Abstract
Differential gene expression largely accounts for the coordinated manifestation of the genetic programme underlying embryonic development and cell differentiation. The 3′ untranslated region (3′-UTR) of eukaryotic genes can contain motifs involved in regulation of gene expression at the post-transcriptional level. In the 3′-UTR of dmrt1, a key gene that functions in gonad development and differentiation, an 11-bp protein-binding motif was identified that mediates gonad-specific mRNA localization during embryonic and larval development of fish. Mutations that disrupt the 11-bp motif leading to in vitro protein-binding loss and selective transcript stabilization failure indicate a role for this motif in RNA stabilization through protein binding. The sequence motif was found to be conserved in most of the dmrt1 homologous genes from flies to humans suggesting a widespread conservation of this specific mechanism.
INTRODUCTION
Assembly and formation of the gonad primordium is the first step towards gonad differentiation and subsequent sex differentiation (1). Primordial germ cells (PGCs) give rise to the gametes that are responsible for the development of a new organism in the next generation. In many organisms, following their specification the germ cells migrate towards the location of the prospective gonadal primordium (2–5). Similar to other vertebrates, the structure of fish gonads is composed of germ cells and associated supporting somatic cells (6). The precursors of the somatic cells originate from cells of the lateral plate mesoderm where the gonadal primordium develops, while germ cells are derived from the germline lineage (7,8). To carry out their highly specialized biological functions, together somatic gonadal primordium and germline cells must establish specialized programs of gene expression. However, the early transcriptional and post-transcriptional regulatory events underlying the differentiation of gonad precursor cells through crucial interactions of somatic and germline cells are barely understood.
The dmrt1 gene is an important regulator of male development in vertebrates (9). It is a highly conserved gene involved in the determination and early differentiation phase of the primordial gonad in vertebrates. In the fish medaka dmrt1bY, a functional duplicate of the autosomal dmrt1a gene on the Y-chromosome, has been shown to be the master regulator of male gonadal development (10,11), comparable to Sry in mammals (12). In males mRNA and protein expression occur before morphological sex differentiation in the somatic cells surrounding PGCs of the gonadal anlage and later on exclusively in Sertoli cells (13). Here it is synexpressed with the autosomal dmrt1a (14,15). However, nothing is known about the mechanism(s) that bring about this highly restricted expression pattern.
The expression of most genes is dynamically regulated temporally and spatially. Spatial organization of cells and subcellular compartments arises in part from the sorting and subsequent localization of proteins and RNA. Evidence has been obtained that regulation occurs at multiple steps on the level of gene expression including transcription, splicing, mRNA transport, mRNA stability, translation, protein stability and post-translational modifications (16,17). Selective advantages could have favored the evolution of regulatory mechanisms at the post-transcriptional level, such as speed of response, reversibility, fine-tuning of protein amounts, coordinated regulation of protein families, potential for spatial control, and efficacy in systems lacking transcriptional control mechanisms. Efficient cell-specific mRNA processing depends on a temporally and spatially orchestrated sequence of protein–protein, protein–RNA and RNA–RNA interactions (16).
Striking examples of localized messengers, transcriptional, post-transcriptional and translational regulations can be found among the maternal mRNAs of fly, fish and frog implicated in the establishment of axial polarity. For example, in the posterior part of the Drosophila embryo, Nanos (Nos) protein represses translation of maternal hb mRNA (18). Conversely, synthesis of both Bcd and Hb proteins in the anterior of the embryo requires that Nos is limited to the posterior (19). The restricted distribution of Nos is generated by selective translation of a subset of nos mRNA that is localized to the germ plasm at the posterior of the embryo coupled with translational repression of nos mRNA distributed throughout the whole embryonic cytoplasm (20). Both posterior localization and translational repression of nos RNA are mediated by the nos 3′-untranslated region (3′-UTR) (21). A nucleotide translational control element (TCE) within the nos 3′-UTR confers repression through formation of two stem-loop structures, whose functions are temporally distinct (22).
The zebrafish nanos1 homologue which is required during germline development (23) has also been shown to be remarkably post-transcriptionally regulated. Here, microRNA miR-430 targets the 3′-UTR of nanos1 during zebrafish embryogenesis in order to confer restriction of mRNA to PGCs (24). This miR-430 target site was shown to reduce poly(A) tail length, mRNA stability and translation, suggesting that differential susceptibility to microRNAs contributes to tissue-specific gene expression (24). Implicit in these mechanisms are the existence of cis-acting signals and trans-acting factors forming mRNA–protein complexes (mRNPs) that account for specificity and selectivity.
While the importance of complex post-transcriptional regulation—like in the case of nanos- has been widely demonstrated for the development of the germline, such mechanisms have not been uncovered so far for the development of the somatic part of the gonad, which determines the development towards testis or ovary. Studying mechanisms regulating localization and translation of gonad-specific genes during early gonad induction, we demonstrate that an 11-nt protein-binding motif located in the 3′-UTR of dmrt1bY mediates gonad-specific mRNA stability during embryonic and larval development. Interestingly, the sequence motif was found to be highly conserved in the homologous genes from flies to humans.
MATERIALS AND METHODS
Fish maintenance and breeding
Medaka were taken from closed breeding stocks of the Carbio strain and kept under standard conditions. Medaka embryos were staged according to Iwamatsu (25).
Whole-mount in situ hybridization
RNA whole-mount in situ hybridization using GFP digoxigenin (DIG)-labeled probe was performed as described for Medaka (26). Briefly, after capped mRNA injection, embryos of the desired stage were fixed with 4% paraformaldehyde and dehydrated with methanol. Anti-sense-DIG-labeled RNA probes were synthesized according to the manufacturers’ instructions (Roche, Meylan). Hybridization and detection with alkaline phosphatase (AP)-coupled anti-DIG antibody (Roche, Meylan) were performed according to Thisse et al. (27).
Plasmid constructs and RNA injections
To obtain RNA transcripts of eGFP the GFP open reading frame (ORF) from pEGFP-N1 (Clontech Laboratories) was inserted (BamH1/Not1) into pCS2+ plasmid (pCS2:GFP). To produce pCS2:GFP:dmrt1bY 3′-UTR, a PCR product containing the entire 3′-UTR of dmrt1bY flanked by Not1 sites, was amplified from medaka testes and inserted into the Not1 sites of pCS2:GFP. Similarly, pCS2:GFP:dmrt1a 3′-UTR, pCS2:GFP:d. rerio dmrt1bY 3′-UTR, pCS2:GFP:h. sapiens 3′-UTR, pCS2:GFP:O. curvinotus dmrt1bY 3′-UTR and pCS2:GFP:fugu 3′-UTR and deletion constructs (Supplementary Figure 2) were constructed the same way. Corresponding RFP plasmids were constructed by replacing the GFP ORF by RFP. Xenopus β-globin constructs were produced by inserting the xenopus β-globin 3′-UTR (Not1/Kpn1) from plasmid pRN3 (28) into pCS2:GFP (pCS2:GFP:xlβ-globin 3′-UTR). For constructing the pCS2:GFP:BOXxlβ-globin 3′-UTR plasmid, the dmrt1bY box was inserted between the Not1 sites. The GFP/RFP 3′-UTR constructs include the mmGFP5/RFP ORF cloned upstream of the 3′-UTR of the zebrafish nanos1 gene (23,29). All constructs were checked by restriction digests, diagnostic PCRs and sequencing. Sertoli cell-specific Ds-Red expressing sox9prom:DsRed transgenic medaka fish (30) was provided by Prof. Tanaka.
Capped RNAs for injections were transcribed from linearized vectors using the SP6/T3/T7 m MESSAGE mMACHINE Kit (Ambion). One nanoliter was injected into the cytoplasm of one-cell stage Medaka embryos as described (31).
Cell culture and transfection
Medaka spermatogonial (Sg3), embryonic stem (MES-1) or fibroblast (OL-17) cells were cultured as described (32–34). Cells were grown to 70–80% confluency in six-well plates and then transfected with 5 µg expression vector using GeneJuice reagent (Novagen) as described by the manufacturer. Luciferase activity was then quantified using the Luciferase Reporter Assay System from Promega and luciferase activity was normalized against mRNA luciferase copy number. Transcriptional differences between luciferase constructs were evaluated statistically by paired Student's test.
Electromobility shift assay
Nearly confluent cells [Medaka spermatogonial (Sg3), mouse Sertoli (TM4) and Medaka embryonic stem (MES-1)] cell lines grown on plates were washed with ice-cold phosphate-buffered saline (PBS) and removed from the plates with 1 mM EDTA in PBS. The cells were centrifuged at low speed and then resuspended in Passive Lysis Buffer (Promega) supplemented with leupeptin (0.2 µg/ml) and aprotinin (10 µg/ml). Binding assays were carried out using the Gel Shift Assay System (Promega) using radiolabeled RNA oligonucleotides: (Y) UGGUUCACGUCUGCUGCAGGUCUCUGACUCU for the native box target sequence and Mut(2)-box UGGUUCACGUUUGGUCGGGATCUCUGACUCU; Mut(3)-box UGGUUCACGUUCUUCACAUGUCUCUGACUCU; Mut(4)-box UGGUUCACGUCUGCUGAGACGCUCUGACUCU; Mut(5)-box UGGUUCACGUCUGCCAUAGGUCUCUGACUCU as competitors.
RESULTS AND DISCUSSION
Dmrt1a/dmrt1bY 3′-UTRs regulate spatial and temporal expression during early Medaka development
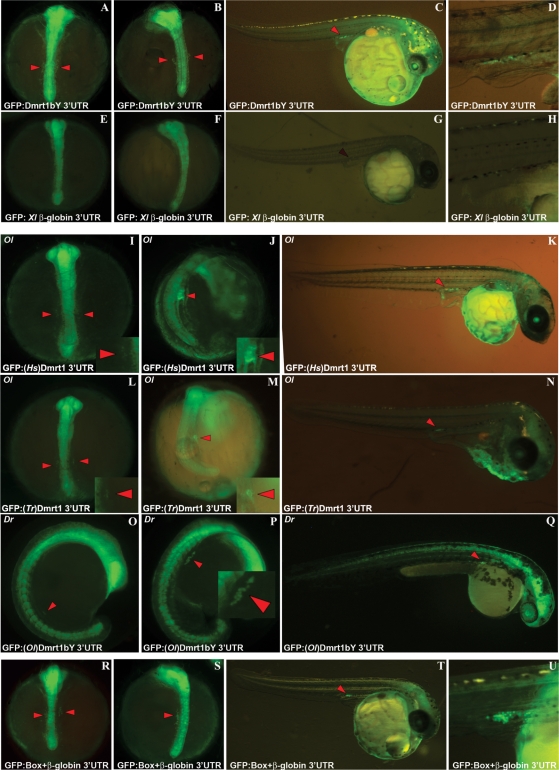
While searching for potentially conserved regulatory sequences in genes involved in gonad induction and formation we analyzed the tightly regulated Medaka dmrt1a and dmrt1bY duplicated gene pair. Postulating that important cis-regulatory motifs required for mRNA regulation in the context of gonad formation might have been retained between the duplicates despite of the processes of co-ortholog gene specialization and subfunctionalization, we noticed that the dmrt1a and dmrt1bY 3′-UTRs appeared more conserved than expected for independently diverging genes. To test whether this conservation of dmrt1a/dmrt1bY UTRs implied common regulatory mechanisms, we examined GFP expression of reporter constructs that contained either the dmrt1a or dmrt1bY 3′-UTR or the Xenopus β-globin 3′-UTR as a control (Figure 1). After injection of the different constructs into one cell stage embryos we initially observed a high and uniformly distributed GFP expression in the whole embryo (Figure 1). After 2 days of development (stages 22–24; 12–16 somites), GFP fluorescence slowly vanished elsewhere except in the primordial gonad area of the fish injected with either dmrt1a or dmrt1bY UTR (Figures 1A, B, C, D compared to 1E, F, G, H).
Figure 1.
A short highly conserved cis-regulatory motif located in dmrt1bY/dmrt1a 3′-UTRs regulates spatial and temporal expression during early development. (A–D) GFP expression of a reporter construct that contains Medaka dmrt1bY 3′-UTR during somitogenesis (A and B) and at hatching stage (C and D). (E–H) GFP expression of a control reporter construct that contains Xenopus β-globin 3′-UTR during somitogenesis (E and F) and at hatching stage (G and H). (I–Q) GFP expression of reporter constructs that contain either human dmrt1 3′-UTR (I–K) or takifugu dmrt1 3′-UTR (L–N) in Medaka embryos during somitogenesis (I, J and L, M) and at hatching stage (K and N). (O–Q) GFP expression of a reporter construct that contains Medaka dmrt1bY 3′-UTR in zebrafish embryos during somitogenesis (O and P) and at hatching stage (Q). (R–U) GFP expression in Medaka embryos of a reporter construct that contains Xenopus ß-globin 3′-UTR in which the Box was inserted. Specific GFP expression in PGCs is indicated (arrow heads).
GFP fluorescence was clearly detectable in the primordial gonad area until more than 7 days after hatching (stage 40; first fry stage, 2.5 weeks after fertilization) (Supplementary Figure 1). In controls with the β-globin 3′-UTR GFP remained ubiquitously expressed throughout the whole embryonic development. Obviously, the dmrt1a/dmrt1bY 3′-UTRs are responsible for specific expression of the GFP protein in the primordial gonad area.
Surprisingly, fusing either human (AJ276801) (Figure 1I, J and K) or takifugu (CAC42778) (Figure 1L, M and N) dmrt1 3′-UTRs to GFP mRNA also drove primordial gonad area-specific fluorescence in Medaka (Figure 1I–N compared to 1A–D). In addition, injection of GFP mRNA fused to Medaka dmrt1bY 3′-UTR in zebrafish resulted in a similar gonadal persistence of GFP fluorescence (Figure 1O, P and Q) indicating a functional cross-species conserved mechanism mediated by cis-regulatory element(s) in these dmrt1 3′-UTRs.
A short, highly conserved cis-regulatory motif located in dmrt1a/dmrt1bY 3′-UTR is responsible for gonadal differential regulation
To delineate the precise RNA sequence and/or secondary structures involved in gonadal-specific fluorescence, GFP expression of a series of reporters (Supplementary Figure 2) that contained deletion mutants of the dmrt1a/dmrt1bY 3′-UTRs was investigated (Supplementary Figure 2B, C and D). As a result a core 11-nt box located in the 5′ region of the Medaka dmrt1a/dmrt1bY UTRs was isolated and shown to be responsible for gonad-specific fluorescence. Consequently, when the box was inserted into the Xenopus β-globin 3′-UTR, GFP expression was identical to GFP::dmrt1a and dmrt1bY 3′-UTR constructs, namely gonad-specific expression (Figure 1R, S, T and U). Conversely, deletion of the box sequence from the dmrt1bY 3′-UTR drastically extinguished gonad-specific expression (Supplementary Figure 2D compared to 2B and C).
Considering the functional conservation of gonad-specific expression seen with other fish and human dmrt1 UTRs we then searched for similar motifs in the 3′-UTR of dmrt1 genes of other organisms (Table 1). The motif was found to be highly conserved in the fish lineage (Oryzias latipes, Oryzias curvinotus, Takifugu rubripes, Tetraodon nigroviridis, Epinephelus coioides and Danio rerio), but as well in the dmrt1 3′-UTR of other vertebrates including man (Mus musculus, Pan troglodytes, Macaca mulatta and Homo sapiens) and most surprisingly even in the ecdysozoan clade. For the doublesex (dsx, the dmrt1 orthologue) of Anopheles gambiae (Table 1), interestingly, the sex-specific differentially spliced anopheles dsx transcript results in a male dsx form where the box is largely conserved while this is not the case for the female splice form (Table 1). This situation is similar for dsx of the olive fruit fly (Bactocera oleae) for which a male-specific splicing leads to the preservation of a highly conserved box in the ORF while this fragment is spliced out in the female form (Table 1).
Table 1.
The dmrt1 3′ UTR cis-regulatory motif is well conserved from ecdysozoans to mammals
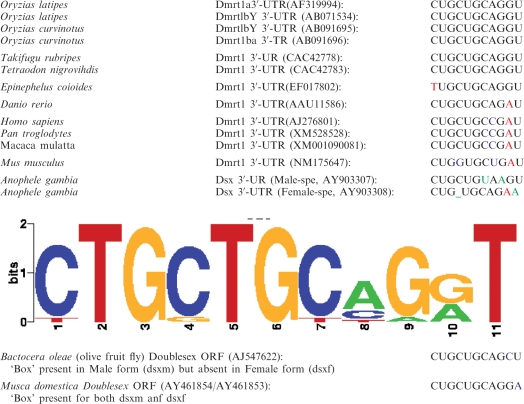 |
Taken together, a short highly conserved cis-regulatory motif located in dmrt1a and dmrt1bY 3′-UTRs (CUGCUGCAGGU) appears to be mostly responsible for differential expression of the transcripts.
The dmrt1 box drives specific stability in the somatic mesoderm anlage of the gonadal primordium as well as in a sub-population of PGCs
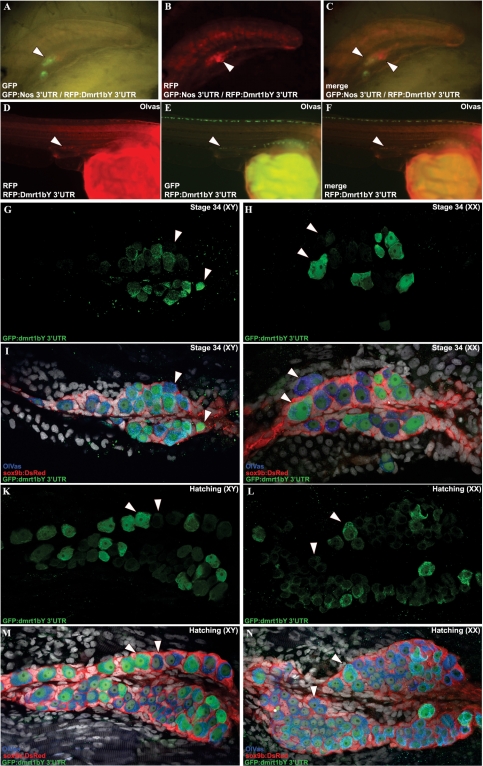
Dmrt1 is expressed in most species specifically in Sertoli cells and PGCs. To find out the contribution of post-transcriptional regulation by this cis-regulatory motif to the restricted expression pattern, Medaka dmrt1bY 3′-UTR was fused to the monomeric RFP mCherry and injected either together with GFP::Nos 3′-UTR into one-cell stage embryos of wild-type Medaka or of the Olvas transgenic strain. The GFP::Nos 3′-UTR construct was previously shown to drive PGC-specific fluorescence in medaka (35–37). Similarly, in Olvas fish the PGCs are marked by GFP expression from stage 25 [18–19 somite stage (25)] onwards (38). RFP expression was compared to germ cell-specific GFP expression due to the nanos 3′-UTR or the vasa promoter (Figure 2A–F). Starting at stages 14–18 fluorescence was exclusively observed from GFP in PGCs (data not shown; Figure 3A). Only by stages 22–24 (nine somite stage) red fluorescence could also be observed with specific gonadal localization (Figure 2D–F). Noteworthy, although at this stage both fluorescences (green and red) were confined to the same embryonic structures, they are clearly expressed from different cell populations (Figure 2A–C). Additionally, around stage 24–26 (16–22 somite stage) cells expressing both fluorochromes could be observed (Figure 2A–C). Subsequently, at hatching stage, gonadal dmrt1bY 3′-UTR-driven GFP expression was then investigated in Sertoli cell-specific RFP expressing sox9prom:DsRed transgenic fish (30) either at stage 34 (Figures 2G, I, H and J) or just after hatching (Figure 2K, L and M) in males and females, respectively. It revealed that although no more supporting cell expression was apparent for these later stages, two different populations of germ cells could be discriminated according to their high or just above background GFP fluorescence expression (arrowheads in Figure 2G, H, K and L). The fact that the RFP reporter gene product was only observed at later stages indicated that mRNAs with the dmrt1bY 3′-UTR are translationally repressed in migrating PGCs and in the somatic gonad precursor cells.
Figure 2.
The dmrt1 box drives specific stability in the somatic mesoderm of the gonadal primordium as well as in a sub-population of PGCs. (A–F) RFP expression from dmrt1bY 3′-UTR containing capped RNA compared to germ cell-specific GFP expression due to the nanos 3′-UTR at stages 24–26 (A–C). (D–F) RFP expression compared to germ cell specific GFP expression achieved with vasa promoter around hatching stage. (G–N) Gonadal dmrt1bY 3′-UTR-driven GFP expression investigated in Sertoli cell specific DsRed expressing sox9prom:DsRed transgenic fish either at stage 34 (G, I and H, J) or just after hatching (K, M and L, N) in males (XY) and females (XX). Arrow heads indicate either putative somatic gonadal precursor cells (C) or different sub-populations of germ cells (A, B and D–N). Blue: DAPI or Olvas; Red: Sertoli cell specific expression (sox9prom:DsRed transgenic fish) and Green: gonadal dmrt1bY 3′-UTR-driven GFP expression.
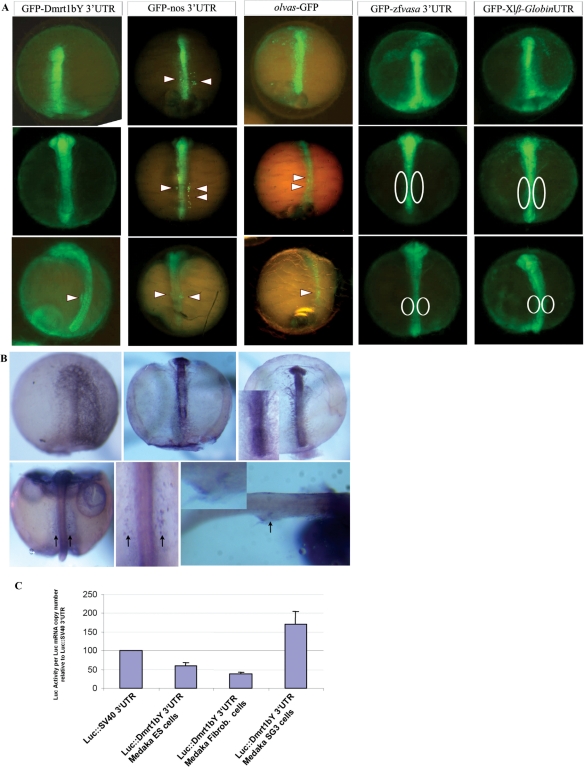
Figure 3.
Tissue-specific and temporal-restricted expression by a combination of dmrt1 3′-UTR induced differential mRNA stability and translational regulation. (A) GFP:dmrt1bY 3′-UTR was injected. As controls, mRNA constructs such as GFP:nos 3′-UTR, olvas:GFP, GFP:zfvasa 3′-UTR and GFP:xlß-globin 3′-UTR were also injected to be able to compare the expression with the pattern of known post-transcriptional mechanisms such as micro-RNA mRNA induced decay, specific PGC translational regulation, and ubiquitous stability, respectively. (B) Subsequently in situ hybridization using an antisense GFP probe was performed at different stages of development to reveal the spatial distribution of the injected GFP:dmrt1bY 3′-UTR RNAs. GFP fluorescence and the distribution of RNA were followed at different stages of development. Arrowheads and circles indicate where the PGCs are or should be located respectively. (C) Luciferase expression in different cell lines transfected with a dmrt1bY 3′-UTR containing construct reveals that translation of the reporter gene was significantly enhanced by the presence of dmrt1bY 3′-UTR in Medaka spermatogonial cells in contrast to either Medaka embryonic stem or fibroblast cells.
Interestingly, such variation and diversity in germ cell-specific Dmrt1 expression is also reported for mouse gonads for which two populations of germ cells could be observed according to Dmrt1 protein presence or absence (39). Consequently, akin to what was observed during medaka gonad formation (Figure 2G–N), it might be possible that mouse Dmrt1+ and Dmrt1− expressing germ cells are the result of a similar mechanism since the ‘box’ motif is also found to be highly conserved in mouse dmrt1 3′-UTR (Table 1).
Embryonic expression of dmrt1 mRNA has been examined in various vertebrates, including mammals (40–42), birds (41), reptiles (41,43) and fish (44). In most cases, dmrt1 is expressed very early in the genital ridge–the structure from which the gonad derives–and later on in germ and Sertoli cells of the male gonadal primordium [see Zarkower et al. (45) for review]. It is then evident that GFP expressing cell are PGCs; likewise are the cells expressing jointly both GFP and RFP. The question remains what the identity of the RFP only expressing cells at early stages (stages 24–26) is. Considering the temporal appearance of these cells and a report identifying and tracing different populations of gonadal precursor cells in medaka (7), it is most likely that these cells are representatives of a subset of gonadal precursors.
Interestingly, like observed for mouse Dmrt1, these results point to the existence of at least two populations of PGCs within the primordial gonad of Medaka with possibly different regulations of dmrt1 in relation to the above described cis-acting element located in its 3′-UTR.
Tissue-specific and temporal-restricted expression is caused by a combination of dmrt1 3′-UTR-induced differential mRNA stability and translational regulation
Independently of promoter-induced transcription striking examples of specifically localized messengers can be found among the mRNAs of fly, fish and frog, most of them being post-transcriptionally (dynamic relocalization and stabilization) and translationally regulated. To test whether dmrt1bY 3′-UTR was more involved in regulating mRNA stability or controlling its translational regulation, embryos were injected with GFP:dmrt1bY 3′-UTR and subsequently analyzed for the spatial distribution of the injected RNA by RNA in situ hybridization at different stages of development (Figure 3). As controls, GFP:nos 3′-UTR, olvas:GFP, GFP:zfvasa 3′-UTR and GFP:xlß-globin 3′-UTR mRNA constructs were also injected in order to compare the expression with the pattern of known post-transcriptional mechanisms such as micro-RNA mRNA-induced decay, specific PGC translational regulation and ubiquitous stability, respectively (Figure 3). GFP fluorescence and the distribution of RNA were followed at different stages of development. Remarkably, GFP expressed from the construct containing the dmrt1bY 3′-UTR, was shown to be quite stable in the whole body, including the primordial gonad area like with the GFP:zfvasa 3′-UTR and GFP:xlß-globin 3′-UTR mRNAs (Figure 3). This is in striking contrast to GFP:nos 3′-UTR construct for which mRNA underwent rapid somatic degradation [Figure 3; (22,24,36,46)]. This pattern of decay is typical for a microRNA-mediated process (24). Although an analogous germ cell-specific fluorescence was observed in germ cells of olvas:GFP injected embryos, in situ hybridizations showed homogenous olvas RNA distribution. This pattern implies germ cell-specific translational regulation (35). Accordingly GFP in situ hybridization after GFP:dmrt1bY 3′-UTR injection revealed that the fused mRNA is first homogenously distributed in the whole embryonic body until stage 24 and only then becomes progressively restricted to the gonad area to be exclusively present in the gonadal primordium around hatching (stage 39) (Figure 3). Hence, it can be inferred that the apparent specific GFP:dmrt1-UTR driven expression in primordial gonad is the result of differential RNA stabilization.
Interestingly these results reflect also the probability of another mechanism affecting mRNAs containing the dmrt1 UTR. The fact that strong fluorescence is observed in the whole body (including PGCs) at early stages suggests that the dmrt1bY box here contributes to enhanced translation. To test whether the dmrt1bY 3′-UTR is able to enhance translation, a plasmid construct containing a thymidine kinase promoter driven luciferase fused either to SV40 3′-UTR or dmrt1bY 3′-UTR was transfected in different cell lines and translation efficiency measured. Translation of the reporter gene was significantly (P < 0.01) enhanced in the presence of dmrt1bY 3′-UTR in Medaka spermatogonial cells in contrast to either Medaka embryonic stem cells or fibroblast cells where protein production was even reduced (Figure 3). The suppression of translation in non-gonad cell types is interesting to note in relation to the non-gonad-specific expression of dmrt1bY transcripts during early Medaka embryogenesis and in adult male spleen (10,14).
In summary, taking into account the endogenous expression of dmrt1a exclusively in the developing gonad and the here observed late gonad-specific translation of injected RFP:dmrt1bY 3′-UTR mRNA, it appears that the UTR mediates a translational regulation process specifically in a subset of PGCs and certain somatic cells of the gonad. From RNA localization studies it is apparent that the later gonad-specific expression is then primarily due to the 11-bp cis-acting dmrt1 motif conferring differential RNA stabilization in primordial gonad cells.
The dmrt1 3′-UTR box specifically binds a protein possibly involved in gonad-restricted expression
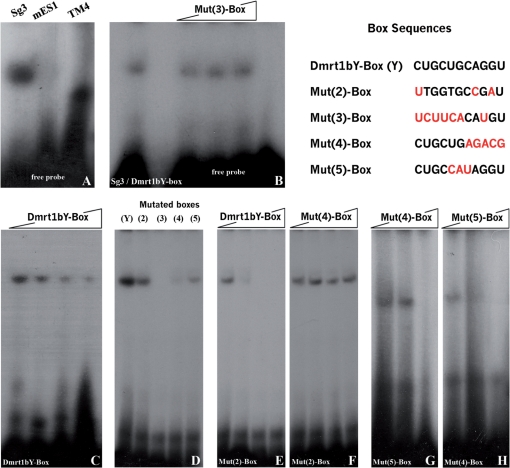
Obviously the 11-mer is involved in a cell-type-specific stability of dmrt1 RNAs. One explanation could be that it is the target for a protein or small ribonucleoproteins particles. To test this hypothesis, electrophoretic mobility shift assays (EMSAs) were performed using the 11-mer box as a probe and different mutated boxes as competitors (Figure 4). In a first step cell extracts from three different cell lines were tested: (i) a medaka spermatogonia cell line, (ii) a medaka embryonic stem cell line and (iii) a mouse sertoli cell line (TM4) (Figure 4A). While no mobility shift was observed for the medaka embryonic stem cell-like line in all conditions tested, a weak shift was apparent for the TM4 cells and a quite robust one for the Medaka spermatogonial cell line (Sg3) (Figure 4A). As controls, using the medaka spermatogonial cell line, a mutated version of the box used as competitor (Mut(3)-Box) did not interfere significantly with the Dmrt1bY-Box binding, indicating the specificity of the interaction (Figure 4B). Furthermore, competition of radioactively with non-radioactively labeled box probes (Dmrt1bY-Box) resulted in progressive loss of the apparent shift (Figure 4C). Next, to better characterize the interaction domain(s) of the box, different point mutations were then introduced to the native dmrt1bY-box (Figure 4D).
Figure 4.
A RNA-binding factor that recognizes the Box could be responsible for primordial gonad restricted stability. (A) Electrophoretic Mobility Shift Assay (EMSA) using cell extracts from three different cell lines (i) a medaka spermatogonial cell line (Sg3), (ii) a medaka embryonic stem cell line (MES-1) and (iii) a mouse sertoli cell line (TM4) and the 11-mer box as a probe shows shift for Sg3 and TM4 cells. (B) A mutated version of the box [Mut(3)-Box] used as competitor (1–1, 1–5 and 1–10 ratios) did not interfere significantly with the binding, indicating the specificity of the binding. (C) As control, using the spermatogonial cell line, competition of radioactively and non-radioactively labeled box probes resulted in progressive loss of the apparent shift (1–1, 1–5 and 1–10 ratios). (D) Different mutated versions of the box [Mut(2–5)-Box] were then tested for binding, and resulted in apparent different binding affinities. (E–H) Relative robustness of the shift was then tested by mean of competition assays among the different mutated versions of the box (E: 1–1, 1–5 and 1–10; F: 1–1, 1–2 and 1–5; G and H: 1–1 and 1–5 ratios).
The first mutated RNA oligonucleotide [Mut(2)-Box] regrouped the three main mutations observed for the different dmrt1 boxes within fish and mammals (see also Table 1). These introduced mutations did not significantly interfere for binding (Figure 4D). Interestingly, this result corroborates in vivo observations showing GFP stabilization after injection of either human or zebrafish dmrt1 3′-UTR containing constructs in medaka (Figure 4D). Nevertheless, this affinity seems to be significantly weaker when competing against the native Dmrt1bY-Box (Figure 4E). Noticeably, Mut(2)-Box competition against another box mutated in its 3′ region [Mut(4)-Box] could not appreciably interfere with the binding (Figure 4F). Similarly, using this box [Mut(4)-Box] and a box mutated in its core region [Mut(5)-Box] alternatively against each other, revealed that although the entire native box is required for efficient binding, its 3′ region is important while the 5′ and core regions are more likely involved in modulating the affinity (Figures 4G and 4H).
In summary these experiments suggest that the 11-mer box is a preferential target for RNA-binding proteins that may be involved in specifically regulating transcript stability.
CONCLUSIONS
The great majority of sequences that direct the subcellular localization, translation and degradation have been found within the 3′-UTRs [see (17,47) for review]. UTRs are highly diverse in sequence, but often contain regulatory motifs that are common to members of the same metabolic family (48). Nevertheless, the analysis of such cis-acting elements has been rather unsatisfying with respect to the identification of common sequences that direct localization of different RNAs (49).
We have identified an 11-nt motif in the Dmrt1 3′-UTR, CUGCUGCAGGU, common to the great majority of Dmrt1 orthologs genes from fly to mammals that is responsible for specific stabilization and translational control of dmrt1 mRNA in the forming primordial gonad of fish and probably of other species. Interestingly, for the first time such stabilizing motif involving cis-regulatory actor(s) has been shown to be not only present in a single organism [see Kloc et al. (49) for review], co-regulating a very specific pool of few synexpressed transcripts (50), but rather to be highly conserved across species for a given transcript family, namely the Doublesex/Mab-3/Dmrt1 genes. The occurrence of such an 11-mer in the 3′-UTR of orthologs genes over a wide range of organisms already indicates that different systems may employ common RNA regulatory mechanisms. Hence, other than a motif involved in specific localization of mRNAs to a well defined subtype of cells, this box would be responsible for mRNA-specific preservation of different subsets of cells all together involved in primordial gonad assembly and formation, namely putative gonadal precuror cells and a specific subclass of PGCs. Consequently, like synexpressed groups reflect the functional compartmentalization of the eukaryote genome (51) and have a striking parallel to the prokaryote operon, our finding might be of particular interest since it might reveal an otherwise hidden logic of cellular regulation where cis-regulatory motifs couple spatial and temporal gene expression in different subset of cells during organogenesis.
Finally, our data indicate that transcript stabilization is achieved by interaction of a specific protein with a cis-acting stability element located in the dmrt1 3′-UTR. Although the identity and the dynamic of action of this stabilizing factor has still to be resolved, our findings point to an obvious level of integrated regulation, namely the presence and the accessibility of this cis-regulatory element. The occurrence of multiple dmrt1-related spliced variants in corals (52), insects (53–55), lizards (56), fish (57,58), chicken (59), mice (60) and human (61) selecting or splicing out parts of the UTRs also argue for such way of regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by a grant of the Rudolf-Virchow-Zentrum for Experimental Medicine (DFG Forschungszentrum) and DFG-Graduiertenkolleg 1048 (Molecular Basis of Organ Development in Vertebrates) through a PhD fellowship to T.W.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol. Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- 2.Santos AC, Lehmann R. Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 2004;14:R578–R589. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 3.McLaren A. Mammalian germ cells: birth, sex, and immortality. Cell Struct. Funct. 2001;26:119–122. doi: 10.1247/csf.26.119. [DOI] [PubMed] [Google Scholar]
- 4.Raz E. Guidance of primordial germ cell migration. Curr. Opin. Cell Biol. 2004;16:169–173. doi: 10.1016/j.ceb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Saito D, Morinaga C, Aoki Y, Nakamura S, Mitani H, Furutani-Seiki M, Kondoh H, Tanaka M. Proliferation of germ cells during gonadal sex differentiation in medaka: Insights from germ cell-depleted mutant zenzai. Dev. Biol. 2007;310:280–290. doi: 10.1016/j.ydbio.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 6.Delvin RH, Nagahama Y. Sex determination and sex differentiation in fish. Aquaculture. 2002;208:191–364. [Google Scholar]
- 7.Nakamura S, Kobayashi D, Aoki Y, Yokoi H, Ebe Y, Wittbrodt J, Tanaka M. Identification and lineage tracing of two populations of somatic gonadal precursors in medaka embryos. Dev. Biol. 2006;295:678–688. doi: 10.1016/j.ydbio.2006.03.052. [DOI] [PubMed] [Google Scholar]
- 8.Kurokawa H, Saito D, Nakamura S, Katoh-Fukui Y, Ohta K, Baba T, Morohashi K, Tanaka M. Germ cells are essential for sexual dimorphism in the medaka gonad. Proc. Natl Acad. Sci. USA. 2007;104:16958–16963. doi: 10.1073/pnas.0609932104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124:447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- 10.Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, Shimizu A, Shan Z, Haaf T, Shimizu N, Shima A, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl Acad. Sci. USA. 2002;99:11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 12.Brennan J, Capel B. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Matsuda M, Kajiura-Kobayashi H, Suzuki A, Saito N, Nakamoto M, Shibata N, Nagahama Y. Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka, Oryzias latipes. Dev. Dyn. 2004;231:518–526. doi: 10.1002/dvdy.20158. [DOI] [PubMed] [Google Scholar]
- 14.Hornung U, Herpin A, Schartl M. Expression of the male determining gene dmrt1bY and its autosomal coorthologue dmrt1a in medaka. Sex Dev. 2007;1:197–206. doi: 10.1159/000102108. [DOI] [PubMed] [Google Scholar]
- 15.Winkler C, Hornung U, Kondo M, Neuner C, Duschl J, Shima A, Schartl M. Developmentally regulated and non-sex-specific expression of autosomal dmrt genes in embryos of the Medaka fish (Oryzias latipes) Mech. Dev. 2004;121:997–1005. doi: 10.1016/j.mod.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 17.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 18.Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- 19.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 20.Deshpande G, Calhoun G, Schedl P. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development. 2004;131:1247–1257. doi: 10.1242/dev.01004. [DOI] [PubMed] [Google Scholar]
- 21.Dahanukar A, Wharton RP. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 1996;10:2610–2620. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- 22.Duchow HK, Brechbiel JL, Chatterjee S, Gavis ER. The nanos translational control element represses translation in somatic cells by a Bearded box-like motif. Dev. Biol. 2005;282:207–217. doi: 10.1016/j.ydbio.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 23.Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 2004;121:605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 26.Kluver N, Kondo M, Herpin A, Mitani H, Schartl M. Divergent expression patterns of Sox9 duplicates in teleosts indicate a lineage specific subfunctionalization. Dev. Genes Evol. 2005;215:297–305. doi: 10.1007/s00427-005-0477-x. [DOI] [PubMed] [Google Scholar]
- 27.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 28.Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 29.Herpin A, Fischer P, Liedtke D, Kluever N, Neuner C, Raz E, Schartl M. Sequential SDF1a and b-induced mobility guides Medaka PGC migration. Dev. Biol. 2008;320:319–327. doi: 10.1016/j.ydbio.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura S, Aoki Y, Saito D, Kuroki Y, Fujiyama A, Naruse K, Tanaka M. Sox9b/sox9a2-EGFP transgenic medaka reveals the morphological reorganization of the gonads and a common precursor of both the female and male supporting cells. Mol. Reprod. Dev. 2008;75:472–476. doi: 10.1002/mrd.20764. [DOI] [PubMed] [Google Scholar]
- 31.Koster R, Stick R, Loosli F, Wittbrodt J. Medaka spalt acts as a target gene of hedgehog signaling. Development. 1997;124:3147–3156. doi: 10.1242/dev.124.16.3147. [DOI] [PubMed] [Google Scholar]
- 32.Hong Y, Liu T, Zhao H, Xu H, Wang W, Liu R, Chen T, Deng J, Gui J. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc. Natl Acad. Sci. USA. 2004;101:8011–8016. doi: 10.1073/pnas.0308668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komura J, Mitani H, Shima A. Fish cell culture: Establishment of two fibroblast-like cell lines (OL-17 and OL-32) from fins of the medaka, Oryzias latipes. In Vitro Cell Dev. Biol. 1988;24:294–298. [Google Scholar]
- 34.Hong Y, Winkler C, Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes) Mech. Dev. 1996;60:33–44. doi: 10.1016/s0925-4773(96)00596-5. [DOI] [PubMed] [Google Scholar]
- 35.Herpin A, Rohr S, Riedel D, Kluever N, Raz E, Schartl M. Specification of primordial germ cells in medaka (Oryzias latipes) BMC Dev. Biol. 2007;7:3. doi: 10.1186/1471-213X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurokawa H, Aoki Y, Nakamura S, Ebe Y, Kobayashi D, Tanaka M. Time-lapse analysis reveals different modes of primordial germ cell migration in the medaka Oryzias latipes. Dev. Growth Differ. 2006;48:209–221. doi: 10.1111/j.1440-169X.2006.00858.x. [DOI] [PubMed] [Google Scholar]
- 37.Saito T, Fujimoto T, Maegawa S, Inoue K, Tanaka M, Arai K, Yamaha E. Visualization of primordial germ cells in vivo using GFP-nos1 3'UTR mRNA. Int. J. Dev. Biol. 2006;50:691–699. doi: 10.1387/ijdb.062143ts. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka M, Kinoshita M, Kobayashi D, Nagahama Y. Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: a useful model to monitor germ cells in a live vertebrate. Proc. Natl Acad. Sci. USA. 2001;98:2544–2549. doi: 10.1073/pnas.041315498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei N, Hornbaker KI, Rice DA, Karpova T, Agbor VA, Heckert LL. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol. Reprod. 2007;77:466–475. doi: 10.1095/biolreprod.106.058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Grandi A, Calvari V, Bertini V, Bulfone A, Peverali G, Camerino G, Borsani G, Guioli S. The expression pattern of a mouse doublesex-related gene is consistent with a role in gonadal differentiation. Mech. Dev. 2000;90:323–326. doi: 10.1016/s0925-4773(99)00282-8. [DOI] [PubMed] [Google Scholar]
- 41.Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- 42.Moniot B, Berta P, Scherer G, Sudbeck P, Poulat F. Male specific expression suggests role of DMRT1 in human sex determination. Mech. Dev. 2000;91:323–325. doi: 10.1016/s0925-4773(99)00267-1. [DOI] [PubMed] [Google Scholar]
- 43.Kettlewell JR, Raymond CS, Zarkower D. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis. 2000;26:174–178. [PubMed] [Google Scholar]
- 44.Marchand O, Govoroun M, D'Cotta H, McMeel O, Lareyre J, Bernot A, Laudet V, Guiguen Y. DMRT1 expression during gonadal differentiation and spermatogenesis in the rainbow trout, Oncorhynchus mykiss. Biochem. Biophys. Acta. 2000;1493:180–187. doi: 10.1016/s0167-4781(00)00186-x. [DOI] [PubMed] [Google Scholar]
- 45.Zarkower D. Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2001;2:175–185. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- 46.Aoki Y, Nagao I, Saito D, Ebe Y, Kinjo M, Tanaka M. Temporal and spatial localization of three germline-specific proteins in medaka. Dev. Dyn. 2008;237:800–807. doi: 10.1002/dvdy.21448. [DOI] [PubMed] [Google Scholar]
- 47.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 48.Mazumder B, Seshadri V, Fox PL. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 49.Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 50.Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3' untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl Acad. Sci. USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- 52.Miller SW, Hayward DC, Bunch TA, Miller DJ, Ball EE, Bardwell VJ, Zarkower D, Brower DL. A DM domain protein from a coral, Acropora millepora, homologous to proteins important for sex determination. Evol. Dev. 2003;5:251–258. doi: 10.1046/j.1525-142x.2003.03032.x. [DOI] [PubMed] [Google Scholar]
- 53.Scali C, Catteruccia F, Li Q, Crisanti A. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J. Exp. Biol. 2005;208:3701–3709. doi: 10.1242/jeb.01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagoshi RN, Baker BS. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- 55.Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 56.Sreenivasulu K, Ganesh S, Raman R. Evolutionarily conserved, DMRT1, encodes alternatively spliced transcripts and shows dimorphic expression during gonadal differentiation in the lizard, Calotes versicolor. Mech. Dev. 2002;119(Suppl. 1):S55–S64. doi: 10.1016/s0925-4773(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 57.Guo Y, Cheng H, Huang X, Gao S, Yu H, Zhou R. Gene structure, multiple alternative splicing, and expression in gonads of zebrafish Dmrt1. Biochem. Biophys. Res. Commun. 2005;330:950–957. doi: 10.1016/j.bbrc.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 58.Huang X, Guo Y, Shui Y, Gao S, Yu H, Cheng H, Zhou R. Multiple alternative splicing and differential expression of dmrt1 during gonad transformation of the rice field eel. Biol. Reprod. 2005;73:1017–1024. doi: 10.1095/biolreprod.105.041871. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y, Lu H, Yu H, Cheng H, Zhou R. Multiple alternative splicing in gonads of chicken DMRT1. Dev. Genes Evol. 2007;217:119–126. doi: 10.1007/s00427-006-0117-0. [DOI] [PubMed] [Google Scholar]
- 60.Lu H, Huang X, Zhang L, Guo Y, Cheng H, Zhou R. Multiple alternative splicing of mouse Dmrt1 during gonadal differentiation. Biochem. Biophys. Res. Commun. 2007;352:630–634. doi: 10.1016/j.bbrc.2006.11.066. [DOI] [PubMed] [Google Scholar]
- 61.Cheng HH, Ying M, Tian YH, Guo Y, McElreavey K, Zhou RJ. Transcriptional diversity of DMRT1 (dsx- and mab3-related transcription factor 1) in human testis. Cell Res. 2006;16:389–393. doi: 10.1038/sj.cr.7310050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.