Abstract
Stress response gene ATF3 plays a pleiotropic role in determining cell fate in response to mitogenic or stress stimuli. An alternate promoter of the human ATF3 gene (designated P1 in this study) has recently been reported, which is located ∼43.5 kb upstream of the previously reported P2 promoter. We showed here that the P1 promoter is highly conserved between human and mouse and is functional in response to various stimuli, whereas the P1 promoter was dominantly induced by serum and the P2 promoter was more efficiently activated in response to TGF-β and oncogenic HRAS. The P1 promoter contains multiple transcriptional start sites, and the different 5′-UTRs markedly affected their translation in response to stress. In human prostate and Hodgkin Reed–Sternberg cancer cells with elevated expression of ATF3, the P1 promoter was constitutively activated and its chromatin structure was modified into active configuration. The differential usage of alternate promoters of the ATF3 gene at both transcriptional and translational level and the modification of chromatin structure may provide a novel mechanism for expressing ATF3 in determining cell fate during stress response and cancer.
INTRODUCTION
An increasing number of studies demonstrate the existence of alternate promoters for human genes and their differential usage is one of important source for generating protein and regulatory diversity (1,2). For example, transcripts from alternate promoters are translated into distinct protein isoforms when their variant 5′ exons contain alternative ATGs, producing proteins with different N-termini or different proteins. For many genes with multiple promoters, however, no variation in the resulting proteins is generated, when a common downstream exon contains the translation initiation site and generates the same open reading frame (ORF). Some of these alternate promoters have different tissue specificity and developmental activity, such as the CYP19 gene encoding the aromatase P450 (3) or glucokinase gene (4). Furthermore, mRNA variants transcribed from the alternate promoter cause difference of their translational efficiencies, since the variant 5′-UTRs regulate their secondary structure of mRNAs, thus affecting its stability and translation. The usage of alternate promoters of genes has also been reported to play role in stress response (5–8).
Activating transcription factor (ATF) 3 is a member of the ATF/CREB family of basic-leucine zipper (b-Zip) type transcription factors. Its mRNA level is low or undetectable in most cells, but is greatly induced by a variety of stress signal (9–11, and references therein). This response has dual effects on cell fate, such as cell cycle arrest and apoptosis (10–14), or cell survival and proliferation (15–21). We have recently reported that ATF3 is induced downstream of c-myc in serum induction (22), and other laboratories showed that the ATF3 expression is elevated in human prostate (23) and Hodgkin Reed–Sternberg cells (24), and more recently in human breast cancer cells (25). Furthermore, transgenic over-expression of ATF3 in basal epithelial cells causes basal cell carcinoma in mouse (26). ATF3 also plays role as a negative regulator of inflammation (27–29). These data support both physiological and pathological role of ATF3 in regulating the stress response of cells to external stimuli.
The human ATF3 gene is mapped to chromosome 1q32.3 region, and is comprised of five exons spanning ∼16 kb (30). The putative gene promoter, designated P2 in this study, contains a TATA motif and many cis-elements for binding of transcription factors responsible for the regulated gene expression (11,21,30–34). More recently, a large scale and genome-wide analysis of 5′-end sequence of full-length cDNAs has identified a novel upstream alternate promoter of the human ATF3 gene as deposited in Genbank (NM001030287) (35). We also identified the transcripts in human and mouse cells that contain various novel 5′-UTR (DDBJ/EMBL/GenBank with accession numbers AB291910, AB291911 and AB291912), but their functional and biological significance remains elusive.
In this report, we investigated the molecular and functional properties of a novel upstream promoter, designated P1, of the human ATF3 gene. It is shown that the P1 promoter is highly conserved between human and mouse and is differentially used at both transcriptional and posttranscriptional level in stress response. More strikingly, the P1 promoter is constitutively activated in human cancer cells with elevated expression of ATF3. This study for the first time describes evidence for the differential usage of alternate promoters of the human ATF3 gene, providing a novel insight into the molecular mechanism by which ATF3 is induced in stress response and cancer.
MATERIALS AND METHODS
Plasmids, antibodies and reagents
Plasmids encoding human HRAS wild-type and oncogenic mutant 12V were purchased from Addgene, and expression plasmids for oncogenic Ras 61L mutant and HA-Suv39 were gifts from Dr Nevins and Dr Jenuwein, respectively. Antibodies used were as follow: rabbit anti-ATF3 (C19, sc-188), anti-H-Ras (F235, sc-29), anti-RNA-polymerase II (N-20, sc-899) from Santa Cruz, anti-β-actin (clone AC-74, A2228), anti-β-tubulin (clone TUB2.1, T4026), anti-HA (clone HA-7) and anti-Flag (clone M2) from Sigma, anti-Leo-1 (A300-175A) from Bethyl, anti-H3-trimethyl-K4 (H3K4me3, ab8580) and anti-H3-trimethyl-K9 (H3K9me3, ab8898) from abcam, anti-acetyl-histone H3 (06-599) from Upstate. Doxorubicin, thapsigargin, tunicamycin, etoposide were from Sigma, methyl methanesulfonate (MMS) and H2O2 were nachalai tesque, recombinant human TGF-β was from PeproTech. Other chemicals were reagents grade.
Cell culture and treatments
Cell lines used in this study are HCT116 human colon carcinoma cell, U2OS human osteosarcoma cell, LNCaP, DU145, PC3 human prostate carcinoma cell (ATCC), L428 and L540 Hodgkin Reed–Sternberg cell (DSMZ, Germany), DAUDI human B cell and MOLT4 human T cell (RIKEN, Japan), HaCaT human keratinocyte (CLS, Germany). RAW264 is a mouse macrophage cell line (RIKEN, Japan). Cells were cultured in adequate media supplemented with 10% calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin in a 5% CO2 atmosphere at 37°C. For induction by serum or various stimuli, cells were cultured in the presence of 0.5% serum for 48 h and induced by 20% serum, 100 μg/ml MMS or various agents. For expression of oncogene HRAS, neomycin-resistant U2OS cells stably expressing mouse ecotropic retrovirus receptor were infected by pMX retroviral vector encoding oncogenic mutants 12V, 61L, or normal HRAS and selected with 2 μg/ml puromycin. For transient expression, plasmid DNA was vortex-mixed with SuperFect (Qiagen) and transfected into cells according to manufacture's instruction.
Cell extract preparation and western blot analysis
Cells treated as indicated were harvested, washed in PBS, and cell extracts were prepared as described (22). The amounts of protein were measured by Lowry method using bovine serum albumin as standard (36). Cell extracts (20 μg protein) were separated on an SDS–PAGE, transferred onto a nitrocellulose membrane, and subjected to western blot using the protocol of ECL kit (Amersham).
RNA isolation and reverse transcription–PCR
Total RNA was isolated by acid-guanidinium method using a kit from Qiagen. For detecting transcripts from P1 or P2 promoters of ATF3 gene, RT–PCR of 1 μg total RNA was performed using a kit from TaKaRa. Primers used were as follow: human P1 transcript, 5′-AGGATGCTCTGCTGTTTCCT-3′ (forward) and 5′-TTAGCTCTGCAATGTTCCTTC-3′ (reverse); human P2 transcript, 5′-TGATGCAACGCTCTCCAAGC-3′ (forward) and 5′-TTAGCTCTGCAATGTTCCTTC-3′ (reverse); mouse P1 transcript, 5′-AACAGGATCTCCCACAGGGT-3′ (forward) and 5′-GACAAAGGGTGTCAGGTTAG-3′ (reverse); mouse P2 transcript, 5′-CAGTGGAGCCAATCGGCTAA-3′ (forward) and 5′-GACAAAGGGTGTCAGGTTAG-3′ (reverse). Measurements of the ATF3 transcripts from the P1 and P2 promoters or GAPDH were also performed by quantitative RT–PCR. To this end, the efficiency of PCR–amplification of mRNA from each promoter was determined by control reaction using the plasmid DNA for each transcript, and used for calibration (Supplementary Figure S1). The data obtained were further normalized with that of GAPDH. Primers used were as follow: ATF3 (P1 transcript), 5′-AGGATGCTCTGCTGTTTCCT-3′ (forward) and 5′-GACAAAGGGCGTCAGGTTAG-3′ (reverse); ATF3 (P2 transcript), 5′-TGATGCAACGCTCTCCAAGC-3′ (forward) and 5′-CAGAAGCACTCACTTCCGAG-3′ (reverse); GAPDH, 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ (forward) and 5′-TTGATTTTGGSGGGATCTCG-3′ (reverse).
Primer extension analysis
Primer (5′-GGGCAAGGTGCTGAAAATCC-3′) complementary to the region from +104 to +123 of the human ATF3 mRNA with novel 5′-UTR sequence in database (NM001030287) was radiolabeled with p32-γ-ATP using T4 polynucleotide kinase, hybridized with PolyA mRNAs from serum-treated HCT116, and extended using AMV reverse transcriptase using a kit from Promega. Extended products were resolved on an 8% polyacrylamide sequencing gel. The sequence ladder was generated from the same primer using a template plasmid DNA containing upstream promoter region of the human ATF3 gene.
5′-Rapid amplification of cDNA ends analysis
Human HCT-116 and mouse RAW264 cells were treated with serum or MMS, and the 5′-ends of the ATF3 mRNA of the P1 promoter were determined by 5′-rapid amplification of cDNA end analysis (5′-RACE) using a kit from Invitrogen. ATF3 cDNA was amplified by PCR using a GeneRacer 5′ primer or GeneRacer 5′ nested primer and 3′ primers specific for ATF3: human ATF3-3′ reverse primer, 5′-CCTGGATGTTGAAGCATCATTTTGA-3′; mouse ATF3-3′ reverse primer, 5′-TGGGATGAAGTGCTGGAACTC-3′. Multiple transcription start sites (TSSs) of the P1 promoter of human and mouse ATF3 genes have been deposited in DDBJ/EMBL/GenBank with accession numbers AB291910 and AB291911, respectively.
RNAi experiments
Mixtures of siRNA oligos (final concentration 100 nM) specific for the 5′-UTR of P1 or P2 transcript isoforms were transfected into HCT116 or LNCaP cells using X-tremeGENE transfection reagent (Roche), or into L428 and L540 Hodgkin Reed–Sternberg cells by electroporation as in ref. (24). HCT116 cells were then cultured in medium with 0.5% serum for 24 h, followed by stimulation with 20% serum or 100 μg/ml MMS. Sequences of siRNAs for the P1 5′-UTR were 5′-AGAGAAAUCCUCCUCUAUAUAGG-3′ (83–105) and 5′-UUUCAGCACCUUGCCCCAAAAUC-3′ (129–151), and those for the P2 5′-UTR were 5′-GCCAGCCUGAGGGCUAUAAAGG-3′ (46–58), and 5′-CUCGCCCGCCGGCCAGACAAACA-3′ (190–212). Control RNAi (siCtl) was a silencer negative control #1 (AM4611) from Ambion.
Luciferase assay
Reporter plasmid, pLuc-P1ATF3 was constructed by subcloning a 2.1-kb fragment of the human ATF3 gene upstream promoter from −2091 to +9 (major TSS as +1) between Kpn1 and HindIII sites of pGL3 vector (Promega). pLuc-P2ATF3 reporter plasmid containing a 1850-bp region of P2 promoter was as described (11). Cells (5 × 104 cells) were transfected with 0.5 μg each reporter plasmid. At 16 h posttransfection, cells were starved with 0.5% serum for 48 h and then stimulated with 20% serum, 50 μg/ml MMS, 20 μg/ml etoposide, 50 μM H2O2, 1 μM doxorubicin, 2 μM thapsigargin, 1 μM tunicamycin or 100 pM TGF-β For treatment with H2O2, cells were exposed to 50 μM H2O2 in PBS for 1 h, and then cultured in fresh medium containing serum. After cell culture for 16 h, extracts were assayed for the luciferase activity as described (11), using a Dual Luciferase Reporter Assay System according to the manufacture's protocol (Promega).
Translation in vitro and in vivo
cDNAs for ATF3 transcripts with various UTRs were subcloned between Nhe1 and BamH1 sites of pcDNA3.1 expression vector. Each DNA (0.1 μg) was transcribed along with 0.2 μg vector encoding HA-Suv39 as an internal control in 10 µl of in vitro TNT assay kit driven by T7 RNA polymerase, followed by translation. ATF3 protein generated was quantitated by western blot and densitometric measurement. For the in vivo translation, the plasmid above (1 μg each) encoding the ATF3 protein tagged with Flag at the C-terminus was transfected into 293 cells and the expression of ATF3 protein was measured by western blot. Total RNA was also extracted, treated with RNase-free DNase (Sigma) and assayed for ATF3 mRNA generated from the transgene by quantitative RT–PCR. Primers used: 5′-CTGCAGAAAGAGTCGGAG-3′ (forward, coding sequence of ATF3) and 5′-TAGAAGGCACAGTCGAGG-3′ (reverse, BGH sequence of the vector).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as described (22) according to the protocol supplied by Upstate. ChIP DNA was detected using standard PCR with the following primer pairs for the different regions of the ATF3 gene: −5-kb region, 5′-TGGACACACACACGGAAACT-3′ (forward) and 5′-GTCACATCTTCCCATCTGATC-3′ (reverse), the P1 promoter region, 5′-AGGATGATGGAAGGCTGTCA-3′ (forward) and 5′-GTCACATCTTCCCATCTGATC-3′ (reverse), +10-kb region, 5′-TATGGGCTTTTGTTCGGGTTC-3′ (forward) and 5′-TCAAAATGCGTGTGTGTGTG-3′ (reverse), +20-kb region, 5′-AATGCCCTCACAGAAACACC-3′ (forward) and 5′-GTTAGGCAGGAAGGGGAAAG-3′ (reverse), +30-kb region, 5′-GGTGTGAATGTGCTTTGTGG-3′ (forward) and 5′-AGAATCGGAATGGCTGTGAG-3′ (reverse), the P2 promoter region (+40 kb), 5′-CGAACTTGCATCACCAGTGC-3′ (forward) and 5′-GGTCGTTTACTCCGTGTTGC-3′ (reverse), +50-kb region, 5′-ACATGTCCATCAGCTTCCAG-3′ (forward) and 5′-CCACTGCTTGTGGATTAAGG-3′ (reverse), +55-kb region, 5′-ACCTGTTCCCCATGGATGTA-3′ (forward) and 5′-TGTGTCAGGGAGCCCAAATA-3′ (reverse).
Statistical analysis
Multiple comparisons were evaluated by ANOVA followed by Scheffe's post hoc test. Data are presented as mean ± SD. Statistical significance was assigned at the level of P < 0.05.
RESULTS
The upstream alternate promoter P1 is conserved between human and mouse
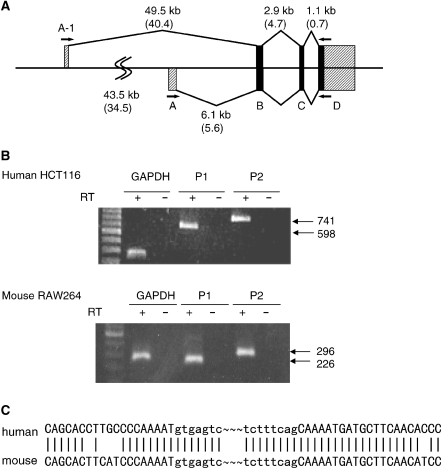
It has recently been demonstrated that the human ATF3 gene contains an alternate promoter (Genbank accession number NM001030287) (35). Figure 1B, upper panel, showed two different products were amplified from human cells by RT–PCR using two different 5′ primers, which located at exon A-1 or canonical exon A (Figure 1A), respectively. These bands were only detected after the synthesis of the first strand cDNA using oligo-dT primer, indicating that they were derived from the matured mRNA. In Figure 1B, lower panel, a specific band was also amplified by RT–PCR of total RNA of mouse RAW264 cells using a 5′ primer located at the ∼34.5-kb upstream of the mouse exon A. Sequence analysis of these human and mouse transcripts revealed that the longer bands correspond to those previously reported (37,38), but the shorter ones contained novel 5′-UTRs representing the transcripts from the upstream promoter (DDBJ/EMBL/GenBank with accession number AB291912 for mouse atf3). In Figure 1C, the sequence comparison of the novel transcripts of human and mouse showed that the junction of the upstream exon A-1 and exon B are nearly identical. Moreover, the nucleotide sequence alignment of the human and mouse P1 promoter shows high homology (Figure 2B). Taken together, these data indicate that the novel P1 promoter is highly conserved between human and mouse.
Figure 1.
Conservation of the upstream alternate promoter of the ATF3 gene in human and mouse. (A) Schematic representation of the structure of the human and mouse ATF3 genes. Exon A is further devided into A-1 and A corresponding to the novel promoter recently identified (35) and that previously reported (27), respectively. (B) Total RNAs from serum-induced human HCT116 or mouse RAW264 cells were assayed for the expression of transcripts containing the exon A-1 or A by RT–PCR. (C) Specific bands in (B) were subcloned, sequenced and the nucleotide sequences of the junction between the exon A-1 and B of human and mouse are shown.
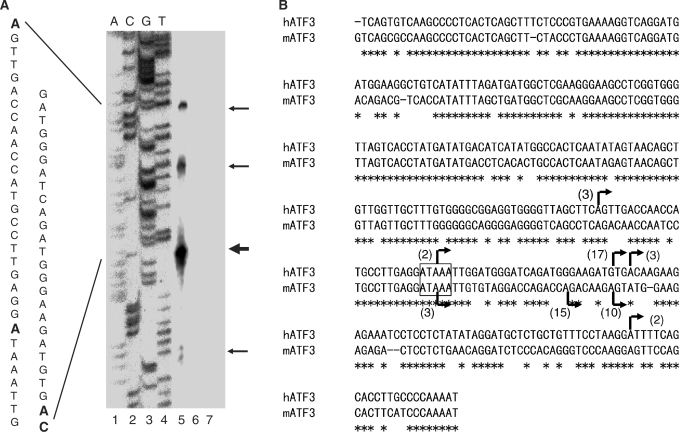
Figure 2.
Structure of the upstream promoter P1 of the ATF3 gene. (A) Primer extension analysis of mRNAs from the P1 promoter of the human ATF3 gene. A primer specific for the P1 transcript was annealed to 1 μg of polyA mRNAs isolated from the serum-treated HCT116 cells (lane 5), 1 μg yeast tRNA (lane 6) or none (lane 7), and the TSSs were determined as in Materials and methods section. Lanes 1–4 represent a sequence ladder. The arrows on the right indicate the TSSs, and the sequence is shown on the left. Bold letters represent multiple TSSs identified. (B) Alignment of the nucleotide sequence of the P1 promoter of the human and mouse ATF3 gene. Arrows indicate the multiple TSSs, and the numbers in parentheses represent the clones analyzed by 5′-RACE of the serum-stimulated cells. Putative TATA box is shown in the box. Multiple TSSs of the human and mouse genes were deposited in DDBJ with accession numbers AB291910 and AB291911, respectively.
The upstream alternate promoter P1 contains multiple TSSs in human and mouse
To determine the TSSs from the P1 promoter of the human ATF3 gene, we first performed a primer extension analysis using mRNA from the serum-stimulated human cells. As shown in Figure 2A, several extended products, one major and at least three minor bands were identified, indicating the presence of multiple TSSs from the P1 promoter. We further performed a 5′-RACE assay to determine the 5′-end(s) of ATF3 transcripts. Figure 2B showed that multiple TSSs determined by 5’-RACE coincided with those by primer extension, indicating the isolation of intact mRNA and the complete reaction by reverse transcriptase. Multiple initiation sites were also observed in the mouse P1 promoter (Figure 2B). From the sequence homology of the P1 promoter and the multiple TSSs between human and mouse, it is speculated that ATAAA sequence at ∼30-bp upstream from the major start site is functional as putative TATA motif. Further, we observed other binding motifs for ATF/CRE, AP1, p53, E2F or NF-kB within the 2-kb upstream region of the P1 promoter (data not shown).
Differential response of the alternate promoters P1 and P2 to various stimuli
Stress response gene ATF3 is activated by not only stress but also mitogenic stimuli such as serum, epidermal growth factor or fibroblast growth factor (16,17). As shown in Supplementary Figure S2, ATF3 mRNA and protein was induced by serum and MMS treatment. Figure 3A showed that both the P1 and P2 transcript isoforms were induced to a similar extent by MMS treatment. In contrast, the P1 transcript was significantly induced by serum, whereas the induction of P2 transcript was more rapid but decreased earlier than the P1 transcript. To clarify the involvement of each promoter, we knocked down the P1 or P2 transcript by RNA interference-mediated gene silencing. As shown in Figure 3B, the knockdown of the P1 or P2 transcript partially suppressed the induction of ATF3 protein in response to MMS, demonstrating the contribution of both promoters. In contrast, in serum-stimulated cells, only the knockdown of the P1 transcript significantly reduced the ATF3 induction, clearly defining the direct contribution of the P1 promoter in serum induction. Next, we addressed if the P1 and P2 transcripts were induced by other stimuli. Figure 3C showed that both transcripts were induced by etoposide, H2O2, doxorubicin, thapsigargin and tunicamycin. ATF3 is also induced by TGF-β (13) or oncogenic HRAS (14), which mediates suppression of cell proliferation, tumor growth or cell senescence. Both the P1 and P2 isoforms were also induced in HaCaT human keratinocytes stimulated with TGF-β or in cells stably expressing oncogenic HRAS 12V or 61L, but not normal HRAS. The induction of the P2 transcript dominated that of the P1 transcript in these cells. Further, we examined the activation of the P1 and P2 promoter by these stimuli using a reporter assay. Figure 3D clearly showed both the P1 and P2 reporter activities were induced in response to these stimuli. Taken together, these data demonstrate that the upstream alternate promoter P1 of the ATF3 gene is activated at the transcriptional level by various stimuli, but differently from the canonical P2 promoter.
Figure 3.
Differential response of the ATF3 alternate promoters to various stimuli. (A) HCT116 cells were serum-starved in the presence of 0.5% serum for 48 h, and then stimulated by 20% serum, or 100 μg/ml MMS. At each time indicated, the ATF3 mRNA isoforms from the P1 (filled circle) or P2 promoter (filled square) was measured. Relative expression of these transcripts was normalized to GAPDH as in Materials and methods section and Supplementary Figure S1. Lower panel shows a regular RT–PCR gel of the representative experiment. (B) HCT116 cells transfected with siRNA oligos control (siCtl), specific for the P1 (siP1) or P2 transcript (siP2) were stimulated with serum or MMS as in Materials and method section. At each time indicated, the expression of the P1, P2 transcript or ATF3 protein was measured. *P < 0.05 compared with control siCtl. (C) HCT116 cells were treated with 20 μg/ml etoposide (Etopo), 50 μM H2O2, 1 μM doxorubicin (Dox), 2 μM thapsigargin (Thap), 1 μM tunicamycin (Tuni) and HaCaT cells were treated with 100 pM TGF-β. At each time indicated, the expression of the P1, P2 transcript or ATF3 protein was measured. U2OS cells stably expressing the normal, oncogenic 12V or 61L mutants of HRAS were also assayed. (D) Cells transfected with pLuc-P1ATF3 (closed) or pLuc-P2ATF3 (open) were treated with various agents as in (C) for 16 h and assayed for luciferase activity. Data represent the fold induction compared to unstimulated cells. For U2OS cells expressing oncogenic HRAS, the fold induction of activity compared to the empty vector is shown. All the data from (A) through (D) represent means with standard error bars of three independent experiments.
Regulation of translation by 5′-UTR of the transcripts from the P1 promoter
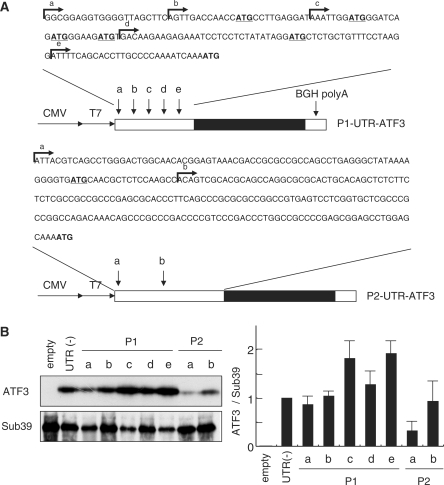
Figure 4A and Supplementary Figure S3 showed that the 5′-UTR of the P1 transcripts has unique structural properties; multiple TSSs, less score of GC content (∼50%), and five upstream AUG (uAUGs) compared to the P2 transcripts; two TSSs, higher GC content (∼70%), and one upstream AUG (uAUGs). Moreover, the 5′-RACE analysis revealed that multiple TSSs of the P1 promoter were differently used in MMS-treated HCT116 cells from serum-stimulated cells; the number ratio of clones obtained were 6:12:12:0 (b:c:d:e) in MMS-cells compared to 3:2:20:2 in serum response as in Figure 2B. To explore the possible effect of different 5′-UTRs on the translational efficiency, we performed a translational reporter assay using the ATF3 expression plasmids containing different 5′-UTR of the P1 and P2 transcripts. Figure 4B showed that most of the P1 and P2 constructs generated the ATF3 protein in vitro with an apparently comparable efficiency, except for the long 5′-UTR P2 transcript. The relative efficiency of the in vivo translation of various reporters in unstimulated 293 cells appeared to be similar to those in vitro (Figure 4C). In contrast, in cells treated by serum or MMS, the translation of the reporters P1-c, d, e and P2-b were significantly increased, whereas the translation of the P1-a and P1-b constructs was barely activated (Figure 4D). This suggests the presence of not only the inhibitory sequence(s) between a and c but also the stimulatory element(s) in the shorter UTR forms, c, d and e. Taken together, these data indicate that the structure of 5′-UTRs of the P1 transcripts as well as the differential use of TSSs significantly affects their translation in response to stress.
Figure 4.
Efficiency of translation of the ATF3 mRNA isoforms from the P1 and P2 promoters. (A) Expression reporter plasmids encoding the ATF3 transcripts with various 5′-UTRs from the P1 (upper panel) and P2 promoters (lower panel) are shown. Arrows a through e indicate the 5′-ends of mRNAs corresponding multiple TSS of the P1 promoter in Figure 2B, and arrows a and b indicate the 5′-ends the P2 transcripts obtained from the database and reference (27). (B) Each plasmid was assayed for the in vitro translation as described in the Materials and methods section. The relative expression of ATF3 to plasmid without UTR is shown. (C) 293 cells were transfected with each expression plasmid, cultured for 24 h, and the ATF3 mRNA and protein generated from the transgene was measured as in the Materials and methods section. Relative efficiency of translation is the ratio of the ATF3 protein normalized to mRNA compared to that without 5′-UTR. (D) 293 cells were transfected with each plasmid, starved with 0.5% serum, followed by stimulation with 20% serum for 24 h or 50 μg/ml MMS for 12 h. After measurement of both ATF3 protein and mRNA generated from the transgene, relative efficiency of translation is shown as in (C). All the data from (B) through (D) represent the means with standard error bar of three independent experiments.
Constitutive activation of the P1 promoter in ATF3-expressing human cancer cells
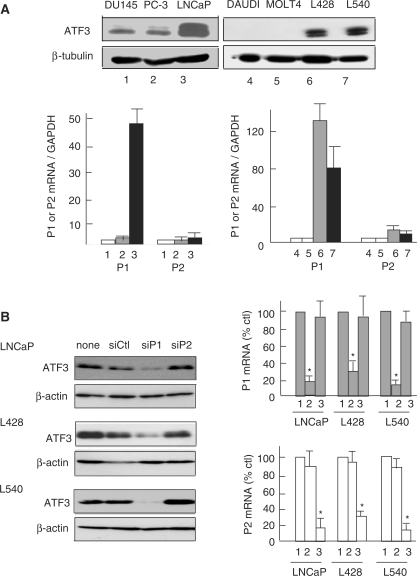
ATF3 is constitutively overexpressed in several forms of human cancer cells such as prostate, Hodgkin disease and mammary tumors (23–25), and the elevated ATF3 expression correlates to the cell proliferation, progression and metastasis (23–25,39). To clarify the functional role of alternate promoters in these cells, we examined the expression of ATF3 from each promoter. Figure 5A showed that, in human prostate cancer LNCaP and Hodgkin Reed–Sternberg L428 and L540 cells, the amount of ATF3 protein was increased as reported (23,24). Most significantly, the P1 transcript was dominantly expressed in these cells compared to DU145 and PC3 or DAUDI and MOLT 4 cells, demonstrating that the P1 promoter is selectively activated. To further clarify the actual involvement of each promoter, we knocked down the P1 or P2 transcripts. Figure 5B clearly showed the knockdown of the P1 transcript drastically suppressed the expression of the ATF3 protein along with the P1 transcript in LNCaP, L428 and L540 cells, whereas the P2 knockdown had only marginally effect. The data strongly support that the P1 promoter has a direct contribution to the elevated expression of ATF3 in these cancer cells. We next performed ChIP assay to examine the association of RNA polymerase II with the ATF3 gene locus. Figure 5C showed that, in LNCaP cells, RNA polymerase II and Leo1, a component of PAF1 complex in the elongating RNA polymerase II (40), were clearly recruited through the P1 promoter to downstream regions, compared to −5-kb region and the control IgG, whereas the Leo1 signal peaked at around +10-kb region. In contrast, DU145 cells that had lower expression of ATF3 did not show significant recruitment of RNA polymerase II. The increased association of RNA polymerase II to the P1 promoter region was also observed in Hodgkin L428 cells (Supplementary Figure S4). These data unambiguously indicate the upstream P1 promoter of the ATF3 gene is constitutively activated in human prostate and Hodgkin Reed–Sternberg cells.
Figure 5.
Constitutive activation of the upstream P1 promoter in human prostate and Hodgkin RS cancer cells. (A) The expression level of ATF3 protein in DU145 (lane 1), PC-3 (lane 2), LNCaP (lane 3), DAUDI (lane 4), MOLT4 (lane 5), Hodgkin RS L428 (lane 6) or L540 (lane 7) cells was measured by western blot (upper panel). In the lower panel, total RNAs from these cells were assayed for the P1 or P2 transcripts as in Figure 3A. Relative expression of the ATF3 transcripts represents the ratio to that of GAPDH. (B) LNCaP, L428 or L540 cells were transfected with control siRNA oligos (siCtl, lane 1), or specific for the P1 (siP1, lane 2) or P2 transcript (siP2, lane 3), cultured for 48 h and assayed for the P1 and P2 transcripts or ATF3 protein. The relative expression of the P1 and P2 transcripts is shown as per cent of the control (siCtl). *P < 0.05 compared with control siCtl. (C) LNCaP (filled circle, open circle) or DU145 (filled triangle, open triangle) cells were fixed with formaldehyde and immunoprecipitated using anti-RNA polymerase II (upper panel) or anti-Leo1 antibody (lower panel) as in Materials and methods section. Immunoprecipitated DNA was measured by quantitative PCR throughout the human ATF3 gene locus and expressed as percent of the input DNA for the control IgG (open) and specific antibodies (closed), respectively. All the data from (A) through (C) represent the means of three independent experiments with standard error bars.
Chromatin modification of the human ATF3 gene in cancer cells
Constitutive activation of the upstream P1 promoter in cancer cells prompted us to examine the chromatin modifications of the ATF3 gene promoter. As shown in Figure 6A, antibodies against the pan-acetylated histone H3 and trimethylated H3K4me3 efficiently immunoprecipitated the P1 promoter region in LNCaP and Hodgkin 428 cells, compared to DU145 or DAUDI cells. In contrast, no significant H3K9 methylation was observed at the ATF3 P1 promoter region in LNCaP or L428 cells. As an inactivated gene control, the ChIP signal of H3K9 methylation at the β-globin gene promoter in L428 cells was also shown (Figure 6A). Figure 6B and Supplementary Figure S5 further showed that the significant pan-acetylation and K4 trimethylation of H3 were detected from the P1 promoter to +10-kb region compared to other region in LNCaP and L428 cells, respectively. Taken together, the P1 promoter of the ATF3 gene in LNCaP and Hodgkin L428 cells has an active chromatin configuration associated with the histone H3 acetylation and H3K4 trimethylation.
Figure 6.
Histone modifications across the human ATF3 gene locus in LNCaP and Hodgkin L428 cells. (A) ChIP assay was performed in LNCaP, DU145, L428, or DAUDI cells using anti-panacetyl H3, anti-trimethyl H3K4 or anti-trimethyl H3K9 antibodies as in Materials and methods section. The P1 and P2 promoter region of the ATF3 gene and the control GAPDH gene was amplified and analyzed by gel electrophoresis. In L428 and DAUDI cells, the promoter region of the human β-globin gene was also examined. (B) Immunoprecipitated DNA by anti-panacetyl H3 or anti-trimethyl H3K4 antibodies of LNCaP (filled circle, open circle), DU145 (filled traingle, open triangle) cells was measured by quantitative PCR throughout the human ATF3 gene locus, and expressed as percent of the input. Data represent the means of three independent experiments with standard error bars for control IgG (open) and specific antibodies (closed), respectively.
DISCUSSION
In the present report, we described the structural and functional properties of the upstream alternate promoter P1 of the human ATF3 gene. The P1 promoter is used differently from the P2 promoter at the transcriptional and translational level in stress response. Moreover, it is constitutively activated in human cancer cells associated with the active chromatin configuration, as a consequence, followed by the elevated expression of ATF3.
Kimura et al. (35) reported that at least 52% of the human genes contain putative alternative promoters, with 3.1 alternative promoters per gene on average, and the genes subject to regulation by alternative promoters are enriched in those encoding signal transduction-related proteins. Indeed, the ATF3 gene encodes a transcription factor that is induced and plays role in various stress response and signaling. The P1 promoter of the ATF3 gene was highly conserved between human and mouse, which localizes ∼43.5-kb and ∼34.5- kb upstream of the previously reported canonical promoter P2 (30), respectively. The P1 promoter has several properties that are conserved between human and mouse. (i) Sequence of the promoter region as well as the junction of exon A-1 and B are highly conserved (Figure 1C and 2B). The putative TATA motif is localized ∼30 base upstream from the major TSS, and other binding motifs for AP1, ATF/CREB, NF-kB, E2F, or p53 are conserved. (ii) Multiple TSSs are present in the human and mouse genes. (iii) The P1 promoter has less stretch of CG dinucleotides, whereas the P2 promoter contains at least three CpG islands within the 2-kb region of the promoter. This structural difference between the P1 and P2 promoters is of intrigue, because there is one CpG-island-containing promoter per 2.6 CpG-less alternate promoters (35). Overall, the structural properties of the P1 promoter strongly support that it is evolutionally conserved between human and mouse, and has distinct control mechanism.
The P1 promoter of the ATF3 gene in human cells was activated by various stimuli including serum, DNA damage and oxidoreductive stress, and the extent of the P1 promoter activation was comparable to or exceeded that of the P2 promoter (Figure 3). Indeed, the 5′-flanking region of the P1 promoter contains several motifs for transcription factors involved in the signaling of stress stimuli, including the binding motifs for ATF/CRE, AP1, NF-kB, E2F, c-Myc or p53. Whereas the significance and implication of these motifs in each stress response was not addressed in this report, it is also possible that a common motif(s) in the ATF3 gene locus confers stress response on both the P1 and P2 promoters. This is an important issue and must await further study.
Various 5′-UTRs of the P1 transcripts contain the features of poorly translated mRNA, that is relatively long 5′-UTR with multiple upstream AUGs (uAUGs) and upstream ORFs (Figures 2B and 4A). These features are often found in mRNAs encoding regulatory proteins like proto-oncogenes or growth factor (41), consistent with biological role of the ATF3 gene. In our translational reporter assay, however, sequential mutations of uAUGs in the 5′-UTR of the P1 transcrips had no significant effect on their translation (data not shown). The assay revealed the presence of the inhibitory region in the longest 5′-UTR as well as the stimulatory element(s) in the shorter 5′-UTRs, implicating a role of trans-acting factor(s) interacting with these element(s) in the 5′-UTR in stress response. The identification of such factor(s) and the corresponding cis-element(s) is an important issue of further study. It should be noted that the present study employed the reporters containing only the different 5′-UTRs, since the P1 and P2 transcripts have the common sequence from the coding through the 3′-UTR (NM_001030287 and NM_001674). However, it is possible that the 3′-UTR may contribute to the regulation, since 3′-UTR is well known to affect the translational efficiency in vivo. More importantly, the 5′-RACE analysis revealed the differential usage of TSSs in MMS- and serum-treated cells. Thus, it is highly likely that the selection or shift of the TSSs within the P1 promoter also regulates the efficiency of translation in vivo.
Early studies showed that the ATF3 induction correlates with cell-detrimental outcome such as cell cycle arrest or apoptosis, thus providing an idea that it is death factor (9–12). However, ATF3 is also implicated in cell proliferation (13,15,18,22), transformation (19), cell survival (20,21) and tumorigenesis (14,23–26). Our present study demonstrated for the first time that the P1 promoter is constitutively activated in human cancer cells with the elevated expression of ATF3 protein. In LNcaP and Hodgkin cancer cells, the chromatin structure of the P1 promoter region is remodeled and fixed to an active open state through the histone modification, including histone H3 acetylation and H3K4 trimethylation (Figure 6). As in Figure 3A, the serum response of the P2 promoter was more rapid but suppressed earlier than the P1 promoter. It is speculated that the P2 promoter responds to serum, but is suppressed by the transcription from the upstream promoter P1 through a mechanism called delayed transcriptional interference (42), while it is also possible that the splicing of the transcripts is differently regulated. At this moment, however, it is not known to what extent these mechanisms could be applied to constitutive activation of the P1 promoter in LNCaP and Hodgkin cancer cells. It is also unlikely that cis-element(s) of the promoter responsible for serum response is the only mechanism by which the P1 promoter is constitutively activated in cancer cells, since the P2 promoter is induced by serum in rat fibroblasts via ATF/CRE motif (22), and the reporter activity driven by the P2 promoter was activated by serum as efficiently as the P1 promoter (Figure 3D).
In conclusion, the present study for the first time described the molecular and functional properties of the upstream alternate promoter P1 of the stress response gene ATF3. Of intrigue, the expression from the P1 promoter is regulated at both the transcriptional and translational levels, and is constitutively activated with altered chromatin structure in human cancer cells. The differential usage and regulation of the alternate promoters of the human ATF3 gene adds an important dimension to the control mechanism by which it plays dual role in determining cell fate during stress response and oncogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [18012015, 18055008] to S.K. in part. Funding for open access charge: Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Nevins and Dr Jenuwein for their gifts of valuable plasmids, Dr Yamaoka for his helpful discussion, Ms Y. Hosaka and A. Nakamura for their technical assistance.
REFERENCES
- 1.Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH-M. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Kamat A, Graves KH, Smith ME, Richardson JA, Mendelson CR. A 500-bp region, ∼40kb upstream of the human CYP19 (aromatase) gene, mediates placenta-specific expression in transgenic mice. Proc. Natl Acad. Sci. USA. 1999;96:4575–4580. doi: 10.1073/pnas.96.8.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magnuson MA, Shelton KD. An alternate promoter in the glucokinase gene is active in the pancreatic β cell. J. Biol. Chem. 1989;264:15936–15942. [PubMed] [Google Scholar]
- 5.Chotani MA, Payson RA, Winkles JA, Chiu IM. Human fibroblast growth factor 1 gene expression in vascular smooth muscle cells is modulated via an alternate promoter in response to serum and phorbol ester. Nucleic Acid Res. 1995;11:434–441. doi: 10.1093/nar/23.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meshorer E, Soreq H. Virtues and woes of AchE alternative splicing in stress-related neuropathologies. Trends Neurosci. 2006;29:216–224. doi: 10.1016/j.tins.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Murray-Zmijew F, Lane DP, Bourdon JC. p53/-63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 8.Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takanahshi S, Takanahsi Y. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J. Biol. Chem. 2008;283:2543–2553. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- 9.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expression. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 10.Yin T, Sandhu G, Wolfgang CD, Burrier A, Webb RL, Rigel DF, Hai T, Whelan J. Tissue-specific pattern of stress kinase activation in ischemic/reperfused heart and kidney. J. Biol. Chem. 1997;272:19943–19950. doi: 10.1074/jbc.272.32.19943. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y, Zhang C, Nawa T, Aso T, Tanaka M, Oshiro S, Ichijo H, Kitajima S. Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2 terminal kinase and promoter response element. Blood. 2000;96:2140–2148. [PubMed] [Google Scholar]
- 12.Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, Zhu X, Mazzacurati L, Li X, Petrik KL, Fornace AJ, Rajasekaran B, Zhan Q. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cell growth. Oncogene. 2002;21:7488–7496. doi: 10.1038/sj.onc.1205896. [DOI] [PubMed] [Google Scholar]
- 13.Kang Y, Chen CR, Massague J. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 14.Lu D, Wolfgang CD, Hai T. ATF3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J. Biol. Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 15.Hsu JC, Laz T, Mohn KL, Taub R. Identification of LRF-1, a leucine-zipper protein that is rapidly and highly induced in regenerating liver. Proc. Natl Acad. Sci. USA. 1991;88:3511–3515. doi: 10.1073/pnas.88.9.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohn KL, Laz TM, Hsu JC, Melby AE, Bravo R, Taub R. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol. Cell. Biol. 1991;11:381–390. doi: 10.1128/mcb.11.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 18.Allan AL, Albanese C, Pestell RG, LaMarre J. Activating transcription factor 3 induces DNA synthesis and expression of cyclin D1 in hepatocytes. J. Biol. Chem. 2001;276:27272–27280. doi: 10.1074/jbc.M103196200. [DOI] [PubMed] [Google Scholar]
- 19.Perez S, Vial E, van Dam H, Castellazzi M. Transcription factor ATF3 partially transforms chick embryo fibroblasts by promoting growth factor-independent proliferation. Oncogene. 2001;20:1135–1141. doi: 10.1038/sj.onc.1204200. [DOI] [PubMed] [Google Scholar]
- 20.Kawauchi J, Zhang C, Nobori K, Hashimoto Y, Adachi TM, Noda A, Sunamori M, Kitajima S. Transcriptional repressor activating transcription factor 3 protects human umbilical vein endothelial cells from tumor necrosis factor-alpha-induced apoptosis through down-regulation of p53 transcription. J. Biol. Chem. 2002;277:39025–39034. doi: 10.1074/jbc.M202974200. [DOI] [PubMed] [Google Scholar]
- 21.Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor ATF3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and akt activation. J. Neurochem. 2003;23:5187–5196. doi: 10.1523/JNEUROSCI.23-12-05187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Hua B, Adachi S, Guney I, Kawauchi J, Morioka M, Adachi TM, Tanaka Y, Nakabeppu Y, Sunamori M, et al. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. EMBO J. 2005;24:2590–2601. doi: 10.1038/sj.emboj.7600742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelzer AE, Baktic J, Haag P, Berger AP, Pycha A, Schafer G, Rogatsch H, Horninger W, Bartsch G, Klocker H. The expression of transcription factor activating transcription factor 3 in the human prostate and its regulation by androgen in prostate cancer. J. Urol. 2006;175:1517–1522. doi: 10.1016/S0022-5347(05)00651-8. [DOI] [PubMed] [Google Scholar]
- 24.Janz M, Hummel M, Truss M, Wollert-Wulf B, Mathas S, Johrens K, Hagemeier C, Bommert K, Stein H, Dorken B, et al. Classical Hodgkin lymphoma is characterized by high constitutive expression of activating transcription factor 3 (ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells. Blood. 2006;107:2536–2539. doi: 10.1182/blood-2005-07-2694. [DOI] [PubMed] [Google Scholar]
- 25.Yin X, DeWille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27:2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- 26.Wang A, Arantes S, Conti C, McArthur M, Aldaz CM, Macleod MC. Epidermal hyperplasia and oral carcinoma in mice overexpressing the transcription factor ATF3 in basal epithelial cells. Mol. Carcinogenesis. 2007;46:476–487. doi: 10.1002/mc.20298. [DOI] [PubMed] [Google Scholar]
- 27.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:143–154. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 28.Rosenberger CM, Clark AE, Treuting PM, Johnson CD, Aderem A. ATF3 regulates MCMV infection in mice by modulating IFN-gamma expression in natural killer cells. Proc. Natl Acad. Sci. USA. 2008;105:2544–2549. doi: 10.1073/pnas.0712182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilchrist M, Henderson WR, Clark AE, Simmons RM, Ye X, Smith KD, Aderem A. Activating transcription factor 3 is a negative regulator of allergic pulmonary inflammation. J. Exp. Med. 2008 doi: 10.1084/jem.20072254. Sep 15, [Epub ahead of print, doi:10.1084/jem.20072254] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang G, Wolfgang CD, Chen BPC, Chen TH, Hai T. ATF3 gene. Genomic organization, promoter, and regulation. J. Biol. Chem. 1996;271:1695–1701. doi: 10.1074/jbc.271.3.1695. [DOI] [PubMed] [Google Scholar]
- 31.Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Gao C, Kawauchi J, Hashimoto Y, Tsuchida N, Kitajima S. Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem. Biophys. Res. Commun. 2002;297:1302–1310. doi: 10.1016/s0006-291x(02)02382-3. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi K, Lee S-H, Kin J-S, Wimalasena Y, Kitajima S, Baek SJ. Activating transcription factor 3 and early growth response-1 are the novel targets of LY294002 in a PI3K independent pathway. Cancer Res. 2006;66:2376–2384. doi: 10.1158/0008-5472.CAN-05-1987. [DOI] [PubMed] [Google Scholar]
- 34.Lu D, Chen J, Hai T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem. J. 2007;401:559–567. doi: 10.1042/BJ20061081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 37.Chen BP, Liang G, Whelan J, Hai T. ATF3 and ATF3deltaZip. Transcriptional repression versus activation by alternatively spliced isoform. J. Biol. Chem. 1994;269:15819–15826. [PubMed] [Google Scholar]
- 38.Drysdale BE, Howard DL, Johnson RJ. Identification of a lipopolysaccharide inducible transcription factor in murine macrophages. Mol. Immunol. 1996;33:989–998. doi: 10.1016/s0161-5890(96)00043-0. [DOI] [PubMed] [Google Scholar]
- 39.Bandyopadhyay S, Wang Y, Zhan R, Pai SK, Watabe M, Iiizumi M, Furuta E, Mohinta S, Liu W, Hirota S, et al. The tumor metastasis suppressor gene Drg-1 down-regulates the expression of ATF3 in prostate cancer. Cancer Res. 2006;66:11983–11990. doi: 10.1158/0008-5472.CAN-06-0943. [DOI] [PubMed] [Google Scholar]
- 40.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shearwin KE, Callen BP, Egan JB. Transcriptional interference - a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.