (38). Also, in a quite early work in 1978, EM pictures revealed a DNA shortening phenomenon when a high concentration of the drug was used (39). Although this shortening effect has been observed, a reasonable explanation was still lacking. Moreover, this kind of structure changes is rarely mentioned in literatures studying the binding mode of cisplatin to DNA. Here we used both single-molecule technique and AFM imaging method to study the mechanism of DNA shortening that occurred at high cisplatin concentration.
First, we incubated λ DNA with cisplatin of high concentration (∼513 μM) for 12 h. Then the solution was introduced into the flow cell. After DNA ligation, we stretched the DNA by the magnetic tweezers. It was observed that almost all DNA molecules exhibited surprisingly short lengths (data not shown). Even the highest force (20 pN) could only stretched most of them to lengths of less than 7 μm. The longest one we observed was 11.4 μm, still much less than λ DNA's contour length (16.5 μm). These results are different from those observed in the previous experiments at low cisplatin concentration, where the contour length was almost invariable. Thus we believe different new structures may be formed at high drug concentration.
To observe the DNA shortening process more quantitatively, we then carried out another experiment. As described in the Materials and methods section, λ DNA was constructed in the flow cell. After drug addition with a final concentration 770 μM, a low force (∼1 pN) was used to stretch the single DNA molecule. At this low force, DNA was in a flexible state. Under this condition, we observed that the DNA extension decreased rapidly to less than 3 μm in about 100 min due to its interaction with cisplatin (Figure 5A). Note that the DNA had been checked to be intact after ligation according to the reasonable persistence length and contour length. However, its extension under ∼1 pN was less than the normal value of 14 μm at the beginning. This should be due to the fact that cisplatin–DNA interaction already occurred to some extent during the process of drug flowing and force adjustment. The experiment was repeated several times while the results were similar.
Figure 5.
Single-molecule experiments under high cisplatin concentration (770 μM). (A) A typical variation of the DNA extension. The external force exerted on the DNA was ∼1 pN. The sharp decrease corresponded to the time of drug flow. (B) Control experiment where DNA was kept stretched by a high force (20 pN) close to its contour length. Although 770 μM cisplatin was added, DNA hardly shortened.
We found that the shortening process as shown in Figure 5A was irreversible: even a high force (20 pN) could only stretch the condensed DNA to a maximum of ∼11 μm, much less than its full contour length of 16.5 μm. As a control, we repeated the experiment while using a high force (20 pN) from the beginning. In this case, we observed that the DNA was unable to shorten in the presence of the high concentration cisplatin (Figure 5B).
The above results indicate that the new structures that cause DNA shortening cannot be formed at a high stretching force (20 pN). But once they are formed at a low force, they cannot be disrupted by a 20 pN high stretching force.
To determine the new DNA structures that formed accompanying DNA shortening, AFM scanning of DNA was carried out at the cisplatin concentration of 770 μM. The measurements were carried out at different incubation times.
After an incubation of 1 h, DNA became curled (Figure 6A), in contrast with the extended structure on mica surface in the absence of drug (Figure 3A). Actually, this kind of coiling was different from that observed at low cisplatin concentration (77 μM, Figure 3). Here, besides local distortions such as kinks, local condensations were also presented, as indicated by arrows. With incubation time increasing, DNA became coiled more and more compactly, possessing more and more local structures (Figure 6B and C). After an incubation of 6 h or longer, the DNA molecules took a shape that was a collection of small aggregates (Figure 6D and E). Finally, after an incubation of 12 h, DNA was condensed to a globule with a typical height of ∼6 nm and a width of ∼100 nm (Figure 6F).
Figure 6.
AFM images of DNA incubated with 770 μM cisplatin. (A) Incubation of 1 h, the arrows indicate the local condensations; (B) 2 h; (C) 4 h; (D) 6 h; (E) 8 h and (F) 12 h. All scale bars are 500 nm.
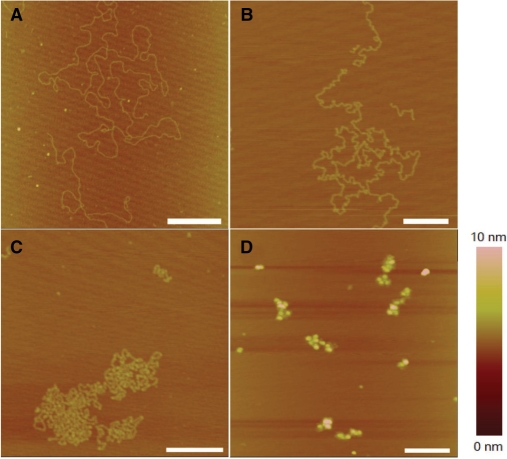
To better understand the change of DNA structures, we also carried out experiments with the method of naturally adsorbing DNA onto mica surface by Mg2+, without combing. Mg2+ provides weak adsorption between DNA and mica, thus DNA can diffuse in 2D on the surface and get to equilibrium state finally (40). It is a useful way to observe DNA molecules without strong influence from the surface. The AFM pictures are presented in Figure 7. By comparing the results of molecular combing with natural adsorption, we find that: (i) for DNA with local kinks at low cisplatin concentrations (77 μM), molecular combing just makes DNA less coiled, without affecting the local kink structures much (see Figures 3B and 7B); (ii) at high cisplatin concentrations (770 μM) and when DNA's morphology becomes more complex (Figures 6A and 7C), DNA appears, with the natural adsorption method, more coiled and more compact while showing no more structures except for local kinks. In combination with the results from molecular combing (Figure 6A), it can be confirmed that long-distance crosslinks do exist (Figure 7C); (iii) at high cisplatin concentrations (770 μM) and for long incubation time (Figures 6D–6F and 7D), the two methods give similar results because the DNA structures are too compact to be opened by molecular combing. In brief, the two AFM methods may let us obtain similar conclusions about the structural changes of DNA in the presence of cisplatin.
Figure 7.
AFM images of λ DNA adsorbed naturally onto mica surface by 5 mM Mg2+. (A) Untreated (free) DNA. (B) DNA treated by 77 µM cisplatin for 6 h. (C) DNA treated by 770 µM cisplatin for 1 h. (D) DNA treated by 770 µM cisplatin for 6 h. All scale bars are 500 nm.
It is worth mentioning that the experimental results in the bulk (Figure 6) are somewhat different from the single molecule results (Figure 5) in terms of the condensation speed. The same concentration of drug makes the DNA condensate more rapidly in the single molecule experiment. The reason should be that the ratio of cisplatin to DNA molecules is higher in the latter case.
Effect of oxaliplatin on DNA structure
To see if the DNA structural changes under the action of cisplatin as observed above are inducible by another similar anti-cancer drug oxaliplatin, we next carried out similar AFM studies with Pt(dach)Cl2. We used Pt(dach)Cl2 instead of oxaliplatin itself in the experiment because it induces the same adduct structure as oxaliplatin while it is more active in DNA-binding in vitro.
Oxaliplatin is the third generation of the platinum anti-cancer drug, which demonstrates anti-tumor activity in cell lines with acquired cisplatin resistance (41). Oxaliplatin has a non-hydrolyzable DACH carrier ligand instead of diammine compared to cisplatin, and produces the same kind of inter- or intra-strand crosslinks as cisplatin, with the 1,2-(GpG) adduct bending the double helix towards the major groove by about 30° (42), somewhat less severe than cisplatin. Here we used AFM imaging to study the structural modification of DNA by Pt(dach)Cl2 and to see if this kind of platinum anti-cancer drug analogous to cisplatin in structure altered the DNA structure in a similar manner as cisplatin.
We incubated DNA with Pt(dach)Cl2 at a concentration of 77 or 770 μM according to the standard procedure. After an incubation of 12 h, AFM imaging was then performed in the same way as for cisplatin. Under the condition of 77 μM drug, DNA looked flexible, with obvious kinks as well as some tiny globules assumed to be micro-loops (Figure 8A). When 770 μM drug was used, DNA was condensed into a compact globule (Figure 8B).
Figure 8.
AFM images of DNA incubated with different concentrations of Pt(dach)Cl2 at 37°C for 12 h. (A) 77 μM drug. (B) 770 μM drug. DNA condensed to a compact globule with a typical size of ∼200 nm and a height of ∼3 nm. The scale bars are 500 nm.
Comparing with previous results of cisplatin, it can be seen that Pt(dach)Cl2 resembles cisplatin in the modification of DNA structures. The difference is that the effect of Pt(dach)Cl2 is less strong than that of cisplatin. Comparing Figure 8A with Figures 3C and D, we can see that DNA is still in a relatively relaxed form after incubation with 77 μM Pt(dach)Cl2 for 12 h. Also, the final globules formed by 770 μM Pt(dach)Cl2 have a height of ∼3 nm and a width of ∼200 nm, thus looser than those formed with 770 μM cisplatin (∼6 nm height and ∼100 nm width, Figure 6F).
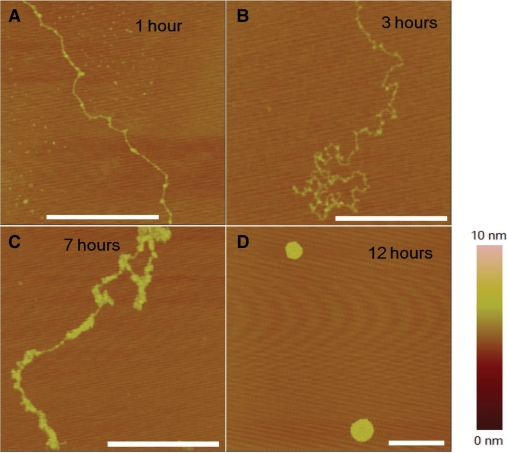
To investigate the mechanism of Pt(dach)Cl2–DNA interaction, we further carried out another experiment to observe the structural evolvement with time. Total 770 μM drug was used. As can be seen, first, some kinks of DNA appeared (Figure 9A). Then the DNA became curled on the mica surface with several visible micro-globules (Figure 9B), then the small structures developed to large aggregates (Figure 9C), and finally the molecule was condensed into a compact globule (Figure 9D), similar to the DNA condensing process in the presence of cisplatin (Figure 6).
Figure 9.
AFM images of DNA incubated with 770 μM Pt(dach)Cl2 with different times. (A) 1 h; (B) 3 h; (C) 7 h; (D) 12 h. The scale bars are 500 nm.
From the above experimental observations, we come to the conclusion that Pt(dach)Cl2 induces kinks to DNA helix, makes DNA more flexible, and leads to formation of micro-loops and bigger structures in the same way as cisplatin. The difference is that the DNA structure changes induced by Pt(dach)Cl2 are comparatively a little weaker, in agreement with the adduct's crystal structure (42).
DISCUSSION
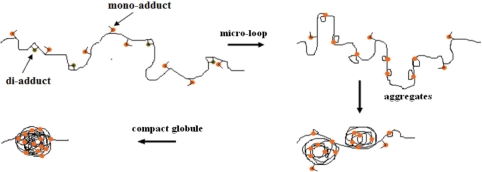
Combining the experimental results obtained with both single molecule and AFM imaging methods, we propose a new model for DNA shortening by cisplatin-binding (Figure 10). As is well known, cisplatin forms both di-adduct such as GpG, ApG and mono-adduct. In the latter case, the mono-adduct is still free to bind to other nucleotides. Since DNA may fluctuate thermally in solution, bases distant from each other may be brought to short distances. Thus if one nucleotide possesses a mono-adduct, a micro-loop might be formed through binding of the mono-adduct to the other distant nucleotide. This kind of micro-loops should be just what we observed at 77 μM cisplatin (Figure 3C and D). When a high concentration of cisplatin is used, these small loops may be further crosslinked, leading to the shortened morphology and finally the rather compact structure (Figure 6). According to the above model, it is expected that crosslinks between different DNA molecules may happen in the same way as internal crosslinks when DNA concentration is high. Indeed, at the DNA concentration of 50 ng/μl as used in our AFM experiments, we have observed such crosslinks (data not shown). But this phenomenon should not affect our conclusion about the effect of cisplatin on DNA structure and condensation.
Figure 10.
Proposed model for DNA shortening by cisplatin. Both di- and mono-adducts are formed. Due to the thermal fluctuation, micro-loops might occur through long-distance crosslinks. Then further distant crosslinks make the DNA condense to a collection of aggregates, and finally, the DNA is compacted to a globule.
Theoretical work on loop formation of DNA reveals that kinks can not only reduce the loop size severely from 550 to 110 bp, but also increase the probability of loop formation dramatically (43). Therefore, those di-adducts by adjacent crosslinks may also play an important role in promoting the loop formation, explaining why so many loops formed in our experiments.
Clearly, when the DNA is stretched by a high force, the thermal fluctuation cannot overcome the tension on DNA, thus it is expected that the probability is low for micro-loops to form. This was supported by the control experiment, where the DNA molecule was kept in a stretched form and 770 μM cisplatin was unable to condense DNA to short length (Figure 5B). The DNA extension remained almost constant during the whole recording time, indicating no particular structure was formed.
Similar kind of DNA-shortening effect has been reported for the DNA-binding protein Fis which bends DNA in a nonspecific manner (44). Single-molecule experiments revealed that the DNA condensation occurred abruptly when DNA tension was reduced to a protein-concentration-dependent threshold of less than 1 pN. The authors suggested that the condensation happened via Fis stabilizing DNA self-crossing generated by thermal fluctuation. Note that, in their case, 9 pN force could entirely disentangle the condensed DNA, which is different from our present case where the micro-loops and big structures formed by cisplatin were difficult to be opened even under a high tension of about 20 pN.
Actually, the 770 μM cisplatin concentration we used is too high and it is only practical for the in vitro experiment. We chose to use such a high concentration in order to accelerate the drug interaction with DNA. According to our studies, there are mainly two factors that influence the formation of loop structures. One is the drug concentration and the other is the incubation time. We identified with AFM that DNA incubated with 15.4 μM cisplatin for even 48 h showed no micro-loops except for local kinks (data not shown). With 77 μM cisplatin and an incubation time of 6 h, there were hardly any micro-loops either (Figure 3B). After an incubation of 12 h, quite a lot of micro-loops were formed, but bigger structures were still absent (Figures 3C and D). However, with 770 μM cisplatin, even 1 h incubation time could produce quite a lot of new structures (Figure 6A).
From our observations, it is obvious that the thermal fluctuation of DNA is quite important for the formation of micro loops and DNA condensation. In the WLC measurement, DNA was under tension during most of the time, thus loop formation was suppressed, resulting in a basically invariable contour length. In addition, for a short DNA (comparable to its persistence length), it is difficult to bend severely, thus is unable to form the loop structures. This should just be what happened in the previous works (38). We assume the formation of micro-loop structures to be an important property for cisplatin–DNA interaction with drug concentration of tens of micromoles per liter and an incubation time of about 12 h. Previously, it was generally believed that cisplatin modified the DNA structures through intrastrand and interstrand crosslinks between adjacent bases, our present study further shows that distant crosslinks should also play important roles in the drug–DNA interaction.
Our AFM imaging experiments with Pt(dach)Cl2 suggest that oxaliplatin interacts with DNA in a similar way as cisplatin. Besides oxaliplatin, there are also several other platinum anti-cancer drugs that resemble cisplatin in structure, such as carboplatin (45), JM118 (46) and ZD0473 (47). They all bind to double helix, make intrastrand and interstrand crosslinks, and distort DNA towards the major groove in a similar manner. Based on the studies of cisplatin and oxaliplatin, it is reasonable to think that these similar platinum anti-cancer drugs modify DNA structures in the same two-mode manner: (i) first, the drug induces interstrand or intrastrand crosslinks of adjacent bases, making DNA more flexible. Thus, in the single molecule analysis using a magnetic tweezers, DNA exhibits a decreased persistence length, and in AFM imaging DNA shows a lot of local kinks. (ii) Then, the drug induces formation of loops through crosslinks of distant nucleotides brought together by DNA thermal fluctuation. At low drug concentrations, DNA exhibits a lot of micro loops, while at extremely high drug concentrations, the structures with micro loops will be further crosslinked to form collection of large aggregates and finally the DNA is condensed to a compact globule.
CONCLUSIONS
Mechanical properties of single λ DNA molecules in the presence of the anti-cancer drug cisplatin were investigated by a magnetic tweezers. Compared with free λ DNA, the modified DNA was more flexible, with its persistence length decreased from ∼52 to ∼15 nm. This behavior is in agreement with the previously known binding mode of cisplatin: forming intrastrand and interstrand crosslinks and distorting the double helix towards the major groove. The permanent static bending causes a decrease of the WLC persistence length. AFM imaging of λ DNA treated by cisplatin of the same concentration revealed more directly the curled structure of DNA with a lot of local kinks. Besides, AFM imaging also showed that the contour length of DNA was not affected obviously by cisplatin.
Importantly, we have found another behavior of cisplatin–DNA interaction: the formation of loop structures due to DNA thermal fluctuation and then DNA condensation. In the single molecule experiments using the magnetic tweezers, the extension of DNA decreased rapidly upon addition of a high concentration cisplatin. The condensed DNA was unable to be stretched back to its contour length even under a high force (20 pN). AFM scanning revealed the processes of DNA condensation. First, micro-loops with a size of ∼20 nm appeared, caused by crosslinks of mono-adducts with distant binding sites due to DNA thermal fluctuation as well as a reduced persistence length. Then large aggregates were formed. Finally DNA was condensed into a large compact globule.
Pt(dach)Cl2 was also analyzed by AFM imaging. Our experiments reveal that Pt(dach)Cl2 (or oxaliplatin) makes DNA flexible and induces DNA loop structures in the same manner as cisplatin, though a little weaker.
We think the occurrence of loop structures may be an important feature of DNA modification by this kind of platinum anti-cancer drugs. The present study should be useful for further understanding of the biological function of the platinum anti-cancer drugs and for the design of new drugs in the future.
FUNDING
This research was supported by the National Natural Science Foundation of China and the Innovation Project of the Chinese Academy of Sciences. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the reviewers’ helpful comments and suggestions.
REFERENCES
- 1.Chaney SG, Campbell SL, Temple B, Bassett E, Wu BY, Faldu M. Protein interactions with platinum-DNA adducts: from structure to function. J. Inorg. Biochem. 2004;98:1881–1559. doi: 10.1016/j.jinorgbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Pinto AL, Lippard SJ. Binding of the antitumor drug cis-diamminedichloroplatinum(II) (cisplatin) to DNA. Bioschim. Biophys. Acta. 1985;780:167–180. doi: 10.1016/0304-419x(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 3.Bruhn SL, Toney JH, Lippard SJ. Biological processing of DNA modified by platinum compounds. Prog. Inorg. Chem. 1990;38:477–516. [Google Scholar]
- 4.Bancroft DP, Lepre CA, Lippard SJ. 195Pt NMR kinetic and mechanistic studies of cis- and trans-diamminedichloroplatinum (II) binding to DNA. J. Am. Chem. Soc. 1990;112:6860–6871. [Google Scholar]
- 5.Johnson NP, Hoeschele JD, Rahn RO. Kinetic analysis of the in vitro binding of radioactive cis- and trans-dichlorodiammineplatinum (II) to DNA. Chem. Biol. Interact. 1980;30:151–169. doi: 10.1016/0009-2797(80)90122-2. [DOI] [PubMed] [Google Scholar]
- 6.Vaisman A, Chaney SG. The efficiency and fidelity of translesion synthesis past cisplatin and oxaliplatin GpG adducts by human DNA polymerase β. J. Biol. Chem. 2000;275:13017–13025. doi: 10.1074/jbc.275.17.13017. [DOI] [PubMed] [Google Scholar]
- 7.Jung YW, Lippard SJ. Multiple states of stalled T7 RNA Polymerase at DNA lesions generated by platinum anticancer agents. J. Biol. Chem. 2003;278:52084–52092. doi: 10.1074/jbc.M310120200. [DOI] [PubMed] [Google Scholar]
- 8.Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutat. Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 9.Takahara PM, Frederick CA, Lippard SJ. Crystal structure of the anticancer drug cisplatin bound to duplex DNA. J. Am. Chem. Soc. 1996;118:12309–12321. [Google Scholar]
- 10.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 11.Gelasco A, Lippard SJ. NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry. 1998;37:9230–9239. doi: 10.1021/bi973176v. [DOI] [PubMed] [Google Scholar]
- 12.Stehlikova K, Kostrhunova H. DNA bending and unwinding due to the major 1,2-GG intrastrand cross-link formed by antitumor cis-diamminedichloroplatinum(II) are flanking-base independent. Nucleic Acids Res. 2002;30:2894–2898. doi: 10.1093/nar/gkf405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatch K, Danilowicz C, Coljee V, Prentiss M. Measurement of the salt-dependent stabilization of partially open DNA by Escherichia coli SSB protein. Nucleic acids Res. 2007;36:294–299. doi: 10.1093/nar/gkm1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pant K, Karpel RL, Williams MC. Kinetic Regulation of Single DNA Molecule Denaturation by T4 Gene 32 Protein Structural Domains. J. Mol. Biol. 2003;327:571–578. doi: 10.1016/s0022-2836(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 16.McCauley M, Hardwidge PR, Maher LJ, Williams MC. Dual Binding Modes for an HMG Domain from Human HMGB2 on DNA. Biophys. J. 2005;89:353–364. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci.USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffar Ali BM, Amit R, Braslavsky I, Oppenheim AB, Gileadii O, Stavans J. Compaction of single DNA molecules induced by binding of integration host factor (IHF) Proc. Natl Acad. Sci. USA. 2001;98:10658–10663. doi: 10.1073/pnas.181029198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amit R, Oppenheim AB, Stavans J. Increased Bending Rigidity of Single DNA Molecules by H-NS, a Temperature and Osmolarity Sensor. Biophys. J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krautbauera R, Popeb LH, Schradera TE, Allenb S, Gaub HE. Discriminating small molecule DNA binding modes by single molecule force spectroscopy. FEBS Lett. 2002;510:154–158. doi: 10.1016/s0014-5793(01)03257-4. [DOI] [PubMed] [Google Scholar]
- 21.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sischka A, Toensing K, Eckel R, Wilking SD, Sewald N, Ros R, Anselmetti D. Molecular mechanisms and kinetics between DNA and DNA ligands. Biophys. J. 2005;88:404–411. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu WB, Wang XL, Zhang XH, Ran SY, Yan J, Li M. Compaction dynamics of single DNA molecules under tension. J. Am. Chem. Soc. 2006;128:15040–15041. doi: 10.1021/ja064305a. [DOI] [PubMed] [Google Scholar]
- 24.Edwardson JM, Henderson RM. Atomic force microscopy and drug discovery. Drug Discov. Today. 2004;9:1359–1364. doi: 10.1016/S1359-6446(03)02905-2. [DOI] [PubMed] [Google Scholar]
- 25.Krautbauer R, Clausen-Schaumann H, Gaub H. Cisplatin changes the mechanics of single DNA molecules. Angew. Chem. Int. Ed. 2000;39:3912–3915. doi: 10.1002/1521-3773(20001103)39:21<3912::AID-ANIE3912>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Park JS, Jeong B, Lee KJ, Hong SC, Hyon JY, Hong S. The elastic property of a single DNA molecule cross-linked by cisplatin: a magnetic tweezers study. J. Korean Phys. Soc. 2006;49:963–967. [Google Scholar]
- 27.Wei M, Cohen SM. Effects of spectator ligands on the specific recognition of intrastrand platinum-DNA cross-links by high mobility group box and TATA-binding proteins. J. Biol. Chem. 2001;276:38774–38780. doi: 10.1074/jbc.M106374200. [DOI] [PubMed] [Google Scholar]
- 28.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 29.Strick TR, Allemand J-F, Bensimon D, Croquette V. Behavior of Supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strick TR, Allemand J-F, Bensimon D, Bensimon A, Croquette V. The Elasticity of a Single Supercoiled DNA Molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 31.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 32.Bouchiat C, Wang MD, Allemand J-F, Strick T, Block SM, Croquette V. Estimating the Persistence Length of a Worm-Like Chain Molecule from Force-Extension Measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abels JA, Moreno-Herrero F, van der Heijden T, Dekker C, Dekker NH. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys. J. 2005;88:2737–2744. doi: 10.1529/biophysj.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZG, Li Z, Zhou HL, Wei G, Song YH, Wang L. Imaging DNA molecules on mica surface by atomic force microscopy in air and in liquid. Microsc. Res. Tech. 2005;66:179–185. doi: 10.1002/jemt.20156. [DOI] [PubMed] [Google Scholar]
- 35.Allen MJ, Bradbury EM, Balhorn R. AFM analysis of DNA-protamine complexes bound to mica. Nucleic Acids Res. 1997;25:2221–2226. doi: 10.1093/nar/25.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan J, Marko JF. Effects of DNA-distorting proteins on DNA elastic response. Phys. Rev. E. 2003;68:011905. doi: 10.1103/PhysRevE.68.011905. [DOI] [PubMed] [Google Scholar]
- 37.Moreno-Herrero F, Seidel R, Johnson SM, Fire A, Dekker NH. Structural analysis of hyperperiodic DNA from Caenorhabditis elegans. Nucleic Acids Res. 2006;34:3057–3066. doi: 10.1093/nar/gkl397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onoa GB, Cervantes G, Moreno V, Prieto MJ. Study of the interaction of DNA with cisplatin and other Pd(II) and Pt(II) complexes by atomic force microscopy. Nucleic Acids Res. 1998;26:1473–1480. doi: 10.1093/nar/26.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macquet JP, Butour JL. Modifications of the DNA secondary structure upon platinum binding: a proposed model. Biochimie. 1978;60:901–914. [Google Scholar]
- 40.Rivetti C, Guthold M, Bustamante C. Scanning Force Microscopy of DNA Deposited onto Mica: Equilibration versus Kinetic Trapping Studied by Statistical Polymer Chain Analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 41.Raymond E, Faivre S, Chaney S, Woynarowski J, Cvitkovic E. Cellular and molecular pharmacology of oxaliplatin. Mol. Cancer Ther. 2002;1:227–235. [PubMed] [Google Scholar]
- 42.Spingler B, Whittington DA, Lippard SJ. 2.4 Å crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 43.Sankararaman S, Marko JF. Formation of loops on DNA under tension. Phys. Rev. E. 2005;71:021911. doi: 10.1103/PhysRevE.71.021911. [DOI] [PubMed] [Google Scholar]
- 44.Skoko D, Yan J, Johnson RC, Marko JF. Low-force DNA condensation and discontinuous high-force decondensation reveal a loop-stabilizing function of the protein Fis. Phys. Rev. Lett. 2005;95:208101. doi: 10.1103/PhysRevLett.95.208101. [DOI] [PubMed] [Google Scholar]
- 45.Knox RJ, Friedlos F, Lydall DA, Roberts JJ. Mechanism of cytotoxicity of anticancer platinum drugs: Evidence that cis-diamminedichloroplatinum(II) and cis-diammine-(1,1-cyclobutanedicarboxylato)platinum(II) differ only in the kinetics of their interaction with DNA. Cancer Res. 1986;46:1972–1979. [PubMed] [Google Scholar]
- 46.Silverman AP, Bu W, Cohen SM, Lippard SJ. 2.4 Å crystal structure of the asymmetric platinum complex {Pt(ammine)(cyclohexylamine)}2+ bound to a dodecamer DNA duplex. J. Biol. Chem. 2002;277:49743–49749. doi: 10.1074/jbc.M206979200. [DOI] [PubMed] [Google Scholar]
- 47.Rogers P, Boxall FE, Allott CP, Stephens TC, Kelland LR. Sequence-dependent synergism between the new generation platinum agent ZD0473 and paclitaxel in cisplatin-sensitive and -resistant human ovarian carcinoma cell lines. Eur. J. Cancer. 2002;38:1653–1660. doi: 10.1016/s0959-8049(02)00107-7. [DOI] [PubMed] [Google Scholar]