Abstract
Here, we report on a significant effect of substitutions on the binding affinity of a series of 2-amino-1,8-naphthyridines, i.e., 2-amino-1,8-naphthyridine (AND), 2-amino-7-methyl-1,8-naphthyridine (AMND), 2-amino-5,7-dimethyl-1,8-naphthyridine (ADMND) and 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND), all of which can bind to cytosine opposite an AP site in DNA duplexes. Fluorescence titration experiments show that the binding affinity for cytosine is effectively enhanced by the introduction of methyl groups to the naphthyridine ring, and the 1:1 binding constant (106 M−1) follows in the order of AND (0.30) < AMND (2.7) < ADMND (6.1) < ATMND (19) in solutions containing 110 mM Na+ (pH 7.0, at 20°C). The thermodynamic parameters obtained by isothermal titration calorimetry experiments indicate that the introduction of methyl groups effectively reduces the loss of binding entropy, which is indeed responsible for the increase in the binding affinity. The heat capacity change (ΔCp), as determined from temperature dependence of the binding enthalpy, is found to be significantly different between AND (−161 cal/mol K) and ATMND (−217 cal/mol K). The hydrophobic contribution appears to be a key force to explain the observed effect of substitutions on the binding affinity when the observed binding free energy (ΔGobs) is dissected into its component terms.
INTRODUCTION
The chemistry of DNA-binding drugs and/or small ligands is of ongoing interest due to their promising functions and biological activities, including their anticancer properties and ability to regulate gene expression (1–4). Besides the anthracycline antibiotics doxorubicin and daunorubicin, many DNA-binding molecules have been developed as effective pharmaceutical agents, especially in cancer chemotherapy (5–7). Another class of DNA-binding molecules is useful stain agents for nucleic acids, and typical of such molecules are ethidium and Hoechst 33258 (8–9). Of particular interest to us is a class of ligands applicable to gene analysis (10,11), especially single-nucleotide polymorphism (SNP) typing (12).
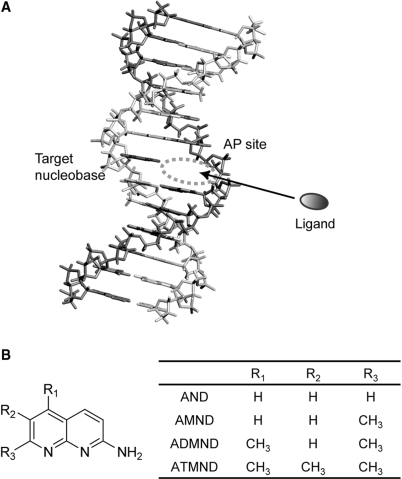
We have recently found a series of aromatic ligands that can selectively bind to a nucleobase opposite an abasic site (AP site) in DNA duplexes, and we have proposed a new strategy of ligand-based fluorescence assay for SNP typing (Figure 1A) (13–23). In contrast to typical DNA binding such as intercalation or groove binding (24,25), it is characteristic of ligands to bind to non-Watson–Crick base-pairing sites in DNA duplexes, where the binding is selectively promoted by a pseudo-base pairing along the Watson–Crick edge of the intrahelical target nucleobases (cf. Figure 7). Successful examples of this class of ligands are the mismatch-binding molecules developed by Nakatani and co-workers, and a surface plasmon resonance (SPR) assay has been proposed based on these molecules for the detection of mismatched base pairs in heteroduplexes (26–28). In our approach, on the other hand, we have paid attention to the AP site as a binding cavity for effective ligand–nucleobase interactions: while naturally occurring AP site is one of the most common forms of DNA damages (29), we have incorporated a chemically stable AP site in a probe DNA so as to orient the AP site toward a target nucleobase (Figure 1A). Such AP sites in the duplex provide a unique binding pocket that allows a direct contact of ligands to nucleobases via hydrogen bonding (cf. Figure 7), where the ligand is further stacked with two nucleobases flanking the AP site. Useful affinity and selectivity for target nucleobases opposite the AP site has been indeed obtained by various kinds of heterocyclic planar compounds, including cytosine selective 2-amino-7-methyl-1,8-naphthyridine (AMND, Figure 1B) (13,14), guanine selective 2-amino-6,7-dimethyl-4-hydroxypteridine (15), adenine selective alloxazine (16) and thymine selective amiloride (17), riboflavin (18). All of these ligands show a complexation-induced fluorescence signaling, and SNP genotype of samples can be clearly distinguished by combining ligands with selectivity for respective target nucleobases.
Figure 1.
(A) Schematic illustration of the ligand binding to nucleotides opposite an AP site in a DNA duplex. For the detection of SNPs, an AP site-containing probe DNA is hybridized with a target DNA so as to place the AP site towards a target nucleotide, by which a hydrophobic binding pocket is provided for aromatic ligands to bind to target nucleotide. (B) Structures of the series of 2-amino-1,8-naphthyridines examined in this work.
Figure 7.
Proposed binding modes of 2-amino-1,8-naphthyridines with cytosine or thymine opposite the AP site in DNA duplexes.
Simple substituent modification of the parent ring was found to be quite effective for tuning the binding affinity and selectivity when designing such AP site-binding ligands. In the case of pteridines capable of selectively binding to guanine, for example, the binding affinity of the parent ligand, 2-amino-4-hydroxypteridine, was 0.62 × 106 M−1 (in 110 mM Na+, pH 7.0, at 5°C) when binding to guanine in an 11-meric AP site-containing DNA duplex (5′-TCCAGXGCAAC-3′/3′-AGGTCGCGTTG-5′, X = AP site (Spacer-C3), G = target) (15,19). In contrast, the pteridine modified with two methyl groups, 2-amino-6,7-dimethyl-4-hydroxypteridine, showed an affinity with one order of magnitude higher (6.2 × 106 M−1) than that of the parent ligand (15). In the case of alloxazines, substitutions at the 7- and 8-position of the ring have been found to significantly affect the binding selectivity (16). While alloxazine showed a useful affinity and selectivity for adenine (1.2 × 106 M−1, in 110 mM Na+, pH 7.0, at 5°C), 7,8-dimethylalloxazine (lumichrome) showed a clear selectivity for thymine over other nucleobases (K11 for T: 1.6 × 107 M−1). These results clearly indicate that the ligand–nucleobase interaction at the AP site is strikingly sensitive to substitutions of the parent ring, and the subtle difference in the ligand structure is crucial to getting desirable binding properties. However, little is known about the details of such a substituent effect on the binding events, and therefore the detailed thermodynamic and/or structural characterization would provide significant knowledge for the rational design of this class of ligands (30–33). This would also offer a valuable insight into the molecular basis of interactions for the further development of various kinds of DNA-binding molecules.
In this work, we report on the effect of substitutions in 2-amino-1,8-naphthyridines on the binding to nucleobases in AP site-containing DNA duplexes, for which a series of 2-amino-1,8-naphthyridines (Figure 1B) was prepared, including 2-amino-1,8-naphthyridine (AND), 2-amino-7-methyl-1,8-naphthyridine (AMND), 2-amino-5,7-dimethyl-1,8-naphthyridine (ADMND) and 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND). We examined their binding characteristics by melting temperature (Tm) measurements, fluorescence spectroscopy and isothermal titration calorimetry (ITC). Interestingly, with increasing number of methyl groups attached to the naphthyridine ring, the binding affinity of 1,8-naphthyridines clearly increases for pyrimidine bases opposite the AP site in DNA duplexes: ATMND having three methyl groups does bind to cytosine with a 1:1 binding constant of 1.9 × 107 M−1 (in 110 mM Na+, pH 7.0, at 20°C), which is indeed two order of magnitude higher than that of the parent AND (0.030 × 107 M−1). The binding affinity for thymine is also enhanced effectively from AND (0.012 × 107 M−1) to ATMND (0.91 × 107 M−1). Such a significant effect of substitutions in 2-amino-1,8-naphthyridines is discussed based on the examination of thermodynamic parameters.
MATERIALS AND METHODS
Materials
A series of 2-amino-1,8-naphthyridines examined here were purchased from Specs, Akos, Maybridge and Enamine for AND, AMND, ADMND and ATMND, respectively. All of the DNAs used in the present study were custom synthesized and HPLC-purified (>97%) by Nihon Gene Research Laboratories Inc. (Sendai, Japan). For the synthesis of abasic (AP) site-containing DNAs, a tetrahydrofuranyl residue (dSpacer) was utilized. The concentration of DNA was determined from the molar extinction coefficient at 260 nm (34). Water was deionized (≥18.0 MΩ cm specific resistance) by an Elix 5 UV Water Purification System and a Milli-Q Synthesis A10 system (Millipore Corp., Bedford, MA, USA). The other reagents were commercially available analytical grade and were used without further purification.
Unless otherwise mentioned, all measurements were performed in 10 mM sodium cacodylate buffer solutions (pH 7.0) containing 100 mM NaCl and 1.0 mM EDTA. Before measurements, the sample solutions were annealed as follows: heated at 75°C for 10 min, and gradually cooled to 5°C (3°C/min), after which the solution temperature was raised again to 20°C (1°C/min).
Melting temperature measurements
Absorbance of DNA was measured at 260 nm as a function of temperature using an UV-vis spectrophotometer Model UV-2450 (Shimadzu Corp., Kyoto, Japan) equipped with a thermoelectrically temperature-controlled micro-multicell holder (8 cells; optical path length = 1 mm). The temperature ranged from 2°C to 92°C with a heating rate of 1.0°C/min. The resulting absorbance versus temperature curve was analyzed by a differential method to determine Tm values.
Determination of binding constants by fluorescence titrations
To determine the 1:1 binding constant (K11), fluorescence spectra of ligands were measured at 20°C with a JASCO FP-6500 spectrofluorophotometer (Japan Spectroscopic Co. Ltd., Tokyo, Japan) equipped with a thermoelectrically temperature-controlled cell holder (quartz cuvette, 3 mm × 3 mm). Typically, in the case of ATMND titration, the ligand concentration was fixed at 500 nM, and the concentration of DNA duplex ranged from 0 to 2.5 μM. The changes in fluorescence intensity at 403 nm (maximum wavelength) were monitored as a function of duplex concentration. The resulting titration curve was analyzed by nonlinear least-squares regression based on a 1:1 binding isotherm (35):
| 1 |
where F and F0 are the observed fluorescence intensities of ligand in the presence and absence of DNA duplexes, respectively, and k (= k11/kL) represents the ratio of proportionality constants connecting the fluorescence intensities and concentrations of the species (1:1 complex: k11, free ligand: kL). The free duplex concentration, [D], can be related to known total concentrations of duplex (D0) and ligand (L0), by the following equation:
| 2 |
Salt dependence of the binding constants
The effect of different NaCl concentrations on the 1 : 1 binding constants was examined at 20°C (pH 7.0) by fluorescence titration experiments, as described above, and analyzed according to the polyelectrolyte theory by Record et al. (36) The observed salt dependence of the binding constants is explained by the following relationship:
where Z is the apparent charge on the ligand, and ψ is the proportion of counterions associated with each DNA phosphate group. The slope (SK) of the plot, which is equivalent to the number of counterions released from DNA upon ligand binding, was obtained from lines of best linear least squares fit, and was used to evaluate the polyelectrolyte contribution (ΔGpe) to the observed binding free energy (ΔGobs) using the relationship (36):
The nonpolyelectrolyte contribution (ΔGt) was then given by the following equation (37):
Isothermal titration calorimetry
ITC experiments were carried out using a Microcal VP-ITC calorimeter (Microcal Inc., Northampton, MA, USA). The Origin software (Microcal) was used for data acquisition and analysis. All solutions were degassed by stirring under vacuum before use. Typically, the reference cell contained deionized water, and a titration was done at 20°C so that 15 μl of ligand solution were added (a total of 17 injections) to 1.43 ml of DNA solution in the sample cell. The injection time was 30 s, and the interval between injections was 300 s. In order to remove any air bubbles in the tip of syringe, the initial injection was set as 5 μl and the resulting peak was neglected in the analysis. The peaks produced during titration were converted into heat output per injection by integration and correction for the cell volume and sample concentration. The heats of dilution for the addition of ligand into buffer solution were determined independently, and the net enthalpy for ligand–DNA interactions was determined by subtraction of the heats of dilution. The data thus obtained were best fitted to a model that assumed a single set of identical binding sites, giving binding enthalpies and stoichiometries.
Heat capacity measurements
ITC titration experiments were carried out at four temperatures between 5°C and 20°C, and the binding enthalpies were determined as described above. From the observed temperature dependence of the binding enthalpy, the change in heat capacity, ΔCp, was determined according to the relationship:
The obtained value of ΔCp was then used to estimate the hydrophobic contribution (ΔGhyd) according to the relationship of Record et al. (38):
Preparation and analysis of PCR products
Asymmetric PCR (52,53) amplification of 107-meric sense or antisense strands of K-ras gene (codon 12) (54) was done with a 20-meric forward primer (5′-GACTGAATATAAACTTGTGG-3′) and a 20-meric reverse primer (5′-CTATT GTTGG ATCAT ATTCG-3′). The reaction solution (100 μl) contained dNTPs (0.2 mM each), 10× PCR buffer (10 μl; TaKaRa), forward primer (300 pmol or 20 pmol), reverse primer (20 pmol or 300 pmol), template (0.5 ng) and Taq (2.5 U; TaKaRa Hot Start Version). PCR conditions: 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 52°C for 30 s and 72°C for 30 s, and then 72°C for 7 min and kept at 4°C. The 107-meric PCR product: sense strand, 5′-GACTG AATAT AAACT TGTGG TAGTT GGAGC TGNTG GCGTA GGCAA GAGTG CCTTG ACGAT ACAGC TAATT CAGAA TCATT TTGTG GACGA ATATG ATCCA ACAAT AG-3′; antisense, 5′-CTATT GTTGG ATCAT ATTCG TCCAC AAAAT GATTC TGAAT TAGCT GTATC GTCAA GGCAC TCTTG CCTAC GCCAN CAGCT CCAAC TACCA CAAGT TTATA TTCAG TC-3′ (N = G, C, A, or T).
After PCR amplification, an aliquot (40 μl) from the PCR product was buffered to pH 7.0 with 100 mM sodium cacodylate containing EDTA (1.6 mM). Then, ATMND (50 nM), and a 20-meric AP site-containing probe oligonucleotide (5.0 μM) were added (for sense strand analysis, 5′-CCT ACG CCA XCA GCT CCA AC-3′; for antisense strand analysis, 5′-GTT GGA GCT GXT GGC GTA GG-3′, X = AP site; dSpacer). Fluorescence spectra of the resulting solutions (50 μl) were then measured at 5°C with a JASCO FP-6500 spectrofluorophotometer equipped with a thermoelectrically temperature controlled cell holder (quartz cuvette: 3 mm × 3 mm); the slits for the excitation and emission monochromators were 5 and 5 nm, respectively.
RESULTS AND DISCUSSION
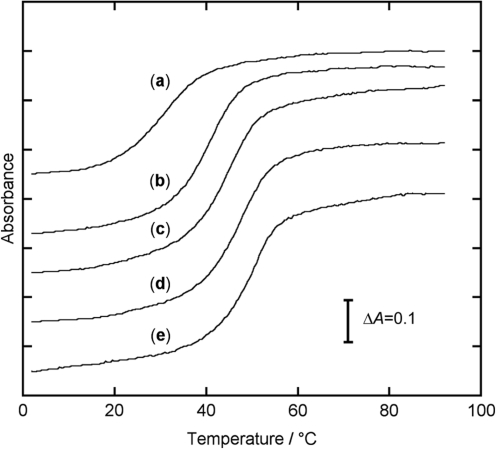
First, we examined the binding of 2-amino-1,8-naphthyridines to cytosine in an 11-meric AP site-containing DNA duplex [5′-TCC AGX GCA AC-3′/3′-AGG TCC CGT TG-5′, X = AP site (dSpacer), C = target] by Tm measurements. As is shown in Figure 2, all melting curves of the duplex give a sigmoidal shape typical for the thermal denaturation of DNA duplexes. In the absence of ligands (curve a), Tm value of the duplex is determined as 30.5°C from the first derivative of the melting curve. In the presence of ligands (curves b–e), an increase in Tm is clearly observed, indicating that each ligand is incorporated into the AP site by the binding to cytosine, which results in an increase in the thermal stability of the DNA duplex. The stabilization by ATMND is the most significant as compared to that of the other three ligands, where the Tm increases by as much as 20.6°C (curve e), and the ΔTm follows in the order of ADMND (+17.4°C) > AMND (+13.6°C) > AND (+10.8°C). It is therefore highly likely that the binding affinity of 2-amino-1,8-naphthyridines strongly depends on the number of methyl groups attached to the mother ring, and ATMND, having three methyl groups, shows the strongest binding affinity among these ligands.
Figure 2.
Thermal denaturation profiles of a 11-meric AP site-containing DNA duplex [5′ -TCC AGX GCA AC-3′/3′-AGG TCC CGT TG-5′, X = AP site (dSpacer), C = target cytosine]. (a) DNA alone, and in the presence of (b) AND, (c) AMND, (d) ADMND and (e) ATMND. [DNA duplex] = 30 µM, [ligand] = 580 µM, in 100 mM NaCl, 1.0 mM EDTA and 10 mM sodium cacodylate (pH 7.0). Absorbance of DNA was measured at 260 nm as a function of temperature, which ranged from 2°C to 92°C with a heating rate of 1.0°C/min. Optical path length = 1 mm.
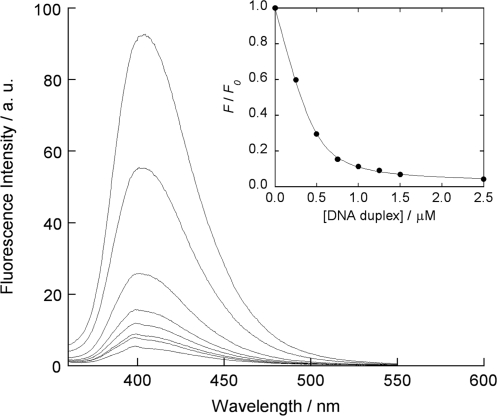
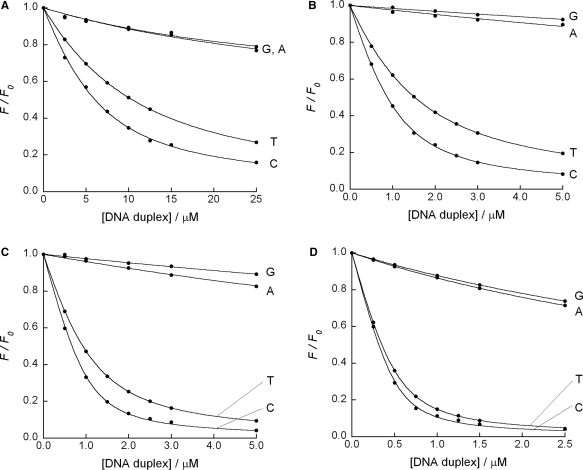
The examination by fluorescence titration experiments clearly supports the above consideration. Figure 3 shows a typical fluorescence response of 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND, 500 nM) to cytosine in a 21-meric AP site-containing DNA duplex [5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site (dSpacer), C = target cytosine], as measured in solutions containing 110 mM Na+ (pH 7.0, at 20°C). While almost no response is observed for a fully complementary duplex (500 nM, 5′-GCA GCT CCC GGG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′), ATMND shows significant quenching in the presence of the AP site-containing duplex, indicating that the binding event is taking place at the AP site. The other three 1,8-naphthyridines (ADMND, AMND and AND) also show fluorescence quenching upon binding to cytosine in the AP site-containing duplex (Supplementary Figure S1). For all ligands, the response is concentration-dependent, which is well analyzed by nonlinear least-squares regression based on a 1:1 binding isotherm (inset of Figure 3, and Supplementary Figure S1). The 1:1 binding constants K11 for cytosine thus obtained are summarized in Table 1.
Figure 3.
Fluorescence reponses of ATMND (500 nM) to 21-meric AP site-containing DNA duplex [5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site (dSpacer), C = target cytosine], measured in solutions buffered to pH 7.0 (10 mM sodium cacodylate) containing 100 mM NaCl and 1.0 mM EDTA. Excitation wavelength, 350 nm; temperature, 20°C. Inset: nonlinear regression analysis of the changes in the fluorescence intensity ratio at 403 nm based on a 1:1 binding isotherm model. F and F0 denote the fluorescence intensities of ATMND in the presence and absence of DNA duplexes, respectively.
Table 1.
Thermodynamic parameters for the 1,8-naphthyridine binding to cytosine in the 21-meric AP site-containing DNA duplexa
| K11 (M−1) | ΔGobs (kcal/mol) | SK | ΔGpe (kcal/mol) | ΔGt (kcal/mol) | ΔHobs (kcal/mol) | TΔSobs (kcal/mol) | |
|---|---|---|---|---|---|---|---|
| AND | 3.0 (±0.2) × 105 | −7.3 (±0.1) | −1.15 | −1.4 | −5.9 | −20.5 ± 0.6 | −13.2 (±0.6) |
| AMND | 2.7 (±0.2) × 106 | −8.6 (±0.1) | −1.31 | −1.7 | −6.9 | −19.8 ± 0.4 | −11.2 (±0.4) |
| ADMND | 6.1 (±0.5) × 106 | −9.1 (±0.1) | −1.36 | −1.8 | −7.3 | −16.7 ± 0.3 | −7.6 (±0.3) |
| ATMND | 1.9 (±0.2) × 107 | −9.8 (±0.1) | −1.42 | −1.8 | −8.0 | −12.8 ± 0.7 | −3.0 (±0.7) |
aK11 (M−1), determined by fluorescence titration experiments (cf. Figures 3 and 4), is the 1 : 1 binding constant in 110 mM Na+ at 20°C ([sodium cacodylate] = 10 mM, [EDTA] = 1.0 mM, [NaCl] = 100 mM, pH 7.0) with the standard deviations obtained from three independent experiments. ΔGobs is the observed binding free energy calculated from ΔGobs = −RTlnK11. SK is the slope of the plot of log K11 versus log [Na+] (cf. Supplementary Figure S3): fitting error is within 0.02. ΔGpe and ΔGt are the polyelectrolyte and nonpolyelectrolyte contributions to the observed binding free energy (ΔGobs) evaluated at 110 mM Na+ (ΔGobs = ΔGpe + ΔGt, ΔGpe = −SKRT ln [Na+]). ΔHobs was directly determined by ITC at 20°C (cf. Figure 5): Errors are the standard deviations obtained from three independent measurements. TΔSobs was calculated from TΔSobs = ΔHobs − ΔGobs. DNA duplex: 5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site (dSpacer), C = target cytosine.
As compared to AND having no methyl groups, the introduction of even one methyl group is effective for increasing the binding affinity, and the resulting ligand, AMND, shows an affinity for cytosine with one order of magnitude higher (K11 = 2.7 × 106 M−1) than that of AND (K11 = 0.30 × 106 M−1). The binding affinity is further enhanced for ADMND having two methyl groups (K11 = 6.1 × 106 M−1), and the affinity does reach 19 × 106 M−1 for ATMND. As for the binding to other three nucleobases (Figure 4), the binding affinity for thymine is also enhanced effectively by the introduction of methyl groups [K11/106 M−1 (n = 3): AND: 0.12 ± 0.01, AMND: 0.98 ± 0.09, ADMND: 2.4 ± 0.2, ATMND: 9.1 ± 0.3], and the binding selectivity for pyrimidines over purines remains unchanged from AND to ATMND. Significantly, the magnitude of binding affinity of ATMND for cytosine is stronger than that of typical intercalators such as ethidium (0.1 × 106 M−1, in 0.2 M NaCl, pH 7.0, at 25°C) (25) or actinomycin (3.8 × 106 M−1, in 0.1 M NaCl, pH 7.0, at 10°C) (39), and is almost comparable to that of groove binders such as distamycin (36 × 106 M−1, in 30 mM NaCl, pH 7.0, at 20°C) (40).
Figure 4.
Fluorescence titration curves for the binding of (A) AND (5.0 µM), (B) AMND (1.0 µM), (C) ADMND (1.0 µM) and (D) ATMND (0.5 µM) to 21-meric AP site-containing DNA duplexes [5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CNC CAG AGG AGC-5′, X = AP site (dSpacer), N = G, C, A or T]. Sample solutions were buffered to pH 7.0 with 10 mM sodium cacodylate, containing 100 mM NaCl and 1.0 mM EDTA. Excitation wavelength, 350 nm; temperature, 20°C. Analysis: AND, 392 nm; AMND, 400 nm; ADMND, 400 nm; ATMND, 403 nm.
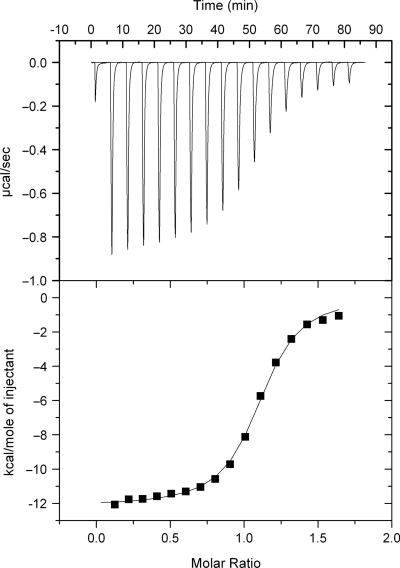
In order to understand such a favorable effect of methyl groups on the binding affinity, thermodynamic parameters for DNA–ligand interactions were examined, focusing on the binding to cytosine in the 21-meric AP site-containing duplex. First, the binding enthalpy was determined by ITC. Figure 5 shows typical ITC data for the binding of 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND) to cytosine in the AP site-containing DNA duplex [5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site (dSpacer), C = target cytosine], obtained in solutions containing 110 mM Na+ (pH 7.0, at 20°C). For all ligands, a large exothermic heat of reaction was observed upon addition of the ligand aliquots into the DNA solution (Supplementary Figure S2). After correction of the dilution heat, the resulting titration curve was best fitted using a model that assumed a single set of identical binding sites, giving the binding enthalpy (ΔHobs). This value was then used to calculate the binding entropy (TΔSobs) using TΔSobs = ΔHobs − ΔGobs, for which ΔGobs (=−RT lnK11) values determined from fluorescence titration experiments were utilized, since ITC cannot be used to obtain an accurate value for K11 under our experimental conditions: this limitation of ITC has been highlighted in literature (24,41).
Figure 5.
ITC data obtained at 20°C for the addition of ATMND aliquots (each 15 µl of 175 µM) into the solution containing DNA duplex [1.43 ml of 20 µM, 5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site (dSpacer), C = target cytosine]. Sample solutions were buffered to pH 7.0 with 10 mM sodium cacodylate, containing 100 mM NaCl and 1.0 mM EDTA. The data were best fitted to a model that assumes a single set of identical binding sites, giving the binding enthalpy (ΔHobs) of −12.8 kcal/mol with a binding stoichiometry (n) of 1.1.
Salt dependence of binding constants was then examined according to the polyelectrolyte theory of Record et al. (36), and the observed binding free energy (ΔGobs = −RT lnK11) was dissected into its polyelectrolyte (ΔGpe) and nonpolyelectrolyte (ΔGt) contributions (37):
The polyelectrolyte contribution arises from a release of counterions from DNA upon ligand binding, while the nonpolyelectrolyte contribution arises from all other molecular interactions. As is shown in Supplementary Figure S3, the binding constant K11 increases with decreasing the salt concentration, and a linear relationship is obtained between log K11 and log [Na+] for all ligand–cytosine interactions (cf. also Supplementary Table S1). The slope (SK) of the linear plot was used to evaluate ΔGpe in 110 mM Na+ using ΔGpe = (−SK)RTln [Na+], followed by calculation of ΔGt (=ΔGobs − ΔGpe).
Thermodynamic parameters thus obtained for the 1,8-naphthyridine-cytosine interactions are summarized in Table 1. While the binding reaction is enthalpy-driven in all cases, the effect of methyl groups can be clearly seen from the comparison of binding enthalpy and entropy. The most favorable gain in binding enthalpy is obtained for AND (ΔHobs = −20.5 kcal/mol), but the loss of binding entropy is also significant (TΔSobs = −13.2 kcal/mol). This results in the weakest binding free energy for AND–cytosine interactions (ΔGobs = −7.3 kcal/mol). By introducing methyl groups to the naphthyridine ring, the enthalpy term becomes destabilized, and this is highly compensated for by less negative values of entropy that result in a more favorable binding free energy. In case of AMND (ΔGobs = −8.6 kcal/mol), the gain in binding enthalpy is somewhat reduced (ΔHobs = −19.8 kcal/mol), whereas the loss of binding entropy is effectively decreased (TΔSobs = −11.2 kcal/mol). This leads to the increased binding affinity of AMND (ΔΔGobs = −1.3 kcal/mol) as compared to AND. Such an effect is more significant for ADMND (ΔGobs = −9.1 kcal/mol, ΔHobs = −16.7 kcal/mol, TΔSobs = −7.6 kcal/mol), and the loss of binding entropy reaches the minimum for ATMND (ΔGobs = −9.8 kcal/mol, ΔHobs = −12.8 kcal/mol, TΔSobs = −3.0 kcal/mol). These results clearly indicate that the introduction of methyl groups effectively reduces the loss of binding entropy, which is responsible for the increase in the binding affinity of 1,8-naphthyridine–cytosine interactions.
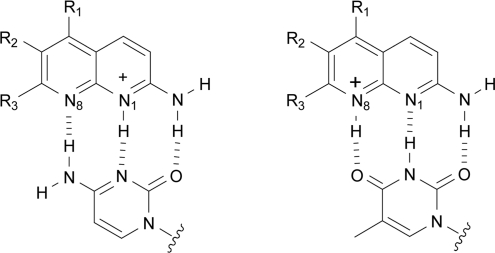
It has been well recognized that, for DNA–ligand interactions, the favorable binding entropy term may be due to the release of structured water from DNA and/or ligand into bulk solvent, and/or due to the release of condensed counterions from DNA. The latter effect has been reasonably observed for positively charged ligands such as ethidium (25), and even for uncharged ligands such as actinomycin (39) or chartreusin (42), the binding has been shown to accompany the release of counterions from DNA. This is generally attributed to lengthening and unwinding of the DNA duplex, both of which increase the phosphate spacing along the helix axis. This results in a decrease in the charge density of the duplex, thereby releasing condensed counterions from DNA. In the present case, 2-amino-1,8-naphthyridines have a positive charge due to the protonation at the N1 moiety when binding to cytosine (cf. Figure 7), and the chemical modification of the naphthyridine ring seems cause some additional effects on the binding-induced release of counterions. As summarized in Table 1, on increasing the number of methyl groups from AND to ATMND, the slope (SK) of the linear plot (SK), which is equivalent to the number of counterions released from DNA upon ligand binding, increases somewhat, providing a more favorable gain from the polyelectrolyte contribution (ΔGpe). In the case of AND, the binding is accompanied by the release of 1.1 counterions, which corresponds to the favorable gain of −1.4 kcal/mol from ΔGpe. In the case of ATMND, 1.4 counterions are released upon binding, and the value of ΔGpe increases to −1.8 kcal/mol. It is therefore likely that the effect of methyl groups is ascribed partially to the increased release of condensed counterions from DNA, which provides a favorable entropy contribution to the overall binding free energy (ΔGobs).
The effect of methyl groups is however more evident for the non-polyelectrolyte contribution (ΔGt). Again, as summarized in Table 1, ΔGt is indeed fundamental in the stabilization of the binding events, and ΔGt clearly increases as the number of methyl groups increases. As compared to AND (ΔGt = −5.9 kcal/mol), the favorable gain from ΔGt is increased by as much as −2.1 kcal/mol for ATMND, which is roughly comparable to the value for increased gain in the overall binding free energy (ΔΔGobs = −2.5 kcal/mol). Thus, the effect of methyl groups on the binding affinity is mainly ascribed to the increased gain from the nonpolyelectrolyte contribution ΔGt, so as to provide a favorable entropic term, probably due to the release of structured water from DNA and/or the ligand itself into bulk solvents.
According to the literatures (24,25,42–44), the nonpolyelectrolyte (ΔGt) contribution is further dissected into four contributions that drive the binding process, and thus the observed binding free energy (ΔGobs) is totally composed of at least five contributions:
where ΔGr+t is the free energy cost resulting from losses in rotational and translational degrees of freedom upon complex formation, ΔGhyd is the free energy for the hydrophobic transfer of the ligand from aqueous solution into the DNA binding site, ΔGconf is the contribution due to conformational transitions in DNA and the ligand, and ΔGint is the contribution from intermolecular DNA–ligand contacts within the binding site. Among these contributions consisting of ΔGobs, it has been well shown that the hydrophobic contribution (ΔGhyd) is a key parameter which is related to the change in surface area that is exposed to solvent upon complex formation, and it is also possible to correlate changes in solvent-accessible surface area (ΔSASA) with the heat capacity changes (ΔCp). This relationship has been successfully shown to hold for a typical DNA–ligand interaction, e.g. the Hoechst 33258 binding to DNA duplex, where the negative change in heat capacity was observed due to the removal of nonpolar surface from bulk solvent upon complexation, and the experimentally determined value for ΔCp was in excellent agreement with the value computed using ΔSASA obtained by two crystal structures (24). We therefore estimated the values of ΔGhyd from the heat capacity change (ΔCp), for which temperature dependence of the binding enthalpy was examined.
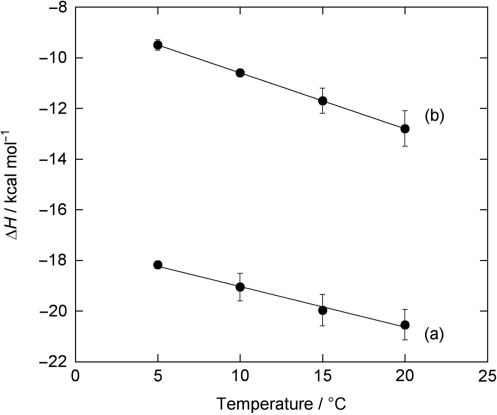
Table 2 summaries the binding enthalpy (ΔHobs) for two typical ligands, AND and ATMND, as determined by ITC measurements at different temperatures. For both ligands, the value for ΔHobs becomes less negative as the temperature is lowered, and ΔCp is determined from the slope of a plot of ΔH versus temperature (Figure 6). The obtained ΔCp is −161 cal/mol K for AND, and −217 cal/mol K for ATMND. These values are then used to estimate hydrophobic contribution (ΔGhyd) using Record's relationship, ΔGhyd = 80 (±10) × ΔCp (38), giving −12.8 (±1.6) kcal/mol and −17.3 (±2.2) kcal/mol for AND and ATMND, respectively. The observed gain from the hydrophobic contribution is significantly different between AND and ATMND (ΔΔGhyd = −4.5 kcal/mol), indicating that ΔGhyd is effectively modulated by the introduction of methyl groups to the naphthyridine ring. This result seems to be very consistent with the observed increase in the binding affinity, due to the reduction of the loss of binding entropy.
Table 2.
Temperature dependence of the observed binding enthalpy (ΔHob) and calculated heat capacity change (ΔCp) for the 1,8-naphthyridine binding to cytosine in the 21-meric AP site-containing DNA duplex
| ΔHobs (kcal/mol)a |
ΔCp (cal/mol K)b | ||||
|---|---|---|---|---|---|
| 20°C | 15°C | 10°C | 5°C | ||
| AND | −20.5 ± 0.6*1 | −20.0 ± 0.6*2 | −19.1 ± 0.5*2 | −18.2 ± 0.1*2 | −161 |
| ATMND | −12.8 ± 0.7*2 | −11.7 ± 0.5*2 | −10.6 ± 0.1*1 | −9.5 ± 0.2*2 | −217 |
aΔHobs was directly determined by ITC experiments. Errors are the standard deviations obtained from at least three independent measurements at each temperature (*1 4 times; *2 3 times). Sample solutions were buffered to pH 7.0 with 10 mM sodium cacodylate, containing 100 mM NaCl and 1.0 mM EDTA. DNA duplex: 5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site (dSpacer), C = target cytosine.
bHeat capacity change calculated from the slope δ(ΔH)/δT obtained by linear least squares fit (cf. Figure 6, r = 0.9906 for AND, r = 0.9999 for ATMND).
Figure 6.
Temperature dependence of the binding enthalpy for 2-amino-1,8-naphthyridine-cytosine interactions: (a) AND and (b) ATMND. Errors are the standard deviations obtained from at least three independent measurements. The linear least squares fit to the data yielded the heat capacity change, ΔCp, of –161 cal/mol K for AND (r = 0.9906), and –217 cal/mol K for ATMND (r = 0.9999), respectively. See Table 2 for further details.
Further parsing of the binding free energy (ΔGobs) was done by consideration of a value for the loss of translation and rotational freedoms (ΔGr+t = TΔSr+t) upon bimolecular complex formation for ligand–DNA binding reactions. While there is some controversy about the value of ΔSr+t (45,46), Spolar and Record (47) have empirically derived a value of ΔSr+t = 50 (±10) cal/mol K, which seems the most appropriate value to use, as has been extensively discussed in literatures (24,25,31). By using this value, we estimate ΔGr+t to be 14.7 kcal/mol at 20°C, with about 20% uncertainly. The remaining two contributions from unfavorable ΔGconf and favorable ΔGint are considered together in this work, and are obtained by subtracting the sum of the other three contributions discussed above (ΔGpe + ΔGr+t + ΔGhyd) from the experimental ΔGobs.
The energetic profiles thus determined for the binding of AND and ATMND are summarized in Table 3. For both ligands, the contribution from the hydrophobic transfer process (ΔGhyd) is very large as compared to the other favorable contributions (ΔGpe, and ΔGconf + ΔGint), and appears to be a key force to explain the observed effect of methyl groups on the binding affinity. In the case of AND, the favorable gain mainly comes from ΔGhyd (–12.8 kcal/mol), and further from ΔGpe (−1.4 kcal/mol) and ΔGconf + ΔGint (−7.8 kcal/mol). The entropic cost of ΔGr+t (14.7 kcal/mol) is overcome by the sum of these favorable contributions. In contrast, in case of ATMND, the large contribution from ΔGhyd (−17.3 kcal/mol) is of sufficient magnitude to overcome the unfavorable contribution from ΔGr+t (14.7 kcal/mol), so that the other favorable gains from ΔGpe (−1.8 kcal/mol) and ΔGconf + ΔGint (−5.4 kcal/mol) effectively contribute to the overall binding free energy. It is therefore highly likely that a major driving force for the 1,8-naphthyridine binding is the contribution from the hydrophobic transfer process (ΔGhyd), which is indeed highly dependent on the chemical modification of the naphthyridine ring with methyl groups.
Table 3.
Energetic profiles for the 1,8-naphthyridine binding to cytosine in the 21-meric AP site-containing DNA duplexa
| ΔGobs (kcal/mol) | ΔGpe (kcal/mol) | ΔGr+t (kcal/mol) | ΔGhyd (kcal/mol) | ΔGconf + ΔGint (kcal/mol) | |
|---|---|---|---|---|---|
| AND | −7.3 | −1.4 | 14.7 | −12.8 | −7.8 |
| ATMND | −9.8 | −1.8 | 14.7 | −17.3 | −5.4 |
aThe estimated contributions to the observed free energy (ΔGobs) from the five sources discussed in the text are given: Binding to the duplex (5′-GCA GCT CCC GXG GTC TCC TCG-3′/3′-CGT CGA GGG CCC CAG AGG AGC-5′, X = AP site; dSpacer, C = target cytosine) in 100 mM NaCl, 1.0 mM EDTA, 10 mM sodium cacodylate, pH 7.0 at 20°C.
The observed nature of the thermodynamic profile, i.e., the large magnitude of negative binding enthalpies, is similar to the profile for typical intercalators such as ethidium (ΔG = −6.7 kcal/mol, ΔH = −9.0 kcal/mol, TΔS = −2.3 kcal/mol, in 0.2 M NaCl, pH 7.0, at 25°C) (25) and daunorubicin (ΔG = −7.9 kcal/mol, ΔH = −9.0 kcal/mol, TΔS = −1.1 kcal/mol, in 0.2 M NaCl, pH 7.0, at 20°C) (31). This is consistent with the intercalation-like binding mode proposed for the 1,8-naphthyridine-cytosine interaction in AP site-containing DNA duplexes (cf. Figure 1). However, it is of interest to note that the values of ΔCp obtained for 1,8-naphthyridines, especially for ATMND (−217 cal/mol K), are indeed larger than the values of such typical intercalators. In the case of ethidium consisting of a hetrocyclic phenanthridine, the value of ΔCp has been estimated to be −139 cal/mol K (25), and even for much larger anthracycline antibiotics, daunorubicin, the ΔCp value has been estimated to be −160 cal/mol K (31). The ΔCp values estimated for 1,8-naphthyridines therefore appear to be somewhat large when considering their relatively smaller molecular size as compared to those of these intercalators. This interesting result may be ascribed to a unique local conformation of AP site-containing DNA duplexes.
As has been reviewed in the literature (48), the existing NMR data show that the local duplex structure strongly depends on the type of adjacent base pairs, the AP residue, and the orphan base, i.e., the base opposite the AP site. While the orphan purine bases, being largely hydrophobic, always stack inside the helix, the position of orphan pyrimidine residues shows more variability (49). When the THF abasic site analog is used, an orphan cytosine residue adopts extrahelical conformations, and tends to be solvent exposed when it is flanked by cytosine residues that have weak stacking ability. In the present study, the THF analogue is used for the AP site, and the target cytosine is flanked by cytosine residues, indicating that the target cytosine base is located outside the helix and exposed to solvent. For the binding of 2-amino-1,8-naphthyridines, it is therefore likely that the DNA duplex must undergo a conformational transition to form the cavity suitable for ligand binding, where the target cytosine stacks inside the helix, and this is followed by the transfer of 1,8-naphthyridines from solution into the binding cavity (AP site). This kind of binding events seem to accompany a considerable change in ΔSASA, providing the relatively large values of ΔCp as compared to those of typical DNA intercalation.
It is also of interest to note that, for the 1,8-naphthyridine-cytosine binding, molecular interactions (ΔGint), arising generally from specific hydrogen bonds, van der Waals contacts and other interactions, are effective contributors to the observed binding free energy (Table 3). Although the sums of values of ΔGint and ΔGconf are estimated in the present study, the sign and magnitude of the values obtained for ΔGconf + ΔGint (AND: −7.8 kcal/mol; ATMND: −5.4 kcal/mol) clearly indicate that the molecular interaction (ΔGint) is overcoming the unfavorable contribution due to conformational transitions in the DNA and the ligand (ΔGconf), and it is a more effective driving force than the polyelectrolyte contribution (ΔGpe: AND, −1.4 kcal/mol; ATMND, −1.8 kcal/mol). This is similar to intercalators such as ethidium (25), but distinct from groove binders such as Hoechst 33258 (24), where molecular interactions (ΔGint) were found to contribute little to the observed binding free energy, and both the hydrophobic (ΔGhyd) and polyelectrolyte (ΔGpe) contributions were found to be the primary driving forces for association.
As for molecular interactions involved in the 1,8-naphthyridine binding, specific hydrogen bonds are crucial for the selective binding to nucleobases in AP site-containing DNA duplexes. As suggested by Nakatani et al. (28,50) and revealed by 15N NMR experiments (51), 1,8-naphthridine (AMND, pKb = 6.8) exists as a tautomeric mixture of N1 and N8 protonated form in acidic solutions, and the N1 protonated form of 1,8-naphthridine binds to cytosine, so that a fully complementary base-pairing is formed via three-point hydrogen bonds (Figure 7). The N8 protonated form seems responsible for the binding to thymine, and this would also allow a fully complementary base-pairing based on three-point hydrogen bonds (Figure 7). These binding modes explain the observed binding selectivity for pyrimidines over purines in AP site-containing DNA duplexes (cf. Figure 4). In the case of ATMND, while the cytosine/thymine selectivity is only moderate, the binding affinity for cytosine is indeed two-order of magnitude higher than those for adenine and guanine (Kd/nM: C, 53 ± 6.0; T, 111 ± 4.1, A, 5800, G, 6000). Similarly, the other three 2-amino-1,8-naphthyridines show the selectivity for pyrimidines over purines, irrespective of the number of methyl groups attached to the naphthyridine ring. Thus, the 1,8-naphthyridine–DNA interaction does involve specific hydrogen bonds, and is not simply promoted by the transfer of ligands from solution into the hydrophobic binding site (AP site), which is another distinct feature, differing from typical DNA intercalation (25).
It should be here noted that the introduction of methyl groups does not contribute to the improvement of the binding selectivity between cytosine and thymine, another important issue from a practical point of view. While ATMND shows some preference for cytosine over thymine, the thermodynamic parameters for the ATMND–thymine interaction is almost comparable with parameters for the binding to cytosine. As summarized in Supplementary Table S2, the binding to thymine is enthalpy-driven (ΔGobs = −9.3 kcal/mol, ΔHobs = −13.6 kcal/mol, TΔSobs = −4.3 kcal/mol), and the values of binding enthalpy and entropy are comparable with the values for ATMND–cytosine interaction (ΔGobs = −9.8 kcal/mol, ΔHobs = −12.8 kcal/mol, TΔSobs = −3.0 kcal/mol). The polyelectrolyte (ΔGpe) contribution is also effective for the thymine binding (ΔGpe = −1.8 kcal/mol), which is consistent with the proposed binding mode by the positively charged (protonated) 1,8-naphthyridine (cf. Figure 7). Thus, a different approach rather than the methylation strategy should be required in order to develop 1,8-naphthyridine-based ligands with the improved binding selectivity between cytosine and thymine.
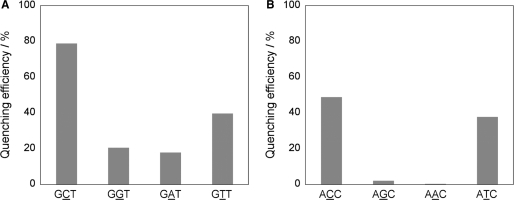
Finally, ATMND was applied to the analysis of single-base mutation present in 107-meric DNAs (K-ras gene, codon 12) (54). After asymmetric PCR amplification (52,53), the products were analyzed at 5°C in a buffer solution (pH 7.0, 100 mM sodium cacodylate) containing 1.6 mM EDTA, 50 nM ATMND and 5.0 μM AP site-containing 20-mer probe DNA. As shown in Figure 8A, in the case of sense strand analysis, a significant fluorescence quenching of ATMND (excitation wavelength: 350 nm, analysis: 403 nm) is observed for the cytosine-containing sequence (GCT: 79%), while the response is relatively moderate for the thymine- or purine-containing sequences (GTT: 40%; GAT: 18%; GGT: 21%). In the case of antisense strand analysis (Figure 8B), a fluorescence response is observed effectively for pyrimidine-containing sequences over purine-containing sequences (ACC: 48%; ATC: 38%; AAC: 1%; AGC: 2%). ATMND would be thus applicable to the detection of cytosine (thymine)-related transversion such as C(T)>G and C(T)>A, for which the simultaneous use of purine-selective ligands would assure the analysis, as has been previously demonstrated for G>A detection (16).
Figure 8.
Fluorescence detection of single-base mutation of PCR products present in (A) sense strand and (B) antisense strand of K-ras gene (107-mer, codon 12). After PCR, the products were analyzed in solutions buffered to pH 7.0 with 100 mM sodium cacodylate containing 1.6 mM EDTA, 50 nM ATMND and 5.0 μM AP site-containing probe DNA. Excitation 350 nm; detection 403 nm. Temperature 5°C.
CONCLUSION
In summary, we have demonstrated a significant effect of the methyl substitutions on the binding of a series of 2-amino-1,8-naphthyridines to pyrimidines in AP site-containing DNA duplexes. Despite the relatively simple modification, the binding affinity of 1,8-naphthyridines clearly increased with increasing the number of methyl groups of the naphthyridine ring, and ATMND having three methyl groups showed the strongest binding affinity of 1.9 × 107 M−1 and 0.91 × 107 M−1, respectively for cytosine and thymine (in 110 mM Na+, pH 7.0, at 20°C). These value were nearly two order of magnitude higher than those of the parent AND having no methyl groups (0.030 × 107 M−1 for cytosine, 0.012 × 107 M−1 for thymine). The obtained thermodynamic parameters for 1,8-naphthyridine-cytosine interactions (ΔGobs, ΔHobs, TΔSobs) indicated that, while the binding was enthalpy-driven for all ligands, the introduction of methyl groups effectively reduced the loss of binding entropy, which was responsible for the increase in the binding affinity of 1,8-naphthyridine–cytosine interactions. From the analysis based on the polyelectrolyte theory, we found that the nonpolyelectrolyte contribution (ΔGt) was fundamental in the stabilization of the binding events, and ΔGt clearly increased with increasing the number of methyl groups. Interestingly, the value of the heat capacity change (ΔCp) was found to be significantly different between AND and ATMND, and the estimated contribution from the hydrophobic transfer process (ΔGhyd) was found to be effectively modulated by the introduction of methyl groups to the naphthyridine ring. Indeed, the obtained energetic profiles revealed that a major driving force for the 1,8-naphthyridine binding was the contribution from the hydrophobic transfer process, which appeared to be a key force to explain the observed effect of methyl groups on the binding affinity.
As has been reported for some DNA-binding molecules, thermodynamic characterization of the binding reactions offers valuable insights into the major driving forces involved in the complex formation, and the obtained thermodynamic data, together with structural characterization, would be a rational basis for the further development of the chemistry of DNA-binding molecules. In particular, the comprehensive examination of a series of structurally related compounds might be an effective approach for this direction. We strongly expect that the results described here represent a piece of such data, especially for the further design of DNA-binding molecules applicable to gene analysis. We are now undertaking further studies on the design and synthesis of this class of ligands including the thermodynamic and structural characterization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Scientific Research (A) (No. 17205009), Scientific Research (B) (No. 20350032) and Exploratory Research (No. 19655022), and for G-COE Project, from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding for open access charge: CREST, JST.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 3.Uil TG, Haisma HJ, Rotz MG. Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities. Nucleic Acids Res. 2003;31:6064–6078. doi: 10.1093/nar/gkg815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spring DR. Chemical genetics to chemical genomics: small molecules offer big insghts. Chem. Soc. Rev. 2005;34:472–482. doi: 10.1039/b312875j. [DOI] [PubMed] [Google Scholar]
- 5.Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 6.Weiss RB. The anthracyclines – will we ever find a better doxorubicin. Semin. Oncol. 1992;19:670–686. [PubMed] [Google Scholar]
- 7.Asche C. Antitumour quinones. Mini Rev. Med. Chem. 2005;5:449–467. doi: 10.2174/1389557053765556. [DOI] [PubMed] [Google Scholar]
- 8.Witter CT, Ririe KM, Andrew RV, David DA, Gundry RA, Balis UJ. The lightcycler™ a microvolume multisample fluorimeter with rapid temperature control. Biotechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 9.Stokke T, Steen HB. Multiple binding modes for Hoechst 33258 to DNA. J. Histochem. Cytochem. 1985;33:333–338. doi: 10.1177/33.4.2579998. [DOI] [PubMed] [Google Scholar]
- 10.Takenaka S, Yamashita K, Takagi M, Uto Y, Kondo H. DNA sensing on a DNA probe-modified electrode using ferrocenylnaphthalene diimide as the electrochemically active ligand. Anal. Chem. 2000;72:1334–1341. doi: 10.1021/ac991031j. [DOI] [PubMed] [Google Scholar]
- 11.Ihara T, Ikegami T, Fujii T, Kitamura Y, Sueda S, Takagi M, Jyo A. Metal ion-directed cooperative DNA binding of small molecules. J. Inorg. Biochem. 2006;100:1744–1754. doi: 10.1016/j.jinorgbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Chicurel M. Faster, better, cheaper genotyping. Nature. 2001;412:580–582. doi: 10.1038/35088146. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimoto K, Nishizawa S, Minagawa M, Teramae N. Use of abasic site-containing DNA strands for nucleobase recognition in water. J. Am. Chem. Soc. 2003;125:8982–8983. doi: 10.1021/ja029786m. [DOI] [PubMed] [Google Scholar]
- 14.Nishizawa S, Yoshimoto K, Seino T, Xu CY, Minagawa M, Satake H, Tong A, Teramae N. Fluorescence detection of cytosine/guanine transversion based on a hydrogen bond forming ligand. Talanta. 2004;63:175–179. doi: 10.1016/j.talanta.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 15.Dai Q, Cui YY, Sato Y, Yoshimoto K, Nishizawa S, Teramae N. Enhancement of the binding ability of a ligand for nucleobase recognition by introducing a methyl group. Anal. Sci. 2006;22:201–203. doi: 10.2116/analsci.22.201. [DOI] [PubMed] [Google Scholar]
- 16.Rajendar B, Nishizawa S, Teramae N. Alloxazine as a ligand for selective binding to adenine opposite AP sites in DNA duplexes and analysis of single nucleotide polymorphisms. Org. Biomol. Chem. 2008;6:670–673. doi: 10.1039/b719786a. [DOI] [PubMed] [Google Scholar]
- 17.Zhao CX, Dai Q, Seino T, Cui YY, Nishizawa S, Teramae N. Strong and selective binding of amiloride to thymine base opposite AP sites in DNA duplexes: simultaneous binding to DNA phosphate backbone. Chem. Commun. 2006;11:1185–1187. doi: 10.1039/b516575j. [DOI] [PubMed] [Google Scholar]
- 18.Nishizawa S, Sankaran NB, Seino T, Cui YY, Dai Q, Xu CY, Yoshimoto K, Teramae N. Use of vitamin B2 for fluorescence detection of thymidine-related single-nucleotide polymorphisms. Anal. Chim. Acta. 2006;556:133–139. doi: 10.1016/j.aca.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimoto K, Xu CY, Nishizawa S, Haga T, Satake H, Teramae N. Fluorescence detection of guanine-adenine transition by a hydrogen bond forming small compound. Chem. Commun. 2003;24:2960–2961. doi: 10.1039/b309229a. [DOI] [PubMed] [Google Scholar]
- 20.Rajendar B, Sato Y, Nishizawa S, Teramae N. Improvement of base selectivity and binding affinity by controlling hydrogen bonding motifs between nucleobases and isoxanthopterin:Application to the detection of T/C mutation. Bioorg. Med. Chem. Let. 2007;17:3682–3685. doi: 10.1016/j.bmcl.2007.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Sankaran NB, Nishizawa S, Seino T, Yoshimoto K, Teramae N. Abasic-site-containing oligodeoxynucleotides as aptamers for riboflavin. Angew. Chem. Int. Ed. 2006;45:1563–1568. doi: 10.1002/anie.200502979. [DOI] [PubMed] [Google Scholar]
- 22.Satake H, Nishizawa S, Teramae N. Ratiometric fluorescence detection of pyrimidine/purine transversion by using a 2-amino-1,8-naphtyridine derivative. Anal. Sci. 2006;22:195–197. doi: 10.2116/analsci.22.195. [DOI] [PubMed] [Google Scholar]
- 23.Morita K, Sato Y, Seino T, Nishizawa S, Teramae N. Fluorescence and electrochemical detection of pyrimidine/purine transversion by a ferrocenyl aminonaphthyridine derivative. Org. Biomol. Chem. 2008;6:266–268. doi: 10.1039/b716682f. [DOI] [PubMed] [Google Scholar]
- 24.Haq I, Ladbury JE, Chowdhry BZ, Jenkins TC, Chaires JB. Specific binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Jenkins TC, Chaires JB. Energetics of DNA intercalation reactions. Biochemistry. 2000;39:8439–8447. doi: 10.1021/bi000474a. [DOI] [PubMed] [Google Scholar]
- 26.Nakatani K, Sando S, Saito I. Scanning of guanine-guanine mismatches in DNA by synthetic ligands using surface plasmon resonance. Nat. Biotechnol. 2001;19:51–55. doi: 10.1038/83505. [DOI] [PubMed] [Google Scholar]
- 27.Hagihara S, Kumasawa H, Goto Y, Hayashi G, Kobori A, Saito I, Nakatani K. Detection of guanine-adenine mismatches by surface plasmon resonance sensor carrying naphthyridine-azaquinolone hybrid on the surface. Nucleic Acids. Res. 2004;32:278–286. doi: 10.1093/nar/gkh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobori A, Horie S, Suda H, Saito I, Nakatani K. The SPR sensor detecting cytosine-cytosine mismatches. J. Am. Chem. Soc. 2004;126:557–562. doi: 10.1021/ja037947w. [DOI] [PubMed] [Google Scholar]
- 29.Demple B, Harrison L. Repair of oxidative damage to DNA - enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 30.Marky LA, Snyder JG, Remeta DP, Breslauer KJ. Thermodynamics of drug–DNA interactions. J. Biomol. Struct. Dyn. 1983;1:487–507. doi: 10.1080/07391102.1983.10507457. [DOI] [PubMed] [Google Scholar]
- 31.Chaires JB. Energetics of drug-DNA interactions. Biopolymers. 1997;44:201–215. doi: 10.1002/(SICI)1097-0282(1997)44:3<201::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 32.Haq I. Part II: the thermodynamics of drug-bipolymer interaction - thermodynamics of drug-DNA interactions. Arch. Biochem. Biophys. 2002;403:1–15. doi: 10.1016/S0003-9861(02)00202-3. [DOI] [PubMed] [Google Scholar]
- 33.Priebe W, Fokt I, Przewloka T, Chaires JB, Portugal J, Trent JO. Exploiting anthracycline scaffold for designing DNA-targeting agents. Methods Enzymol. 2001;340:529–555. doi: 10.1016/s0076-6879(01)40441-1. [DOI] [PubMed] [Google Scholar]
- 34.Puglisi JD, Tinocco I. Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 35.Conners KA. Binding Constants. New York: Wiley; 1987. [Google Scholar]
- 36.Record MT, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on binding and conformational equilibrium of proteins and nucleic-acids – roles of ion association or release, screening and ion effects on water activity. Q.Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 37.Chaires JB. Dissecting the free energy of drug binding to DNA. Anticancer Drug Des. 1996;11:569–580. [PubMed] [Google Scholar]
- 38.Record MT, Jr, Ha JH, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and herical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 39.Bailey SA, Graves DE, Rill R, Marsch G. Influence of DNA-base sequence on the binding energetics of actinomycin-D. Biochemistry. 1993;32:5881–5887. doi: 10.1021/bi00073a022. [DOI] [PubMed] [Google Scholar]
- 40.Rentzeperis D, Marky LA, Dwyer TJ, Geierstanger BH, Pelton JG, Wemmer DE. Interaction of minor-groove ligand to an AAATT/AATTT site –correction of thermodynamic characterization and solution structure. Biochemistry. 1995;34:2937–2945. doi: 10.1021/bi00009a025. [DOI] [PubMed] [Google Scholar]
- 41.Ladbury JE, Wright JG, Sturtevant JM, Sigler PB. A thermodynamic study of the Trp repressor-operator interaction. J. Mol. Biol. 1994;238:669–681. doi: 10.1006/jmbi.1994.1328. [DOI] [PubMed] [Google Scholar]
- 42.Barcelo F, Capo D, Portugal J. Thermodynamic characteriztion of the multivalent binding of chartreusin to DNA. Nucleic Acids Res. 2002;30:4567–4573. doi: 10.1093/nar/gkf558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazur S, Tanious FA, Ding D, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. A thermodynamic and structural analysis of DNA minor groove complex formation. J. Mol. Biol. 2000;300:321–337. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
- 44.Haq I, Jenkins TC, Chowdhry BZ, Ren J, Chaires JB. Parsing free energies of drug-DNA interactions. Methods Enzymol. 2000;323:373–405. doi: 10.1016/s0076-6879(00)23374-0. [DOI] [PubMed] [Google Scholar]
- 45.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical thermodynamic basis for computation of binding affinities: a critical review. Biophys. J. 1997;71:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holtzer A. The critic correction and related fallacies. Biopolymers. 1995;35:595–602. doi: 10.1002/bip.360350605. [DOI] [PubMed] [Google Scholar]
- 47.Spolar RS, Record MT. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–783. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 48.Lukin M, Santos C. NMR structures of damaged DNA. Chem Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 49.Cuniasse P, Fazakerly GV, Guschlbauer W, Kaplan B, Sowers LC. The abasic site as a challenge to DNA-polymerase – a nuclear-magnetic-resonance study of G, C AND T opposite a model abasic site. J. Mol. Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- 50.Suda H, Kobori A, Zhang J, Hayashi G, Nakatani K. N,N’-bis(3-aminopropyl)-2,7-diamino-1,8-naphthyridine stabilized a single pyrimidine bulge in duplex DNA. Biorg. Med. Chem. 2005;13:4507–4512. doi: 10.1016/j.bmc.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimoto K, Nishizawa S, Koshino H, Sato Y, Teramae N, Maeda M. Assignment of hydrogen-bond structure in a ligand-nucleobase complex inside duplex DNA: combined use of quantum chemical calculations and 15N NMR experiments. Nucleic Acids Symp. Ser. 2005;49:255–256. doi: 10.1093/nass/49.1.255. [DOI] [PubMed] [Google Scholar]
- 52.Innis MA, Myambo KB, Gelfand DH, Brow MAD. DNA sequencing with thermus-aquaticus DNA-polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc. Natl Acad. Sci. USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiviniemia M, Nurmi J, Turpeinen H, Lovgren T, Ilonen J. A homogeneous high-throughput genotyping method based on competitive hybridization. Clin. Biochem. 2003;36:633–640. doi: 10.1016/s0009-9120(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 54.Bos JL. Genetic mechanisms in tumor initiation and progression.10. the ras gene family and human carcinogenesis. Mutat. Res. 1988;195:255–271. doi: 10.1016/0165-1110(88)90004-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.