Abstract
The HIV-1 TAR RNA represents a well-known paradigm to study the role of dynamics and conformational change in RNA function. This regulatory RNA changes conformation in response to binding of Tat protein and of a variety of peptidic and small molecule ligands, indicating that its conformational flexibility and intrinsic dynamics play important roles in molecular recognition. We have used 13C NMR relaxation experiments to examine changes in the motional landscape of HIV-1 TAR in the presence of three ligands of different affinity and specificity. The ligands are argininamide, a linear peptide mimic of the Tat basic domain and a cyclic peptide that potently inhibits Tat-dependent activation of transcription. All three molecules induce the same motional characteristics within the three nucleotides bulge that represents the Tat-binding site. However, the cyclic peptide has a unique motional signature in the apical loop, which represents a binding site for the essential host co-factor cyclin T1. These results suggest that all peptidic mimics of Tat induce the same dynamics in TAR within this protein binding site. However, the new cyclic peptide mimic of Tat represents a new class of ligands with a unique effect on the dynamics and the structure of the apical loop.
INTRODUCTION
Many of RNA functional roles depend on its ability to change structure upon binding of small molecules, peptides or proteins (1–4). This observation suggests that many RNAs possess intrinsic dynamic features which expand their functional potential beyond what their limited chemical repertoire would otherwise allow. This behavior is evident in the HIV-1 TAR RNA (Figure 1), one of the first systems where the role of conformational change on RNA function became clear (5–7). TAR is a conserved stem-loop found at the 5′-end of all viral transcripts. It folds immediately after transcriptional initiation off the HIV promoter to provide a structure that recruits the HIV protein Tat and the cyclin T1/cdk9 kinase complex to promote efficient transcriptional elongation (6). The importance of the Tat–TAR interaction for viral replication has led to considerable interest in discovering new inhibitors of viral replication that function by disrupting the formation of this complex. Many different ligands, ranging from small molecules to peptide mimetics (5,8–11) have been produced that bind to TAR at the Tat-binding site. The structures of these complexes have confirmed that TAR dramatically changes its structure in order to promote ligand binding (5,6,12). However, none of these compounds have so far progressed even to the pre-clinical stage of drug development.
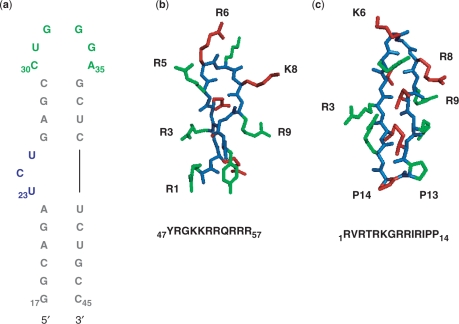
Figure 1.
(a) Sequence and secondary structure of HIV-1 TAR RNA; residues are colored showing the cyclin T1-binding site in green, the Tat-binding site in blue and the remaining double helical regions in gray. (b) Sequence of the 11-mer Tat-derived peptide corresponding to the Tat basic domain. (c) Cyclic peptide inhibitor of Tat–TAR complex. Both peptides are shown with side chains colored in alternating green and red with the peptide backbone colored in blue.
Binding of Tat and the subsequent recruitment of cyclin T1 and transcriptional activation depend on the ability of TAR to alter its structure in response to ligand binding. A single amino acid (argininamide) induces the same conformational change in TAR that is observed in much larger peptide mimics of Tat protein (6,13). The binding site of Tat is a three nucleotide UCU bulge (U23–U25) that, once bound to argininamide, causes the U23 residue to move near to the A27 and U38 base pair, while C24 and U25 become extrahelical (6).
Given the importance of conformational change to the function of TAR, the dynamics of TAR have been investigated in its ligand-free state using both conventional NMR relaxation measurements and residual dipolar couplings. Multiple studies of the interaction of TAR with metal ions and small molecular weight ligands have suggested that different ligands occupy distinct spots in a trajectory separating two extreme conformational states (14,15). However, it is still not clear what role motion may play in ligand binding and how dynamics relate to complex stability because only ligand-free TAR and its complex with arginineamide have been systematically investigated.
In this article, we systematically examine how the dynamics of TAR change in the presence of ligands of different molecular weight, affinity and specificity by measuring 13C NMR relaxation and R1ρ power dependence for the bases and riboses in the RNA. In addition, we employ the elongation-method technique to establish that inter-helical motions that occur around the bulge in free TAR are quenched in all of the complexes. We investigate how RNA dynamics at different sites correlate with activity for ligands that bind weakly (argininamide with a Kd=1 mM) (5) or more potently (a nM-binding linear peptide mimic of Tat protein, Figure 1b) or both more potently and more specifically (a nM-binding cyclic peptide mimic of Tat protein), Figure 1c. Relaxation parameters (16) (T1, T1ρ, NOE) dependent on the underlying internal motions present in free TAR and in these three distinct complexes are interpreted using the model-free formalism (17,18). Combined, these measurements and their analysis are sensitive to internal motions on the ns–ps and ms–µs timescale, and to global motions on the ns timescale. The results confirm that unbound TAR is highly dynamic in both the bulge and apical loop. Interestingly, ligand binding does not uniformly rigidify the complex and indeed some sites even become more flexible in the bound forms of TAR. While all complexes display very similar motional properties at the Tat-binding site, the cyclic peptide has unique dynamic features in the apical loop that may correlate with its increased specificity. Thus, ligand binding to TAR leads to the formation of complex motional landscapes that are specific for different classes of ligands.
MATERIALS AND METHODS
Sample preparation
All RNAs used in this study were prepared enzymatically via the phage T7 RNA polymerase in vitro transcription method (19,20) using 13C–15N-labeled nucleotides purchased from Isotec and in house purified polymerase. DNA templates were purchased form IDT and included 2′-O-methyl groups attached to the last two residues at the 5′-end of the template in order to reduce further addition of nucleotides past the template end. The RNAs were purified by denaturing gel electrophoresis followed by electro-elution and ethanol precipitation. Finally, microdialysis was performed in a 10 mM potassium phosphate buffer (pH 6.6) with 0.01 mM EDTA. In addition to preparing fully labeled samples, data on free TAR were also collected on two partially labeled samples containing either GU or AC-labeled nucleotides to reduce spectral overlap.
Argininamide was purchased directly from Sigma; the cyclic L22 peptide used in this study was synthesized as described (21); the 11-mer Tat derived peptide (Tat11), 47YGRKKRRQRRR57, was synthesized in house with a PS3 peptide synthesizer using standard fmoc chemistry on an arginine resin (22). The peptide was then cleaved from the resin and purified. After purification, the peptide was characterized using electrospray MS and quantized using UV–VIS spectroscopy (tyrosine ε275=1405 cm−1 mol−1).
Complex formation
The three different complexes of TAR with argininamide, with the Tat-derived 11-mer peptide and with the cyclic peptide (Figure 1) were prepared by slowly titrating the desired amount of ligand into a TAR sample and running an HSQC after each addition to observe the progression in complex formation. The cyclic peptide, argininamide and the linear Tat 11-mer peptide were added to separate TAR RNA solutions until complexes of 1.0, 3.5 and 1.2 molar equivalents were reached, respectively. These ratios were chosen to ensure saturation of the complex given their respective binding constants.
NMR relaxation experiments
All data collection was performed on a Bruker Avance-500 instrument in 10 mM phosphate buffer at pH 6.6 containing 99.9% D2O and at 25°C. The data collected for the free TAR sample were obtained by using a HCN TXI triple resonance probe. The data for the remainder of the samples were collected using a cryoprobe.
The assignments of free TAR and TAR bound to argininamide were taken from previous studies (5,8); assignments for TAR bound to the two different peptides were obtained by collecting 2D NOESY and 3D NOESY-HSQC spectra which were analyzed using previously published methods (23–25).
T1, T1ρ and heteronuclear NOE experiments (26) were recorded as a series of 2D NMR spectra where the value of the relaxation delay, τ, was varied parametrically. The two different spectral regions probed in this study required collection of separate sets of T1/T1ρ experiments. The data for the bases (C6, C8) were collected with the 13C frequency set at 144.5 ppm, while the sugars (C1′) data were collected with the 13C frequency set at 93 ppm; the spectral width was 24 ppm for both datasets. T1 experiments were collected with relaxation delays of 10 ms, 20 ms, 40 ms, 80 ms, 160 ms, 320 ms and 500 ms; T1p experiments used relaxation delays of 4 ms, 8 ms, 12 ms, 24 ms, 32 ms and 48 ms. The experiments with the shortest delay times were repeated for each data set in order to test the reproducibility of the data. The heteronuclear NOE experiments required two experiments for each spectral region. One experiment was performed with initial proton saturation while the other was performed without it. The experiments that utilized proton saturation used a relaxation delay of 2.5 s followed by a 2.5 s saturation period; the experiments without saturation used a single relaxation delay time of 5 s.
Elongated TAR
Intensities of 13C–1H resonances for C1′–H1′, C6–H6 and C8–H8 cross peaks were measured at 25°C using standard non-constant time HSQC experiments on the elongated TAR construct described below (27,28). Peak intensities were normalized for each labeled nucleotide independently. Stem 1 of TAR was elongated by 20 AU base pairs and then two GC base pairs were added at the end of the stem-loop to promote transcription. To reduce spectral crowding, only G and C were 13C/15N labeled by using the appropriate mix of labeled and unlabeled nucleotides.
Power dependence of T1ρ
Power dependence experiments were collected similarly to the previously described T1ρ experiments by changing only the spin-lock power. T1ρ data sets were collected and analyzed for C1′, C6 and C8 resonances at 500 MHz using various spin-lock field strengths of 1.2 kHz, 2.6 kHz, 3.5 kHz, 4.5 kHz and 6.6 kHz by modifying the power level of the spin-lock pulse. Delays of 4 ms, 8ms, 12 ms, 16 ms, 24 ms and 28 ms were used in place of longer delays to prevent sample heating.
Data analysis
All NMR data were processed using NMR pipe (29). The Sparky relaxation fit software was then used to fit the relaxation data to single exponential decay curves, as previously described (30), to obtain values for T1 and T1ρ. Using the power dependence data, Rex values were estimated as the difference between R1ρ (w1→0) and R1ρ (w1→∞), the infinite spin-lock field.
The primary relaxation data were analyzed semi-quantitatively using the Model-Free approximation as implemented in Model-Free 4.15 (17,18) and were found to be 5.9 ns for free TAR; 6.4 ns for the arginine complex; 7.2 and 7.6 ns, respectively, for the linear and cyclic peptide complex. After determining global rotational correlation times, the relaxation parameters were fit to one of five different models (30,31), in order to calculate order parameters for each individual residue. Parameters were varied for each of the five models as follows: (i) S2; (ii) S2 and τe (the effective internal correlation time for fast motions); (iii) S2 and Rex; (iv) S2, Rex and τe; (v) Sf2, Ss2 (the order parameter terms for longer and shorter timescale motions respectively), and τe.
For this analysis, chemical shift anisotropies of 133 ppm and 144 ppm were used for C8 of Guanine and Adenine, respectively; a value of 212 ppm was used for C6 of Uracil and Cytosine; 40 ppm was used for all sugar resonances regardless of nucleotide type (32,33). A bond length of 1.09 Å was assumed for C1′–H1′ bonds, while the assumed bond length for base resonances was 1.104 Å (32,33). We assumed all chemical shift tensors were axially symmetric (η=0) and that the axis of symmetry of the chemical shift tensor remained collinear with the C–H bond. The small errors introduced by the aforementioned assumptions have been discussed at great length in previous publications (16,30).
RESULTS
We executed a comprehensive study of the dynamics of TAR bound to three different ligands and compared these results with the free RNA. The T1, T1ρ and NOE experiments were recorded and analyzed as described in the Methods section for the C1′, C6 and C8 sites for free TAR, and TAR bound to: (i) argininamide (Arg–TAR), (ii) an 11-mer peptide derived from the basic binding region of TAR and (iii) a cyclic peptide recently discovered with unprecedented specificity and activity in viral replication (unpublished results) (Figure 1). The cyclic structure is closely related to a family of peptides that we characterized as inhibitors of the Tat–TAR interaction in BIV (11,21,34). Based on the similarity between BIV and HIV Tat and TAR, we identified the structure of Figure 1c as a potent (Kd=5 nM) and selective inhibitor of the Tat–TAR interaction in vitro that also inhibits viral replication with sub-µM potency in infected human lymphocytes.
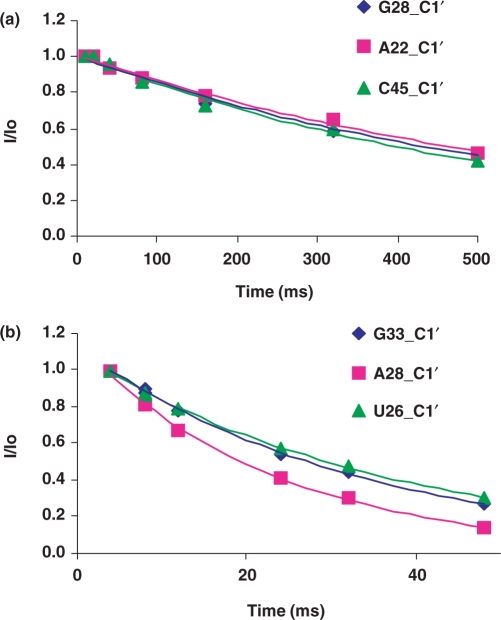
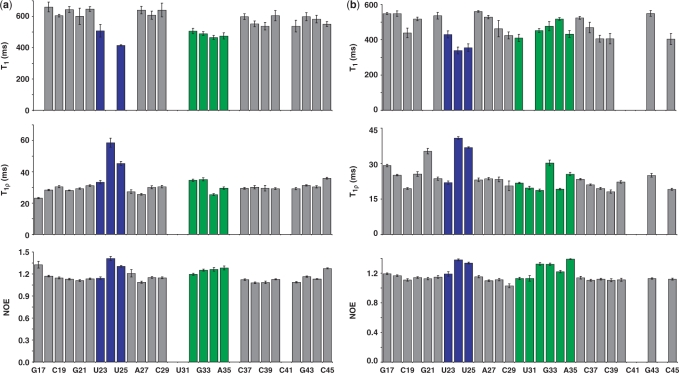
The assignments of free TAR and of TAR bound to argininamide were taken from previous studies (5,8); assignments for TAR bound to the two different peptides were obtained by collecting 2D NOESY and 3D NOESY-HSQC spectra, which were analyzed using previously published methods (23–25). Typical T1 and T1ρ relaxation decay curves for representative residues of the Arg–TAR complex are shown in Figure 2. All relaxation decay curves were well fit by single exponentials, without any evidence for multi-exponential relaxation. Some nuclei were nonetheless excluded from the analysis of the relaxation data because the corresponding resonances were overlapped in the spectra. The 13C relaxation properties for all of the purine C8 (14/14) and nearly all of the pyrimidine C6 (11/15) and ribose C1′ (23/29) could be measured and analyzed reliably for free TAR. For the TAR–Arg complex, the relaxation properties for 12 of the 14 C8 purines, 12 of the C6 pyrimidines and 21 of the C1′ ribose resonances were measured and analyzed. Regarding the TAR complex with the linear Tat peptide, relaxation properties were measured and analyzed for 12 C8 purines, 10 C6 pyrimidines and 22 C1′ ribose resonances. Finally, the relaxation properties of all 14 purine C8′s, 12 pyrimidine C6′s and 25 C1′ ribose resonances for the TAR–cyclic peptide complex, were measured and analyzed. Figure 3 displays one set of relaxation parameters for the riboses and the bases of the cyclic peptide–TAR complex, respectively. Due to the large amount of data that would have to be reported, the majority of the observed relaxation times obtained from the analysis of these decay curves and the heteronuclear NOE are reported in the Supplementary Data.
Figure 2.
Representative primary relaxation data for TAR RNA; normalized peak intensities are plotted as a function of relaxation delay time, τ, for 13C relaxation experiments carried out at 500 MHz on the Arg–TAR complex. (a) 13C T1; (b) 13C T1ρ; the curves through the data represents the best fit using the standard expression for mono-exponential decay.
Figure 3.
(a) Relaxation parameters (T1, T1ρ and heteronuclear NOE) for C1′ resonances plotted as a function of residue position in the cyclic peptide TAR complex; for reference, the Tat-binding site (the UCU bulge) is in blue and the apical loop in green. (b) Relaxation parameters (T1, T1ρ and heteronuclear NOE) for C6 and C8 resonances plotted as a function of residue position in the cyclic peptide TAR complex. All relaxation times for the other complexes are reported as Supplementary Data.
The NMR structures for TAR and two of the TAR–ligand complexes studied here (5,6,8,21) (corresponding to Arg and the cyclic peptide, there is no structure for the Tat peptide–RNA complex because of considerable residual dynamics in the peptide itself) indicate the two double helical sections form ideal A-form helices. These helices are rigid and without significant internal motion (6,30); hence, the average values of T1, T1ρ and the heteronuclear NOE observed in the helical regions of TAR in the different complexes provide a reference point to assess the presence of internal motion in other residues (35). Since the helices are assumed to undergo only global motions, the observed changes in average relaxation times between complexes arise from changes in overall correlation time due to increased molecular weight and/or from altered hydrodynamic properties of the RNA when TAR changes structure upon ligand binding.
The average relaxation values for the C1′, C6, and C8 sites of the helical regions for free TAR and the three TAR–ligand complexes are shown in Table 1. Although the molecular size of the TAR–Arg complex is similar to that of free TAR, it is well known (6) that binding argininamide to TAR changes the overall shape of the RNA. This conformational change may very reasonably account for the small increases in average relaxation times (15% for sugars and 24% for bases) reported in Table 1. The increases in relaxation times observed for the linear and cyclic peptides reflect the increased molecular weight and size of these complexes.
Table 1.
The average relaxation times (ms) and heteronuclear NOE values for the helical residues of unbound TAR and its three complexes
| Site | TAR | Arg–TAR | Tat11–TAR | L22–TAR |
|---|---|---|---|---|
| T1 (ms) | ||||
| C1′ | 536 ± 13 | 619 ± 35 | 602 ± 38 | 604 ± 27 |
| C6 | 354 ± 14 | 441 ± 16 | 468 ± 18 | 429 ± 25 |
| C8 | 442 ± 22 | 551 ± 13 | 556 ± 14 | 529 ± 16 |
| T1ρ (ms) | ||||
| C1′ | 37.2 ± 0.4 | 29.1 ± 0.6 | 26.1 ± 0.6 | 29.5 ± 0.7 |
| C6 | 25.2 ± 0.2 | 18.8 ± 0.5 | 17.6 ± 0.3 | 20.2 ± 0.7 |
| C8 | 31.0 ± 0.3 | 24.5 ± 0.7 | 22.0 ± 0.3 | 25.3 ± 0.7 |
| NOE | ||||
| C1′ | 1.23 ± 0.02 | 1.14 ± 0.02 | 1.18 ± 0.03 | 1.13 ± 0.01 |
| C6 | 1.16 ± 0.02 | 1.13 ± 0.02 | 1.09 ± 0.02 | 1.10 ± 0.02 |
| C8 | 1.18 ± 0.02 | 1.18 ± 0.01 | 1.15 ± 0.02 | 1.14 ± 0.01 |
In this analysis, residues experiencing internal motions are identified by comparing relaxation time to the average values shown in Table 1. Residues exhibiting motion on a fast (ns–ps) timescale are expected to yield relaxation times, T1 and T1ρ, which are smaller and larger than those average values for a rigidly tumbling molecule, respectively. In addition, ps–ns motions will result in larger than average NOE values, due to the dependence of the NOE enhancement on the correlation time. Motion on the μs–ms timescale (conformational exchange) can also be identified from the relaxation data (yielding lower than average T1ρ) and can be confirmed by measuring the power dependence of T1ρ. Based on these considerations, noteworthy trends that emerge from the data are presented in the next section.
All three ligands induce similar changes in dynamics within the bulge loop, but the cyclic peptide induce unique dynamics within the apical loop
The results obtained for free TAR are consistent with previous studies (27,36,37), but it is worth recapitulating them here because they provide a necessary reference. Based on the relaxation data tabulated in the Supplementary Data and the principles described in the previous section, we conclude that several ribose residues in free TAR (U23, U31, G32, G33, G34, A35, G43 and C45) exhibit fast motions on the ps–ns timescale. U23 is part of the Tat-binding site and is critical for ligand binding, while C24 and U25 act primarily as spacers (8). U31, G32, G33, G34 and A35 are all parts of the apical loop, the binding site of Cyclin T1 (38). G43 experiences increased dynamics uncharacteristic of helical residues, for which we have no explanation, while C45 is the terminal 3′ residue and experiences end-fraying motions that are well known to occur in DNA and RNA. In addition, several of these residues (C30, U31, G34 and A35) experience slower motions (µs); these are also observed for A22, which is adjacent to the Tat-binding site and C24 within the bulge. Base resonances within the Tat-binding site (U23, C24) and the apical loop (G32, G33, G34 and A35) also have relaxation times consistent with the presence of fast internal motions; unfortunately, the remaining residues in the Tat-binding site and apical loop could not be resolved from other resonances. Of the bases with well-resolved resonances, G34 and A35 experience slower motions as well (µs). Altogether, these data confirm that the loop bases have inherently higher flexibility and are unlikely to participate in any hydrogen bonding interactions with other nucleotides.
Upon formation of each of the three complexes we have studied, residues in the Tat-binding site experience considerable changes in dynamics (Supplementary Data and Figure 3). While both the base and ribose of U23 become more rigid in each complex, the base and ribose of C24 become more mobile on the ns–ps timescale compared to free TAR. U25 exhibits considerable motion in the complexes on the ns–ps timescale as well; however, comparison with the free RNA cannot be made for this nucleotide because it is overlapped in free TAR. These results are consistent with the structures of TAR showing that U25 and C24 become extra-helical and structurally unconstrained upon binding of argininamide or any Tat-related peptide (5,6,39). In addition, the power dependence data demonstrate that the slower motions experienced by the C24 ribose are quenched in all complexes.
Ligand-specific changes in dynamics are clearly evident in the apical loop as well (Supplementary Data and Figure 3). However, while Arginineamide and the linear Tat peptide complexes have similar dynamic profiles in the loop, the cyclic peptide differs from the other two ligands and from free TAR. In the apical loop, all bases that are resolved in the free form of TAR (G32, G33, G34 and A35) become more rigid on the ns–ps timescale upon ligand binding, with the exception of the A35 base in the Arg complex which retains its flexibility. For the Arg and linear Tat peptide complexes, the ribose of C30 retains the slow motions observed in the free form of TAR, while U31 becomes rigid on the ns–ps timescale and continues to exhibit motions on the µs timescale. While both G32 and G33 retain some flexibility within their riboses, they become more rigid than in the free form; however, G32 and G33 retain more flexibility in the cyclic peptide complex compared to the other complexes. Interestingly, the cyclic peptide complex retains slower motions for nucleotide A35, although these motions are quenched in the Argininamide and linear Tat peptide complexes. Thus, the RNA riboses in the TAR complex with the cyclic peptide are more mobile than the other complexes for residues G32, G33 and, on a different time scale, for A35. G32 retains motion on a ps–ns time scale in all free and bound forms of TAR. Intriguingly, the C30 (resolvable in the Arg complex only) and U31 bases experience motions in both the Arg and linear peptide complexes that are not evident in the cyclic peptide complex.
Model-Free analysis of the 13C relaxation data
In order to obtain more quantitative insight into the changes in TAR dynamics occurring upon binding of each ligand, the primary relaxation data were analyzed with Model-Free to determine order parameters S2, which describe local flexibility for each individual site. Despite the well-known limitations of this approach (30,40,41), it represents a well-accepted first approximation to the description of dynamics in proteins and, more recently, of RNA as well (16,27,30).
The Model-Free analysis was performed for the eight sets of relaxation data, for the bases and riboses, in order to provide quantitative descriptions of the dynamic changes upon ligand binding. The relaxation data for non-terminal helical base pairs were first used to determine an overall rotational correlation time, τm, for free TAR as well as the three complexes. Structures that are not spherical require additional parameters describing the asymmetric diffusion tensor, such as the angles describing the axis of symmetry of the diffusion tensor, in addition to the overall global tumbling time. This is a simple task if a 3D structure is available. Upon analysis of the data using two different models (isotropic and axially symmetric rotational diffusion based on the available structures), the parameters extracted for both the sugar and base resonances of free TAR, Arg–TAR and cyclic peptide–TAR showed a difference of less than 5%. Thus, the axially symmetric model was used when a structure was available (free TAR, Arg–TAR and cyclic peptide–TAR); in the case of the complex with the Tat-derived linear peptide, for which no structure is available, the isotropic model was employed instead.
Global correlation times were calculated using Model-Free 4.15 (31) with only non-terminal helical residues used in the calculation. Free TAR was calculated to have a rotational correlation time of 5.9 ns; this is similar to RNAs of similar size; correlation times of 6.0 ns and 4.3 ns were obtained for the U1A protein-binding site (30) and for HIV-2 TAR (36), respectively. Binding of Arg (τm=6.4 ns) does not increase the mass substantially, but induces a significant conformational change, which may reasonably explain the slightly increased correlation time. Binding of the cyclic and linear peptides results in correlation times of 7.2 ns and 7.6 ns, respectively; these values are close to what is expected from the increase in mass.
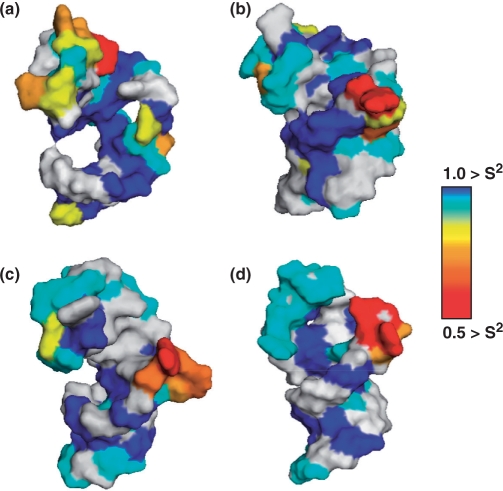
The complete results of this analysis for all nucleotides and all complexes are reported in the Supplementary Data. In order to represent the substantial set of motional parameters obtained from this analysis graphically and concisely, residues have been color-coded according to their observed S2 values on the structure of TAR and its complexes in Figure 4.
Figure 4.
S2 values determined by Model-Free analysis are displayed as a surface representation of the structure of TAR for the C1′, C6 and C8 sites. Residues that could not be analyzed due to spectral overlap are shown in light grey. Sites exhibiting local motion are colored based upon their S2 value and the corresponding color in the above scale: (a) free TAR; (b) Arg–TAR; (c) linear peptide–TAR and (d) cyclic peptide–TAR complex.
For free TAR, the S2 values for riboses with double helices (excluding terminal residues) range from 0.88 to 0.98, with an average of 0.93, while for the bases S2 values are between 0.89 and 0.97 with an average of 0.95. S2 values for the Arg–TAR complex range from 0.86 to 0.94 for the ribose and 0.79 to 0.98 for the bases, with averages of 0.93 and 0.91, respectively. For the linear peptide–TAR complex, S2 values for the riboses ranged from 0.89 to 0.98 with an average of 0.93 and from 0.83 to 0.99 for the bases with an average value of 0.97. S2 values for the cyclic peptide–TAR complex range from 0.92 to 0.99 for the ribose and 0.86 to 0.99 for the bases, with average of 0.95 and 0.94, respectively. Thus, very similar values of S2 are calculated for all complexes, within experimental uncertainty, for all helical residues.
Quantitative analysis of the changes in dynamics in TAR bound to the three ligands
Changes in ns–ps dynamics of TAR upon binding to each of the three ligands are represented graphically in Figure 4. In all cases, the bulge region displays significant increases in flexibility for the two spacer residues, C24 and U25. In contrast, the loop region of all three complexes becomes increasingly rigid, with the cyclic peptide complex showing the greatest decrease in flexibility in this region.
Consistent with the qualitative analysis of relaxation times reported earlier, U23 becomes more motionally rigid in all three TAR complexes. The C24 base and ribose both become more flexible in each of the complexes; the S2 values change from 0.7 and 0.9 in free TAR to 0.36–0.5 and 0.4–0.6 in the complexes. Although U25 cannot be analyzed in free TAR, both its base and ribose are flexible in the bound forms of TAR, with S2 of about 0.50 for the base and of between 0.64 (both peptides) and 0.76 (Arg) for the ribose. These results are consistent with the structure of the TAR complexes, since binding of each of the three ligands pushes C24 and U25 outside of the helical structure of TAR (6,39).
The ns–ps dynamics of the apical loop are significantly more complex than those observed at the bulge. Furthermore, a large proportion of the resolvable residues in the apical loop undergo conformational exchange on the µs time scale resulting in a decrease in the transverse relaxation time. The occurrence of conformational exchange has previously been shown to influence the order parameters extracted from Model-Free (30). In such cases, a qualitative analysis of the T1, T2 and NOE ratio (as shown earlier in the article) is more accurate than Model-Free (30). Therefore, the qualitative discussion presented above remains valid for most loop residues. However, the order parameters of G32 and G33 are not influenced by conformational exchange as they do not experience it. We observe that the order parameters for G32 and G33 of free TAR (0.69 and 0.74 for base and ribose) increase for the cyclic peptide complex (0.81 and 0.81), the Arg complex (0.83 and 0.91) and the complex with the linear Tat peptide (0.87 and 0.92).
Conformational exchange
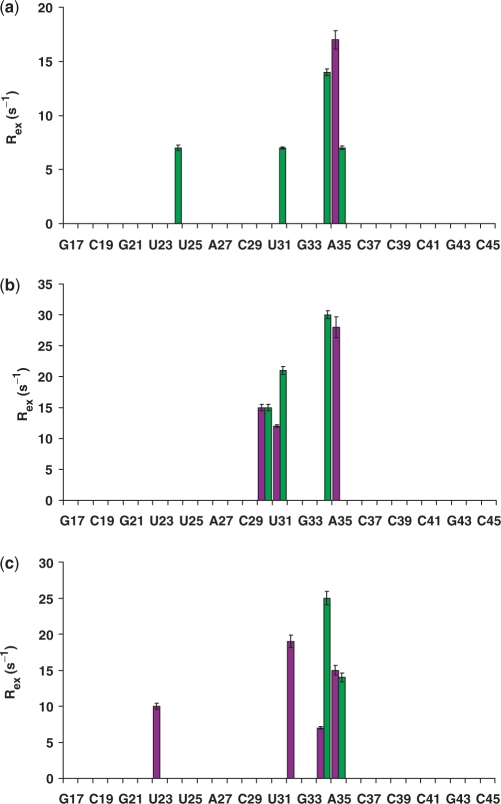
The data on the power dependence of T1ρ identify several sugar residues in free TAR that experience conformational exchange (Figure 5). Consistent with the qualitative and Model-Free analysis of the relaxation data, these motions primarily occur in the bulge C24 (7 s−1) and within the loop [U31 (7 s−1), G34 (14 s−1) and A35 (7 s−1)]. Unlike the relaxation data, however, no evidence of conformational exchange was found from the power dependence studies for residue C30. Power dependence studies probe a limited timescale, as slow as ∼300 µs with the spin-lock powers used in this study, and it is likely that C30 experiences instead motion outside of this timescale. Interestingly, for the Arg complex, power dependence of T1ρ was evident for the C30 ribose, implying that motions on slightly different timescales may occur for this residue in these two states of TAR. In addition to C30, conformational exchange is also present for U31 (21 s−1) and G34 (30 s−1) when Arg is bound. In the cyclic peptide complex, only residues G34 (25 s−1) and A35 (14 s−1) exhibit a clear dependence of T1ρ on the spin-lock field. Significantly, exchange rates for both complexes are 2–3-fold higher than in free TAR.
Figure 5.
Rex (s−1) as determined through T1ρ power dependence experiments. Riboses are displayed in green; bases in purple for: (a) unbound TAR; (b) Arg–TAR and (c) cyclic peptide–TAR complex.
Only two base resonances experience conformational exchange in free TAR. Power dependence data indicate that A35 (15 s−1) undergoes conformational exchange. The reduction in T1ρ is masked by fast motions and not immediately obvious from the primary relaxation data. Conversely, there is no evidence of conformational exchange for the base of G34 from the power dependence data. However, as with the C30 ribose, it is likely that the intermediate motions are outside of the range probed by the spin-lock experiments described here. Consistent with these observations, conformational exchange for the G34 base has been previously described in the literature (37).
The T1ρ power dependence data indicate that the same nucleotides in different complexes experience distinct motions on the time scale probed by these experiments (Figure 5). In the Arg complex, the bases of residues C30 (15 s−1) and U31 (12 s−1) undergo conformational exchange; these motions are absent in the cyclic peptide complex. Relaxation data suggest that residue G32 exhibits µs–ms motion in both the Arg and cyclic peptide complexes; however, the power dependence of T1ρ is only observed in the second case (19 s−1), implying that these slower motions occur on different timescales. In both complexes, A35 undergoes conformational exchange (28 s−1 for Arg, 15 s−1 for the cyclic peptide). Conformational exchange is also observed for U23 (10 s−1) and G34 (7 s−1) in the cyclic peptide complex (Figure 5). Thus, TAR is dynamically rich in the µs–ms time scale, and different ligands have distinct motions in this regime.
Ligand binding quenches inter-helical motions
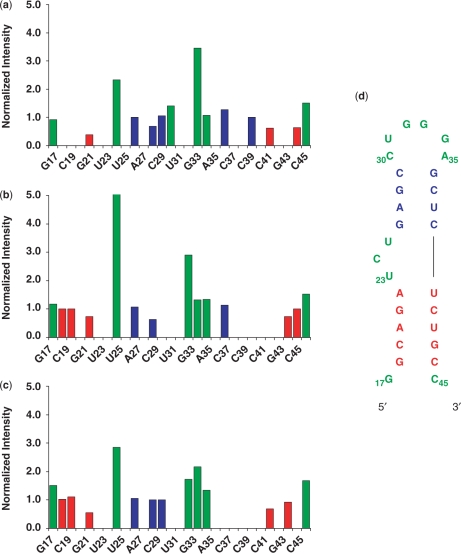
Ligand-free TAR undergoes interhelical motions centered around the bulge that cannot be discerned by relaxation measurements because they occur at rates comparable to the global rotational correlation time (14,27,42,43). Binding of Arg quenches these motions (42). While it is reasonable to assume that binding of either the linear or cyclic peptides will have the same effect, we nonetheless examined helix-elongated TAR RNAs bound to these two peptides to verify this hypothesis. To execute these experiments, the stem 1 helix of TAR was elongated by an additional 22 base pairs: 20 AU base pairs plus two GC base pairs at the end to promote transcription. NMR data were only collected and analyzed on samples labeled with 13C and 15N labeled only at guanidine and cytosine residues to render the elongated stem invisible by NMR. Since variations in intensities of peaks in HSQC spectra report on the overall dynamics of a particular labeled site (28,35), intensities in HSQC spectra for each residue were normalized for each nucleotide (G and C) independently. We analyzed exclusively residues that do not exhibit internal motion, since their presence would affect the apparent intensity of that resonance and could falsely indicate the presence of domain motions.
Figure 6 displays three of the six data sets (the remaining data are in the Supplementary Data) that were collected on the elongated TAR constructs. In free TAR, the resonances located in stem 1 (G21, C41 and C44) have intensities significantly lower than any of the resonances located in either stem 2 or in the flexible regions of TAR (loop, bulge and terminal ends). Data for the riboses (data not shown) reflect a similar trend, with peaks arising from stem 2 being about twice as intense as those observed in stem 1. These results agree with previous studies (27,28) showing that helix 2 experiences significant reorientational motions about the TAR helical axis. As we anticipated, binding of both linear and cyclic peptides reduces the difference in peak intensity between the two stem regions significantly (Figure 6b and c). The average differences in peak intensities between stem 1 and stem 2 for the bases and riboses of the linear peptide–TAR complex were 0.12 and 0.06, respectively, and 0.08 for the bases and 0.01 for the sugars for the cyclic peptide complex. As previously observed for arginine (27,28), these data are consistent with a significant reduction in interhelical motions upon ligand binding, since the difference in peak intensity between the two helices in each of these complexes is significantly lower than in the bound form. Consistent with this conclusion, when RDCs were measured for the cyclic peptide complex for use in structural refinement; no evidence was found for interhelical motions. Namely, the two helical regions could be described with a single set of orientational parameters demonstrating that they are rigidly related to each other.
Figure 6.
The normalized peak intensities of cross-peaks in 13C–1H HSQC spectra versus sequence of the elongated TAR construct. Stem 1 of wt-TAR was elongated by 20 AU base pairs and two GC base pairs. The elongated stem is not shown; stem 1 is in red, stem 2 is blue and the loop, bulge and terminal ends experiencing local motions are in green: (a) TAR bases; (b) Tat-TAR bases; (c) cyclic peptide-TAR bases and (d) HIV-1 TAR RNA sequence.
DISCUSSION
HIV-1 TAR RNA has been the subject of many studies aimed at discovering specific inhibitor of its activity, but until now it has not been possible to identify compounds with the potency and specificity required to elicit strong antiviral activity (5,10,21,34,44). It has been recently convincingly suggested that these small molecules bind to TAR by capturing pre-existing conformations that are sampled perhaps rarely in free TAR (3,10,14,37). The objective of the present work was to provide insight into how dynamics of TAR changes upon ligand binding, and if changes in dynamics correlate with the affinity or specificity of the interaction.
We have measured T1, T1ρ, the NOE ratio and T1ρ power dependence for most C1′, C6 and C8 sites for three complexes of HIV-1 TAR RNA using 13C NMR relaxation techniques. We have studied intrinsic motional properties and changes in dynamics that occur upon binding of three ligands of different affinity and specificity. Arginine binds weakly, with mM dissociation constant, but induces the same conformation in TAR that is observed in much more potent ligands (5). In contrast, the linear 11-mer Tat-derived peptide binds strongly (22); however, because it lacks any stable secondary structure, it also binds non-specifically to other RNAs (5). Finally, the cyclic peptide has a highly ordered structure, binds with much greater selectivity and, perhaps as a consequence, has unprecedented antiviral activity amongst Tat inhibitors (unpublished results). We observe that all three ligands induce essentially the same dynamics within the three nucleotide bulge that constitutes the site of interaction with Tat, and that is different from what is observed for free TAR. However, the much more specific cyclic peptide alters the dynamics of the apical loop, the binding site for the essential human co-factor Cyclin T1, in a way that is distinct from the other ligands and from unbound TAR.
Binding of each of the three ligands alters the dynamics of the Tat-binding site to a similar extent
The primary relaxation data and their analysis using Model-Free indicate that many residues in free TAR are inherently flexible. These observations are consistent with other studies that have probed motions in free TAR (27,36,37) and will therefore not be discussed at length; they have been measured to provide a baseline to discuss changes in motion upon ligand binding. We observe both fast motions (ns–ps) as well as conformational exchange (ms–µs) in a number of bases and sugars located in both the bulge loop where Tat binds and within the apical loop where cyclin T1 binds. These results indicate that the apical loop of free TAR experiences considerable motions on multiple time scales, as originally suggested in the first NMR study of this hairpin (7).
All three ligands induce similar changes in dynamics within the Tat-binding site, although they differ from each other. This result surprised us; the linear Tat-derived peptides, like all other Tat peptide and peptoid mimics (45) studied by us in the past (5,6), remains highly flexible and devoid of stable secondary structure, while the cyclic peptide is well-structured and forms a rigid β-hairpin in the presence and absence of TAR (21,34). U23 becomes more rigid, while C24 and U25 (which are both increasingly extrahelical upon ligand binding) become highly mobile. Slower motions in the μs time scale found in the ribose of C24 in the free form of TAR disappear in all three complexes. Furthermore, interhelical motions centered on the bulge in free TAR (28,42) are essentially quenched in each of the three complexes.
Binding greatly reduces interhelical motions of TAR
The data presented in this study demonstrate that binding of each of the three ligands significantly reduces the interhelical motions of TAR, as reported just for arginine in a previous study (42). Perhaps the decrease in rigidity in C24 and U25 helps compensate for entropic losses associated with the rigidification of U23 and the freezing of the interhelical motions (28,42), in addition to providing a structure conducive to binding. These results agree well with other observations regarding local dynamics in the area of the bulge, where binding was seen to increase rigidity on the ns–ps timescale (except for the extrahelical C24 and U25), while also quenching the conformational exchange present at C24. The loss of these motions is important as the bulge region acts as a flexible hinge between the two stems in the unbound form of TAR.
The precise location of the dynamic hinge allowing for the remarkable relative flexibility of the two halves of TAR remains unclear (27,28,46). Both the base (S2=0.84) and especially the ribose (S2=0.63) of U23 have order parameters consistent with the presence of ps–ns motions in free TAR, as does the base of C24 (S2=0.70). Although the corresponding ribose is rigid on a ns–ps time scale, power dependence studies indicate the sugar of C24 experiences a high degree of conformational exchange which is not seen for U23. The riboses of U40 (S2=0.85), A22 (S2=0.89) and G26 (S2=0.89) also display lower than average order compared to other helical residues. Thus, the ribose of C24 experiences slow conformational exchange and is surrounded by a highly flexible U23 residue and C24 base, and both are near helical sites with increased disorder compared to the remainder of the molecule. Upon binding to any of the ligands in this study, C24 becomes conformationally stable (no exchange), while at the same time extrahelical and therefore flexible on a different time scale (ns–ps). The base of U23 loses its flexibility and U40 rigidifies as well upon binding (27), as also do the riboses of A22 and G26.
Dynamic changes in the apical loop are ligand specific
In contrast to the behavior at the bulge, we observed different changes in apical loop dynamics for the three complexes. Both riboses (U31, G32 and G33) and bases (G32, G33, G34 and A35) become increasingly rigid on the ns–ps timescale and ligand binding induces µs motions for the G32 base in all complexes. These results are surprising; while the linear peptide may very well have a footprint that extends to the apical loop, binding of Arg should only occur near the UCU bulge. We do not attribute these changes to the quenching of interhelical domain motions. In the free TAR, domain motions occur at a rate comparable to rigid rotational diffusion and are therefore not discernable in the relaxation experiments (16,27,47). In both complexes, these motions are simply quenched. It is possible that a secondary binding site forms for Arg in the apical loop of TAR, resulting in the stabilization of several apical loop residues. Alternatively or in addition, changes in the structure and dynamics of the bulge loop are transmitted to the apical loop by an unknown molecular mechanism.
The analysis of the relaxation times and power dependence data indicate nevertheless that arginine and the linear Tat peptide mimic induce similar changes in apical loop dynamics. The C30 (resolved only in the Arg complex) and U31 bases experience motions in both the Arg and linear peptide complexes that are not evident in the cyclic peptide complex. In addition, the A35 base of both complexes remains highly flexible, likely as a result of the A35 flipping completely outside of the loop in the bound structure. This result contradicts observations made for the cyclic peptide complex, where the A35 base is rigid. This last complex nonetheless retains more flexibility than the other complexes for several ribose residues. Thus, the linear Tat-derived peptide binds to the apical loop of TAR in a manner closer to Arg than to the cyclic peptide, despite the similarity in size and affinity to the latter. These dynamic differences are also reflected in the structure of the corresponding complexes. Interactions with the apical loop of linear Tat-peptides were never stable enough to lead to well-defined loop conformation or intermolecular contacts. In contrast, the cyclic peptide interacts in a well-defined manner with the apical loop, leading to the observation of numerous intermolecular interactions and a new loop structure (unpublished results).
While the cyclic peptide significantly rigidifies the apical loop on the ns–ps timescale, it does not quench motions that occur on the ms–µs timescale and that are observed in the relaxation and especially in the power dependence data. For example, the timescale of motion for the G32 residues in the apical loop for this complex differs from the other complexes (G32 exchanges at a high rate, 19 s−1). We interpret these results as indicative of a significantly different structure and dynamic profile of the TAR loop when in complex with the cyclic peptide, potentially a very important feature with regards to inhibition of the activity of TAR in recruiting the cdk9 kinase to the HIV-1 promoter.
CONCLUSIONS
We have observed that binding of ligands of different affinity and specificity to TAR alters its conformational dynamics over a range of timescales. Regardless of the nature of the ligand, all complexes induce a much more rigid conformation for residues that make critical contacts with TAR (U23 and the nearby double helical residues), while increasing flexibility at bulge residues C24 and U25. This result surprised us, because linear peptides bind to TAR in a disordered conformation without forming a stable secondary structure, while the cyclic peptide forms a well-defined and rigid β-hairpin conformation. Thus, ligand binding does not simply eliminate motions, but instead leads to the formation of a new motional landscape that appears to be common to all peptidic ligands that mimic Tat. These changes in structure and dynamics also lead to the quenching of the interhelical motions observed in the ligand-free form of TAR. Examination of the apical loop where cyclin T1 binds indicates that the complexes with Arg and with the linear Tat mimic were very similar to each other, but distinct from the significantly more specific cyclic peptide. The unique profile in loop dynamics, together with the observation of unique intermolecular interactions with the apical loop, identifies the cyclic peptide as representative of a new class of ligands capable of interacting with the apical loop of TAR differently from other previously known peptide mimics of Tat. The unprecedented activity of this cyclic peptide in inhibiting viral replication may at least in part be due to its ability to interfere not just with binding of Tat to TAR, but also to the interaction of cyclic T1 with the TAR apical loop.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by Grants to Gabriele Varani from the National Science Foundation (MCB 0642253) and the National Institutes of Health (AI070090). Funding for open access charge: MCB 0642253.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mrs Amy Davidson for the assignments and structure of TAR bound to the cyclic peptide peptide and Dr Greg Olsen for his help in the preparation and purification of the Tat peptide.
REFERENCES
- 1.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 2.Williamson JR. Induced-fit in RNA-protein recognition. Nature Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hashimi HM. Dynamics-based amplification of rna function and its characterization by using NMR spectroscopy. Chem. Biol. Chem. 2005;6:1506–1519. doi: 10.1002/cbic.200500002. [DOI] [PubMed] [Google Scholar]
- 4.Draper DE. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 5.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 6.Aboul-ela F, Varani G. Recognition of HIV-1 TAR RNA by Tat protein and Tat-derived peptides. J. Mol. Struct. 1998;423:29–39. [Google Scholar]
- 7.Jaeger JA, Tinoco I., Jr An NMR study of the HIV-1 TAR element hairpin. Biochemistry. 1993;32:12522–12530. doi: 10.1021/bi00097a032. [DOI] [PubMed] [Google Scholar]
- 8.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faber C, Sticht H, Schweimer K, Rosch P. Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B. J. Biol. Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 10.Murchie AI, Davis B, Isel C, Afshar M, Drysdale MJ, Bower J, Potter AJ, Starkey ID, Swarbrick TM, Mirza S, et al. Structure-based drug design targeting an inactive RNA conformation: exploiting the flexibility of HIV-1 TAR RNA. J. Mol. Biol. 2004;336:625–638. doi: 10.1016/j.jmb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Leeper TC, Athanassiou Z, Dias RL, Robinson JA, Varani G. TAR RNA recognition by a cyclic peptidomimetic of Tat protein. Biochemistry. 2005;44:12362–12372. doi: 10.1021/bi0510532. [DOI] [PubMed] [Google Scholar]
- 12.Long KS, Crothers DM. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry. 1999;38:10059–10069. doi: 10.1021/bi990590h. [DOI] [PubMed] [Google Scholar]
- 13.Brodsky AS, Williamson JR. Solution structure of the HIV-2 TAR-argininamide complex. J. Mol. Biol. 1997;267:624–639. doi: 10.1006/jmbi.1996.0879. [DOI] [PubMed] [Google Scholar]
- 14.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1268. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 16.Shajani Z, Varani G. NMR studies of dynamics in RNA and DNA by 13C relaxation. Biopolymers. 2007;86:348–359. doi: 10.1002/bip.20650. [DOI] [PubMed] [Google Scholar]
- 17.Lipari G, Szabo A. Model-free approach to the interpretation on nuclear magnetic relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 18.Lipari G, Szabo A. Model-free approach to the interpretation on nuclear magnetic relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 19.Milligan JF, Groebe DR, Witherell WG, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase and synthetic DNA templates. Meth. Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 21.Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Structure-guided peptidomimetic design leads to nanomolar β-hairpin inhibitors of the Tat-TAR interaction of bovine immunodeficiency virus. Biochemistry. 2007;46:741–751. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 22.Olsen GL, Edwards TE, Deka P, Varani G, Sigurdsson ST, Drobny GP. Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR. Nucleic Acids Res. 2005;33:3447–3454. doi: 10.1093/nar/gki626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varani G, Tinoco I., Jr RNA structure and NMR spectroscopy. Q. Rev. Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 24.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 25.Varani G, Abou-ela F, Allain FHT. NMR investigation of RNA structure. Progr. Nucl. Magn. Spectr. 1996;29:51–127. [Google Scholar]
- 26.Yamazaki T, Muhandiram R, Kay LE. NMR experiments for the measurement of carbon relaxation properties in highly enriched, uniformly 13C, 15N-labeled proteins: applications to 13C carbons. J. Am. Chem. Soc. 1994;116:8266–8267. [Google Scholar]
- 27.Hansen AL, Al-Hashimi HM. RNA dynamics by carbon relaxation and domain elongation. J. Am. Chem. Soc. 2007;129:16072–16082. doi: 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaption. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Shajani Z, Varani G. 13C NMR relaxation studies of RNA base and ribose nuclei reveal a complex pattern of motions in the RNA binding site for human U1A protein. J. Mol. Biol. 2005;349:699–715. doi: 10.1016/j.jmb.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Mandel AM, Akke M, Palmer AG., III Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 32.Stueber D, Grant DM. 13C and 15N chemical shift tensors in adenosine, guanosine dihydrate, 2'-deoxythymine, and cytidine. J. Am. Chem. Soc. 2002;124:10539–10551. doi: 10.1021/ja012485c. [DOI] [PubMed] [Google Scholar]
- 33.Ying J, Grishaev A, Bryce DL, Bax A. Chemical shift tensors of protonated base carbons in helical RNA and DNA from NMR relaxation and liquid crystal measurements. J. Am. Chem. Soc. 2006;128:11443–11454. doi: 10.1021/ja061984g. [DOI] [PubMed] [Google Scholar]
- 34.Athanassiou Z, Dias RL, Moehle K, Dobson N, Varani G, Robinson JA. Structural mimicry of retroviral tat proteins by constrained beta-hairpin peptidomimetics: ligands with high affinity and selectivity for viral TAR RNA regulatory elements. J. Am. Chem. Soc. 2004;126:6906–6913. doi: 10.1021/ja0497680. [DOI] [PubMed] [Google Scholar]
- 35.Shajani Z, Varani G. Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry. 2007;46:5875–5883. doi: 10.1021/bi602658x. [DOI] [PubMed] [Google Scholar]
- 36.Dayie KT, Brodsky AS, Williamson JR. Base flexibility in HIV-2 TAR RNA mapped by solution 15N, 13C NMR relaxation. J. Mol. Biol. 2002;317:263–278. doi: 10.1006/jmbi.2001.5424. [DOI] [PubMed] [Google Scholar]
- 37.Dethoff EA, Hansen AL, Musselman C, Watt ED, Andricioaei I, Al-Hashimi HM. Characterizing complex dynamics in the TAR apical loop and motional correlations with the bulge by NMR, MD and mutagenesis. Biophys. J. 2008;95:3906–3915. doi: 10.1529/biophysj.108.140285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karn J. Tackling Tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 39.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 40.Schurr JM, Babcock HP, Fujimoto BS. A Test of the model-free formulas. Effects of anisotropic rotational diffusion and dimerization. J. Magn. Reson. Ser. B. 1994;105:211–224. doi: 10.1006/jmrb.1994.1127. [DOI] [PubMed] [Google Scholar]
- 41.Palmer AG, III, Williams J, McDermott A. Nuclear magnetic resonance studies of biopolymer dynamics. J. Phys. Chem. 1996;100:13293–13310. [Google Scholar]
- 42.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. Argininamide binding arrests global motions in HIV-1 TAR RNA: Comparison with Mg+2-induced conformational stabilization. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg+2-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J. Mol. Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis B, Afshar M, Varani G, Murchie AI, Karn J, Lentzen G, Drysdale M, Bower J, Potter AJ, Starkey ID, et al. Rational design of inhibitors of HIV-1 TAR RNA through the stabilisation of electrostatic “hot spots”. J. Mol. Biol. 2004;336:343–356. doi: 10.1016/j.jmb.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 45.Hamy F, Felder ER, Heizmann G, Lazdins J, Aboul-ela F, Varani G, Karn J, Klimkait T. An inhibitor of the Tat/TAR RNA interaction that effectively suppresses HIV-1 replication. Proc. Natl Acad. Sci. USA. 1997;94:3548–3553. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musselman C, Al-Hashimi HM, Andricioaei I. iRED Analysis of TAR RNA reveals motional coupling, Long-range correlations, and a dynamical hinge. Biophys. J. 2007;93:411–422. doi: 10.1529/biophysj.107.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shajani Z, Varani G. 13C Relaxation studies of the DNA target sequence for HhaI methyltransferaase reveal unique motional properties. Biochemistry. 2008;47:7617–7625. doi: 10.1021/bi7020469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.