Abstract
Mitochondrial (mt) tRNAMet has the unusual modified nucleotide 5-formylcytidine (f5C) in the first position of the anticodon. This tRNA must translate both AUG and AUA as methionine. By constructing an in vitro translation system from bovine liver mitochondria, we examined the decoding properties of the native mt tRNAMet carrying f5C in the anticodon compared to a transcript that lacks the modification. The native mt Met-tRNA could recognize both AUA and AUG codons as Met, but the corresponding synthetic tRNAMet lacking f5C (anticodon CAU), recognized only the AUG codon in both the codon-dependent ribosomal binding and in vitro translation assays. Furthermore, the Escherichia coli elongator tRNAMetm with the anticodon ac4CAU (ac4C = 4-acetylcytidine) and the bovine cytoplasmic initiator tRNAMet (anticodon CAU) translated only the AUG codon for Met on mt ribosome. The codon recognition patterns of these tRNAs were the same on E. coli ribosomes. These results demonstrate that the f5C modification in mt tRNAMet plays a crucial role in decoding the nonuniversal AUA codon as Met, and that the genetic code variation is compensated by a change in the tRNA anticodon, not by a change in the ribosome. Base pairing models of f5C-G and f5C-A based on the chemical properties of f5C are presented.
INTRODUCTION
In 1981, the complete nucleotide sequence of the human mitochondrial (mt) genome was reported (1). Variations from the universal genetic code were suggested to exist in this organelle, based on comparisons of the DNA sequences of several mt protein genes and the actual amino acid sequences of the corresponding protein products. For example, a termination codon, UGA, and the AUA Ile codon in the universal genetic code are read as Trp and Met, respectively, and the AGA and AGG Arg codons become termination codons in human mitochondria. These ‘nonuniversal’ codons have also been observed in the mt genomes of many other organisms (2–5), and species-specific variations also exist in some codons (6,7).
The genetic code variation for methionine, first found in human mitochondria, is also observed in other mammalian mitochondria in which the genomes possess only a single tRNAMet gene with the CAT anticodon. In addition, there had been no reports suggesting the import of cytoplasmic tRNAs into mammalian mitochondria (8), unlike the import observed in many lower eukaryotic and plant mitochondria. Therefore, the single tRNAMet in mammalian mitochondria should recognize both the AUA and AUG codons as Met, serving as both the elongator and initiator tRNA. Although the anticodon CAU of tRNAMet cannot base pair with the AUA codon, according to the conventional Watson–Crick pairing (9), there have been several observations that the C–A pair at the wobble position can be formed in vivo as well as in vitro under certain limited conditions (10,11). Since the AUA codon occurs much more frequently than the AUG codon in mammalian mt genomes, it had been assumed that some special mechanism must exist in the mitochondrial translation system to allow the AUA codon to be translated as Met. In 1994, we found a novel modified nucleoside, 5-formylcytidine (f5C), in the first position of the anticodon of mt tRNAMet from bovine liver (12) and the nematode, Ascaris suum (13) (Figure 1A). This modified nucleoside was a promising candidate to permit tRNAMet to translate both the AUA and AUG codons through its ability to pair with not only G but also with A in the third position of the codon. The tRNAsMet from squid, fruit fly (14,15), frog, rat and chicken mitochondria (16) also possess f5C in the wobble position.
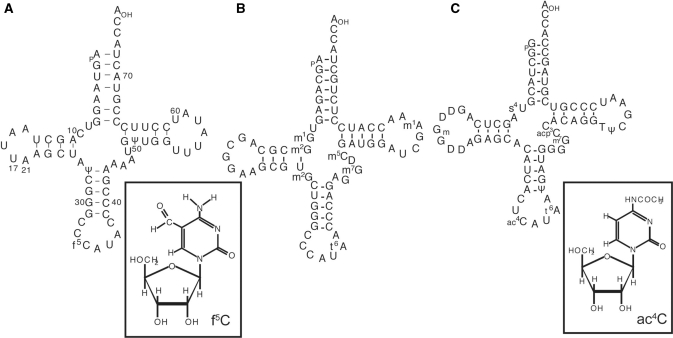
Figure 1.
Nucleotide sequences of tRNAs used in this study and chemical structures of the modified nucleotide in the wobble position. (A) Bovine mt tRNAMet and 5-formylcytidine, (B) bovine cytoplasmic initiator tRNAMet and (C) E. coli elongator tRNAMet and 4-acetylcytidine. The numbering of the residues in the cloverleaf structure of the mt tRNA conforms to that in the previous report (71).
The simplest and clearest way to determine whether f5C in the wobble position of mt tRNAMet actually recognizes A at the third position of the codon, would be to demonstrate that mt tRNAMet with the anticodon f5CAU decodes the AUA codon while the tRNA without this modification is restricted to decoding AUG in an efficient mitochondrial in vitro translation system. For these experiments, it is not feasible to simply substitute the well-characterized Escherichia coli in vitro translation system, because some factors necessary for translation cannot be exchanged between mitochondria and E. coli protein biosynthetic systems. For example, the binding of mt aminoacyl-tRNA to the A-site of ribosomes is efficiently carried out only by the mt elongation factor Tu and Ts complex (mt EF-Tu/Ts) and not by bacterial EF-Tu (17). Although mt elongation factor G (EF-G) can work with both mt and E. coli ribosomes, E. coli EF-G works only on E. coli ribosomes (18). A mitochondrial translation system was reported previously, but it was not useful for estimating the codon recognition, because its activity was much lower than that of the E. coli system (17,19).
In the current work, we have constructed an improved in vitro mitochondrial translation system with enough activity to test codon recognition activity. This system consists of mt aminoacyl-tRNAs, partially purified mt EF-Tu/Ts complex, mt EF-G, and mt ribosomes. Using this system we demonstrate that the modified nucleoside f5C of tRNAsMet is crucial for the recognition of the nonuniversal AUA codon as Met.
MATERALS AND METHODS
Chemicals
[14C]-Phenylalanine (16.6 GBq/mmol), [14C]-isoleucine (11 GBq/mmol) and [35S]-methionine (37 TBq/mmol) were purchased from Amersham. The [35S]-methionine used here was diluted to 200 GBq/mmol with nonradioactive methionine. Poly(U), pyruvate kinase (PK), spermine and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) were obtained from Sigma. The oligoribonucleotides, (AUN)6, (AUN)11 (N = A, C, G) and half fragments of tRNAMet, were synthesized with an automatic DNA synthesizer, ABI 381A, and were deprotected as described previously (20). T4 polynucleotide kinase (PNK) was obtained from TOYOBO, T4 RNA ligase from TAKARA, RNase T1 from Sankyo, RNase U2 from Sigma, RNase PhyM, and RNase CL3 from Boehringer Mannheim.
Partial purification of PheRS, MetRS and IleRS
Mitochondria were prepared from fresh bovine liver as reported (18,21). Crude mitochondrial extracts were prepared and fractionated by chromatography on DEAE-Sepharose fast flow (Pharmacia), with a 5–300 mM KCl gradient as described (22). The fractions containing the desired aminoacyl-tRNA synthetase (aaRS) activities were pooled and dialyzed against 60 volumes of Buffer A [10 mM potassium phosphate (pH 7.2), 50 mM KCl, 10% glycerol, 6 mM 2-mercaptoethanol and 0.2% CHAPS] with three buffer exchanges. Further purification was performed on a ceramic hydroxyapatite column (Bio-RAD) with a gradient of 10–300 mM potassium phosphate in Buffer A. The mt aaRS activities were defined by the amino acid acceptance of partially purified E. coli tRNAMet or tRNAIle.
The extract of E. coli strain A19, harvested at the mid-log phase (0.6A600), was prepared as described (23) and applied to a DEAE-Sepharose column at 50 mM NH4Cl. Fractions eluted with 400 mM NH4Cl were pooled and used as E. coli aaRS after dialysis against the initial buffer (23).
Preparation of tRNA and aminoacyl-tRNA
Amino acid-specific mt tRNAs were purified by a hybridization method, using DNA probes complementary to the 3′-end 30 bases of the tRNA genes, as previously reported (24,25). Escerichia coli tRNAPhe, tRNAsMet and tRNAsIle were purified by DEAE-Sepharose chromatography under two different pH conditions and by reverse phase chromatography (RPC-5), as described (26,27). Unlike tRNAMetf or tRNAIle1, tRNAMetm and tRNAIle2 could not be purified completely after RPC-5, so these tRNAs were isolated from the enriched fraction of RPC-5 by 8% native polyacrylamide gel electrophoresis (data not shown). Aminoacylation was performed as described (19). The charged mt tRNA was recovered from the reaction mixture by the acid–guanidine–phenol–chloroform method to improve the yield slightly (28). Sequences of the all tRNAs prepared in this study were confirmed by Donis–Keller's method (29–32) and the modified nucleotides were identified by the method of Kuchino et al. (31).
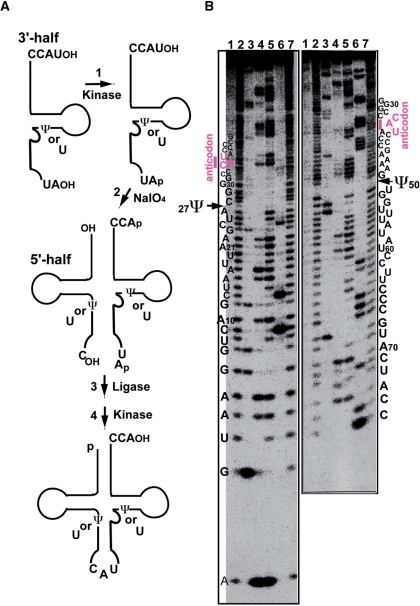
Synthetic tRNAMet variants were prepared by four steps (Figure 3A), as previously reported (33). The process consists of 5′-phosphorylation of the 3′-half fragment by T4 PNK (step 1), removal of the 3′-teminal nucleotide of the 3′-half fragment by NaIO4 treatment (step 2), ligation of the 5′-half and 3′-half fragments by T4 RNA ligase (step 3), and 5′-phosphorylation and 3′-dephosphorylation of the ligated tRNA by T4 PNK (step 4). The 5′-half and 3′-half fragments have sequences identical to the respective sequences of native tRNAMet, except for f5C and pseudouridine (Ψ). Two species of synthetic tRNAMet were prepared. One does not possess any modification. The other possesses only Ψ at the positions 27 and 50 (Figure 3B). The sequences of the fragments were confirmed by Donis–Keller's method before and after ligation (29,30,32).
Figure 3.
Preparation of synthetic tRNAMet variants. (A) Process of synthesizing mt tRNAMet by a combination of chemical synthesis and ligation (33). The synthetic tRNAs have sequences identical to the respective sequences of native tRNAMet, except for f5C and pseudouridine (Ψ). (B) Sequencing analysis using the Donis–Keller′s method of the synthetic mt tRNAMet labeled with [32P] at the 5′-end (left) and the 3′-end (right), respectively (30,32). Electrophoresis was performed on a 15% polyacrylamide–7 M urea–10% glycerol gel. Lanes: control without ribonuclease (RNase) (lane 1); limited alkaline hydrolysis (lanes 2 and 7); digestion by RNase T1 (specific for G: lane 3), RNase U2 (specific for A: lane 4), RNase PhyM (specific for A and U: lane 5) and RNase CL3 (specific for C: lane 6). As indicated by the arrows, Ψ was not digested by RNase PhyM (12). The numbering of the residues in the cloverleaf structure of the mt tRNA conforms to that in the previous report (71).
The modified nucleotide f5C on mt tRNAMet was reduced to 5-hydroxyl-cytidine by NaBH4. Purified mt tRNAMet (0.5A260 unit), was dissolved in 0.1 ml of 0.5 M Tris–HCl buffer containing 5 mM MgCl2 and 100 mM NaBH4 (pH 7.6), and incubated on ice for 30 min in the dark. After the addition of 0.2 ml of 0.5 M sodium acetate (pH 5.0), the tRNA was recovered by ethanol precipitation. The location and identification of the modified nucleotides were determined according to by the method of Kuchino et al. (31,34). The reduced mt tRNAMet partially digested with formamide was labeled with [γ-32P]ATP at the 5′-end, and the resulting [32P]-labeled oligonucleotides were separated by polyacrylamide gel electrophoresis. They were eluted from the gel and digested with nuclease P1 to produce 5′-[32P]-labeled mononucleotides, which were analyzed by two-dimensional thin layer chromatography (2D-TLC) on Avicel SF plate (Funakoshi Co.). The standard 5′-mononucleotides (pA, pG, pU and pC) were prepared by digestion of crude tRNA mixture with nuclease P1 and detected by UV absorption.
General components used in the translation assay
Mitochondrial EF-G and EF-Tu/Ts were prepared from bovine liver mitochondria, as described (22). Escherichia coli EF-Tu/Ts was prepared by standard methods (23). Ribosomes were prepared from E. coli strain A19 and mitochondria as reported (35,36), and were stored in the following ribosome buffers. E. coli 70S ribosome buffer: 20 mM Hepes-KOH (pH 7.6), 6 mM Mg(OAc)2, 30 mM NH4Cl and 6 mM 2-mercaptoethanol; mt 55S ribosome buffer: 20 mM Tris-HCl (pH 7.6), 20 mM MgCl2, 80 mM KCl and 6 mM 2-mercaptoethanol.
To check the purity of the mitochondrial ribosomes, samples were analyzed by sucrose density gradient centrifugation using linear sucrose gradients (38 ml, 6–38% in the mt ribosome buffer). Gradients were prepared using the Gradient Mate model 117 (BioComp), according to the user's manual. The ribosomes (80 A260 units), dissolved in 0.6 ml of the ribosome buffer without sucrose, were layered onto the gradients. The centrifugation was carried out at 20 000 r.p.m. for 16 h in an SW28 rotor using an XL-7 rotor (Beckman) at 4°C. Fractions (0.45 ml) were collected from top to bottom, using the Piston Gradient Fractionator (BioComp).
Ribosome binding assay
Reaction mixtures (40 μl) contained 50 mM Tris–HCl (pH 7.5), 1 mM dithiothreitol (DTT), 1 mM spermine, 8 mM MgCl2, 40 mM KCl, 2.5 mM phosphoenolpyruvic acid (PEP), 0.5 mM GTP, 1 mM methionine, 2.5 U/ml PK, 14 U of mt EF-Tu/Ts, 0.1 A260 (3.2 pmol, 0.08 μM) mt ribosomes and 2.6 pmol [35S]-Met-tRNA (22 000 c.p.m./pmol). The units are defined as described previously (37). The amount of the (AUN)6 oligonucleotide (N means A, C or G) is indicated in the figures. Incubation was carried out at 30°C for 15 min. The reaction was stopped by dilution with chilled wash buffer (5 ml), containing 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2 and 40 mM KCl. The amount of [35S]Met-tRNA bound to the ribosomes was determined essentially as reported previously (19).
In vitro translation system
For optimization of mt poly(U)-dependent poly(Phe) synthesis, reaction mixtures (20 μl) contained 50 mM Tris–HCl (pH 8.5), 1 mM DTT, 1 mM spermine, 7 mM MgCl2, 30 mM KCl, 0.5 mM GTP, 2.5 mM PEP, 2 U/ml PK, 0.125 mg/ml poly(U), 11 U of mt EF-Tu/Ts, 12 U of mt EF-G, 0.04 A260 (1.2 pmol, 0.1 μM) mt ribosomes and 7 pmol [14C]Phe-tRNA (928 c.p.m./pmol). In the case of E. coli ribosomes (4 U, 0.05 A260), the concentration of MgCl2 was reduced to 6 mM, the 30 mM KCl was replaced by 60 mM NH4Cl, and E. coli EF-G was used. The concentration of spermine was reduced to 0.2 mM for E. coli tRNA. The polymerization reactions were carried out at 37°C, and were stopped by the addition of 4 ml of 5% trichloroacetic acid (TCA), followed by deacylation of the peptidyl-tRNAs by heating at 95°C for 10 min. After the solutions were poured onto nitrocellulose membranes (Advantec, A045B025A), the membranes were washed with 20 ml of 1% TCA and dried. The amount of label remaining on the membrane was measured with a liquid scintillation counter (Packard Co. TRI-CARB).
The reaction mixtures (12 μl) for the in vitro (AUN)-translation system contained 50 mM Tris–HCl (pH 8.5), 1 mM DTT, 0.6 mM spermine, 7 mM MgCl2, 30 mM KCl, 0.5 mM GTP, 2.5 mM PEP, 2 U/ml PK, 1 mM methionine, 1 mM isoleucine, 0.1 mg/ml mRNA [oligo(AUN)11], 14 U of mt EF-Tu/Ts, 12 U of mt EF-G, 0.04A260 (1.2 pmol, 0.1 μM) mt ribosomes and 1.2 pmol aminoacyl-tRNA ([35S]Met-tRNA or [14C]Ile-tRNA).
RESULTS
Construction and optimization of an in vitro mitochondrial translation system using E. coli tRNA
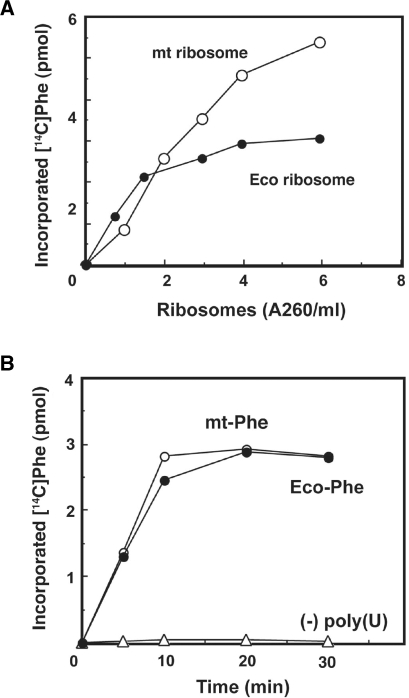
An efficient mitochondrial (mt) translation system was required to characterize the universal and nonuniversal codon recognition by mitochondrial tRNA. Mitochondrial ribosomes and translational factors were prepared as described previously (18). As indicated in Figure 2A, both E. coli and mt ribosomes catalyze poly(Phe) synthesis using E. coli Phe-tRNA and the mt EF-Tu/Ts. The optimum concentration of spermine depended on the tRNA species: the mt tRNA preferred a higher concentration (0.8–1.5 mM) of spermine than E. coli tRNA did (0.2 mM). The poly(U)-dependent poly(Phe) synthesis activity in the complete mt system using mt ribosomes was equal to that obtained using E. coli Phe-tRNA, under the respective optimized conditions described in the Materials and methods section (Figure 2B).
Figure 2.
In vitro translation system prepared from bovine liver mitochondria. (A) Dose–response of poly(Phe) synthesis, as a function of the amount of mitochondrial (open circle) or E. coli (filled circle) ribosomes, using E. coli Phe-tRNA and mt EF-Tu/Ts. The reactions were carried out at 37°C for 8 min. (B) Poly(U)-dependent poly(Phe) synthesis activity of the complete mitochondrial system under the optimized conditions for Phe-tRNA from mitochondria (open circle) and E. coli (filled circle), respectively, as described in the Materials and methods section.
Preparation of mitochondrial tRNAMet with or without f5C, and other nonmitochondrial tRNAsMet
To elucidate whether f5C is necessary for mt tRNAMet to decode the AUA codon, three species of mt tRNAMet were prepared. The native tRNAMet, possessing two pseudouridines (Ψ) at positions 27 and 50 as well as f5C at position 34 (Figure 1A), was prepared from bovine liver mitochondria as reported (12). A synthetic tRNA without any modified nucleoside, and a synthetic tRNA with two Ψ's but no f5C were obtained by enzymatic ligation of the 3′-half and 5′-half fragments with or without Ψ's (Figure 3A), which were both chemically synthesized (33). The sequences of the native and synthetic tRNAsMet were confirmed by Donis–Keller's method (Figure 3B) (29,30). The methionine acceptance activities of these synthetic tRNAs, catalyzed by the partially purified mt MetRS, gave values comparable to those of the native tRNAMet (native tRNAMet, Km = 0.31 μM; synthetic tRNAMet with and without Ψ, Km = 0.37 μM). The kinetic parameters were comparable to those of human mt MetRS (38). We also purified E. coli tRNAMetm (anticodon ac4CAU) and bovine cytoplasmic initiator tRNA (anticodon CAU) from E. coli cells and bovine liver, respectively, as reported (12,26), which were both used as references for examining the decoding properties of mt tRNAMet.
Codon-dependent ribosomal binding of mitochondrial tRNAMet analogs
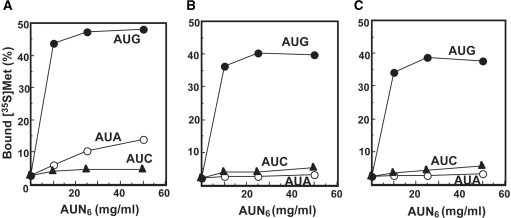
Ribosome binding experiments were carried out using these mt tRNAs and chemically synthesized mRNAs. Synthetic polyribonucleotides comprising six repeats of AUN codon triplets (N is A, C or G) were used to provide efficient binding. As shown in Figure 4, ∼50% of the input native Met-tRNA (1.6 pmol) bound to ribosomes in the presence of the AUG codon. Similar binding was obtained with the Met-tRNAsMet lacking f5C, regardless of the presence or absence of the two Ψ residues. This observation is expected since the anticodon of the synthetic tRNA (CAU) is the complement for the AUG codon. These data also argue that the formylation of C34 does not interfere with pairing with the G residue in the third position of the codon. The native Met-tRNA was also capable of reading the AUA codon although it was less efficiently recognized than the AUG codon. The synthetic Met-tRNAMet that did not carry the f5C modification could read the AUG codon effectively (Figure 4B and C). However, this tRNA could not read the AUA codon at all (Figure 4B and C). These data strongly argue that the recognition of AUA as a methionine codon requires the modification of C34 by the formylation of C5. Comparison of the two synthetic Met-tRNAsMet indicates that the conversion of U to Ψ at positions 27 and 50 is not important for recognition of the AUG codon (Figure 4B and C). These results are in line with the recent report that a chemically synthesized RNA anticodon stem-loop fragment with modified bases f5C and Ψ confers affinity for the AUA and AUG codons on E. coli 70S ribosomes (39).
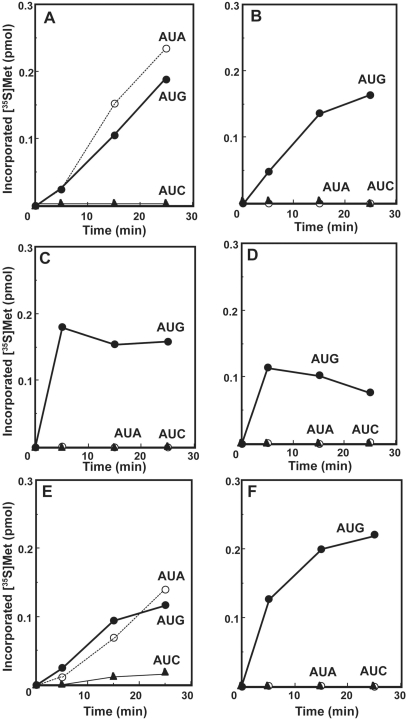
Figure 4.
Codon-dependent ribosome binding assays of bovine mt tRNAMet variants. (A) Native mt tRNAMet purified from bovine liver mitochondria. (B) Synthetic mt tRNAMet with no modified bases. (C) Synthetic mt tRNAMet with two Ψ's. Oligonucleotides AUG6 (filled circle), AUA6 (open circle) and AUC6 (filled triangle) were used as mRNA. The presented values are the averages of three independent experiments and were reproducible within ± 1.2%.
In vitro translation assay using tRNAsMet with various anticodons
To further examine the role of the f5C modification in the translation of AUA codons, the in vitro mitochondrial translation system was used with a message carrying eleven repeats of the AUN codon triplets (AUN)11, to allow the translation products to be efficiently precipitated with TCA. Unlike poly(Phe) synthesis, the background level of Met incorporation in the absence of mRNA increased with concentrations of spermine >2 mM. Therefore, 0.6 mM spermine was included in the reaction mixture to reduce the background. Mitochondrial EF-Tu was used in all of the assay systems shown in Figure 5, except for Figure 5F. Thus, we adjusted all the conditions of the translation assay so that the AUA codon could be recognized by mt tRNAMet efficiently.
Figure 5.
The codon recognition abilities of tRNAsMet possessing different anticodons. Time courses of oligo(AUN)-dependent oligo(Met) synthesis in an in vitro translation system with mitochondrial (A–D) or E. coli (E and F) ribosomes. (A and E) Native bovine mt tRNAMet purified from bovine liver mitochondria. (B) Synthetic mt tRNAMet with two Ψ's. (C) Bovine cytoplasmic initiator tRNAMet. (D and F) E. coli elongator tRNAMet. Oligonucleotides AUA11 (open circle), AUC11 (filled triangle) and AUG11 (filled circle) were used as mRNA. The incorporation of [35S]Met was normalized by subtracting the value without mRNA, which was <2% of the input c.p.m. (about 0.03 pmol).
Native Met-tRNAMet was active in oligo(Met) synthesis with both AUG and AUA codons (Figure 5A), which is in agreement with the ability of this Met-tRNA to bind both codons in the ribosome binding assays. As expected, the native Met-tRNA was not able to read the near-cognate AUC codon. In contrast, the synthetic Met-tRNA carrying the two Ψ residues is active with the (AUG)11 template but not with the (AUA)11 template (Figure 5B). These data are in agreement with the ribosome binding experiments that indicate that the f5C modification is required to read the AUA codon.
Two control Met-tRNAs were also used in these experiments. First, the bovine cytoplasmic initiator Met-tRNAMeti with the CAU anticodon (Figure 5C) and secondly the E. coli elongator Met-tRNAMetm with the ac4CAU anticodon (Figure 5D) were tested in (AUN)11-dependent oligo(Met) synthesis with mt ribosomes. Both of these Met-tRNAs translated AUG codons as Met. However, neither of them could read the AUA codon. The failure of the E. coli elongator tRNAMet (tRNAMetm) to translate the AUA codon (Figure 4D) is reasonable, because AUA is an isoleucine codon in E. coli, and ac4C at position 34 of tRNAMetm is known to serve in preventing the misreading of the AUA codon (40). The mitochondrial ribosomes and mt EF-Tu/Ts used in these experiments work more rapidly with the more canonical cytoplasmic initiator and bacterial elongator than they do with the mitochondrial Met-tRNA. This difference may reflect the somewhat unusual structures of the mitochondrial tRNAs that are generally shorter than canonical tRNAs and that lack some of the common features of these tRNAs (Figure 1).
We noticed that the bovine cytoplasmic initiator tRNAMeti could read the AUG codon in this system (Figure 5C). This observation suggests that mt EF-Tu can recognize bovine cytoplasmic initiator tRNAMet, unlike the case of yeast initiator tRNAMet. This is probably because the bovine cytoplasmic initiator tRNAMeti has no 2′-phosphoribosyl residue in the T-stem (Figure 1B), which is known as the antideterminant of yeast initiator tRNA toward the cytoplasmic EF-1α (41,42). Further, the single tRNAMet gene in mammalian mitochondria provides both the initiator and elongator tRNAs for methionine and mt EF-Tu has presumably evolved to be somewhat flexible in its ability to accommodate Met-tRNAs with some features generally observed in initiator tRNAs.
Both bovine cytoplasmic initiator tRNAMet and mt tRNAMet have C at position 33, just 5′-adjacent to the anticodon. This position is usually occupied by U, to form the U-turn structure in the anticodon loop (43). However, this unusual base replacement does not appear to play a role in the recognition of the AUA codon (Figure 5C). In the first position of the anticodon, the bovine cytoplasmic initiator tRNAMet has an unmodified C, which is unable to pair with A at the third letter of the AUA codon, and thus it cannot be a candidate for recognizing AUA codons in the mt system.
Codon recognition ability of mitochondrial tRNA on E. coli ribosomes
To examine whether the ability of the native Met-tRNAMet to read the AUA codon was a property of the tRNA alone or was dependent also on the use of mitochondrial ribosomes, the decoding of (AUG)11 and (AUA)11 was examined on E. coli ribosomes (Figure 5E). These data clearly show that the native mt Met-tRNAMet decodes AUA on bacterial ribosomes indicating that this decoding is a property of the tRNA and not of the ribosome. Thus, it is evident that the translation of the AUA codon for Met is achieved by the strict recognition of the AUA codon by the tRNA anticodon containing f5C34, irrespective of the origin of the ribosomes. Mitochondrial Met-tRNAMet with E. coli EF-Tu/Ts did not work on either mt or E. coli ribosomes using any oligo (AUN)11 (data not shown). This observation is consistent with previous results obtained with mt Phe-tRNAPhe, which was not effectively delivered to the ribosomal A-site with E. coli EF-Tu (17). In a control experiment (Figure 5F), the clear discrimination of the E. coli elongator Met-tRNAmMet against the AUA codon on E. coli ribosomes was confirmed.
To verify the accurate discrimination of the AUN codon box, the codon recognition abilities of mt tRNAIle, E. coli tRNAIle1 and E. coli tRNAIle2 were tested using both mt and E. coli ribosomes (see Supplementary Material). As expected, all of the tRNAsIle recognized the AUC codon, but not the AUG codon. In addition, tRNAIle2, possessing the anticodon k2CAU (k2C = 2-lysylcytidine), recognized both AUA and AUC codons. This modified base is known to switch both the amino acid acceptance (from Met to Ile) and codon recognition (from AUG to AUA codons) of the tRNA (44). Again the recognition profile was independent of the source of the ribosomes used in agreement with the previous data.
Investigation of a role of the modified nucleotide f5C in mitochondrial tRNAsMet
All of the tRNAs used in this study, except for mt tRNAMet, have N6-threonylcarbamoyladenosine (t6A37) adjacent to U36, the first position of the anticodon (45). It has been verified that t6A37 provides additional stability to codon–anticodon interactions through a cross-strand stack above the first base pair in the ribosomal A-site (46). Bacterial initiator tRNAsMet possess an unmodified A37 because the initiator tRNA has to enter only to the ribosomal P-site during translation initiation. Although mt tRNAMet also possesses an unmodified A37, it plays a dual role as an initiator and an elongator tRNA in mitochondria. Therefore, the unique modification such as f5C may have a significant advantage in stabilizing codon–anticodon base pairs.
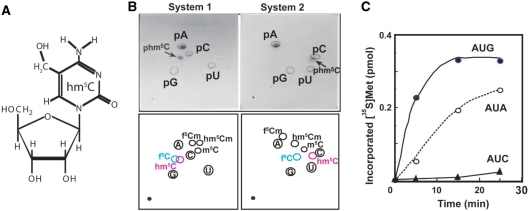
Proton and nitrogen nuclear magnetic resonance (NMR) studies of deoxycytosine derivatives, f5C, 5-hydroxymethylcytosine (hm5C), and 5-hydroxycytosine, show that the 4-amino group and the oxygen group of the substituent at C-5 form an intrabase hydrogen bond indicated by significant changes in both 1H and 15N chemical shifts (47). Further, it was reported that the Klenow fragment incorporates dAMP against f5C more frequently than cytidine (48). These results suggest that the intrabase hydrogen-bonding of C5-substituent derivatives may introduce electronic effects on cytosine, enabling the modified cytidine to base pair with A. To examine the codon recognition ability of another C5-substiuent, f5C of mt tRNAMet was reduced by sodium borohydride to form hm5C (Figure 6A). The sequence of the reduced mt tRNAMet was confirmed by the Donis–Keller's method (29,30,32) and reduction of f5C to hm5C34 was confirmed by the two-dimensional thin-layer chromatography (2D-TLC) in the two solvent systems (34,49,50). As shown in Figure 6B, f5C at the position 34 was clearly reduced to hm5C. The reduced tRNAMet was efficiently aminoacylated by mt MetRS (Km = 0.46 μM), indicating that the reduced tRNAMet retained the normal ternary structure required for the biochemical activity. The hydroxymethyl group was not an anti-determinant for recognition by mt MetRS. The codon recognition ability of Met-tRNA with the anticodon hm5CAU was examined in the mt homologous system described above. As shown in Figure 6C, Met-tRNA with the anticodon hm5CAU recognized both the AUG and AUA codons. This observation suggests that a common chemical property of f5C and hm5C is responsible for base pairing with A.
Figure 6.
Reduction of 5-formylcytidine to 5-hydroxylcytidine (hm5C) of mt tRNAMet. (A) The chemical structure of hm5C. (B) 2D-TLC analysis of the nucleotide at the anticodon wobble position of mt tRNAMet after reduction (upper panels), and diagrams of chromatographic mobility of modified cytosine 5′-monophospates (lower panels) (49,50). The detailed procedure is described in Materials and Methods section. The solvent used for the first dimension in both systems was isobutyric acid/concentrated ammonia/water (66:1:33 v/v/v). For the second dimension, 2-propanol/HCl/water (70:15:15 v/v/v) and ammonium sulfate/0.1 M sodium phosphate (pH 6.8)/1-propanol (60 g:100 ml:2 ml) was used in the systems 1 and 2, respectively. (C) The codon recognition ability of the reduced mt tRNAMet (anticodon hm5CAU) on mt ribosomes. The incorporation of [35S]-Met was normalized by subtracting the value without mRNA.
DISCUSSION
Mitochondrial translation system using various tRNA species
We reconstituted an in vitro mitochondrial translation system, which had sufficient efficiency to test the codon recognition ability of Met-tRNAMet. The addition of spermine significantly improved translational efficiency. The optimum concentration of spermine was different with the mitochondrial and E. coli tRNA species, suggesting that spermine contributes to the structural stability of mt tRNA. Spermine has been observed in tRNA crystal structures (PDB ID: 1evv, 1tn2 and 2tra), which is assumed to contribute to structural stability (51–53). Spermine may play a more important role in stabilizing the structures of mt tRNAs since these tRNAs lack may of the stabilizing features observed with canonical tRNAs. After optimization of mt poly(U)-dependent poly(Phe) synthesis, AUN-dependent oligo(Met) synthesis was improved by decrease of mRNA concentration and addition of nonradioactive Met. The former was effective on the AUA-dependent Met-incorporation but had no effect on the AUG-dependent incorporation. On the other hand, the latter was useful for decrease of the background level.
The codon recognition pattern of each tRNA on mt ribosomes was similar to that on E. coli ribosomes. However, the efficiency of peptide synthesis was not always the same at least with tRNAPhe, tRNAMet and tRNAIle. The efficiency of poly(Phe) synthesis by E. coli tRNAPhe in the mt system was the same as that in the E. coli system (Figure 2B), while that of oligo(Met) or oligo(Ile) by E. coli tRNAMet or tRNAIle in the mt system was lower than that in the E. coli system (Figure 5D and F, and Supplementary Figure 3). Mitochondrial tRNAMet or tRNAIle could work on mt ribosomes better than on E. coli ribosomes (Figure 5A and E, and Supplementary Figure 2). We could not find conditions under which E. coli tRNAMetm worked efficiently with mt EF-Tu/Ts on 70S ribosomes (data not shown). In contrast, E. coli tRNAPhe could work well with any combination of EF-Tu and ribosomes (Figure 2). These results suggested that the combination of tRNA, EF-Tu and ribosomes from heterogeneous sources would cause subtle differences in their interactions with one another when the ternary complex of aminoacyl-tRNA, EF-Tu and GTP associates with ribosomes. Although mt EF-Tu shares high homology and similar structural domains with those of E. coli (54,55), a slightly different affinity between EF-Tu and rRNA or ribosomal proteins would probably affect the peptide synthesis efficiency. This heterogeneous system might be useful to clarify what happens in the ribosome through the entry of the ternary complex. Although the efficiency differs, according to the tRNA species or the combination of EF-Tu/Ts and ribosomes, this assay system would be useful to verify the codon recognition ability of mt tRNAs that have unusual, unstable structures.
The role of the modified nucleotide f5C in the expanded codon recognition
Mammalian mitochondria face the challenge of decoding both AUG and AUA in both the P-site and the A-site using a single tRNAMet. The work described here indicates that a critical feature of tRNAMet in the decoding process is the modified residue f5C in the first position of the anticodon. This work represents the first experimental evidence in an organelle that a modified nucleotide in the anticodon expands the codon recognition ability of a tRNA. Our results clearly demonstrate that native mt Met-tRNAMet with the anticodon f5CAU, can read the AUA codon in addition to the AUG codon. The putative interactions in the f5C-G and f5C-A base pairs can be modeled by base replacements of C34 with f5C in the anticodon of tRNAMetf (PDB: 2J00), and, by the replacement of G3 of the methionine codon with adenine, respectively (Figure 7). The f5C-G pair clearly forms similar interactions to those observed with a normal C–G pair (Figure 7A and B).
Figure 7.
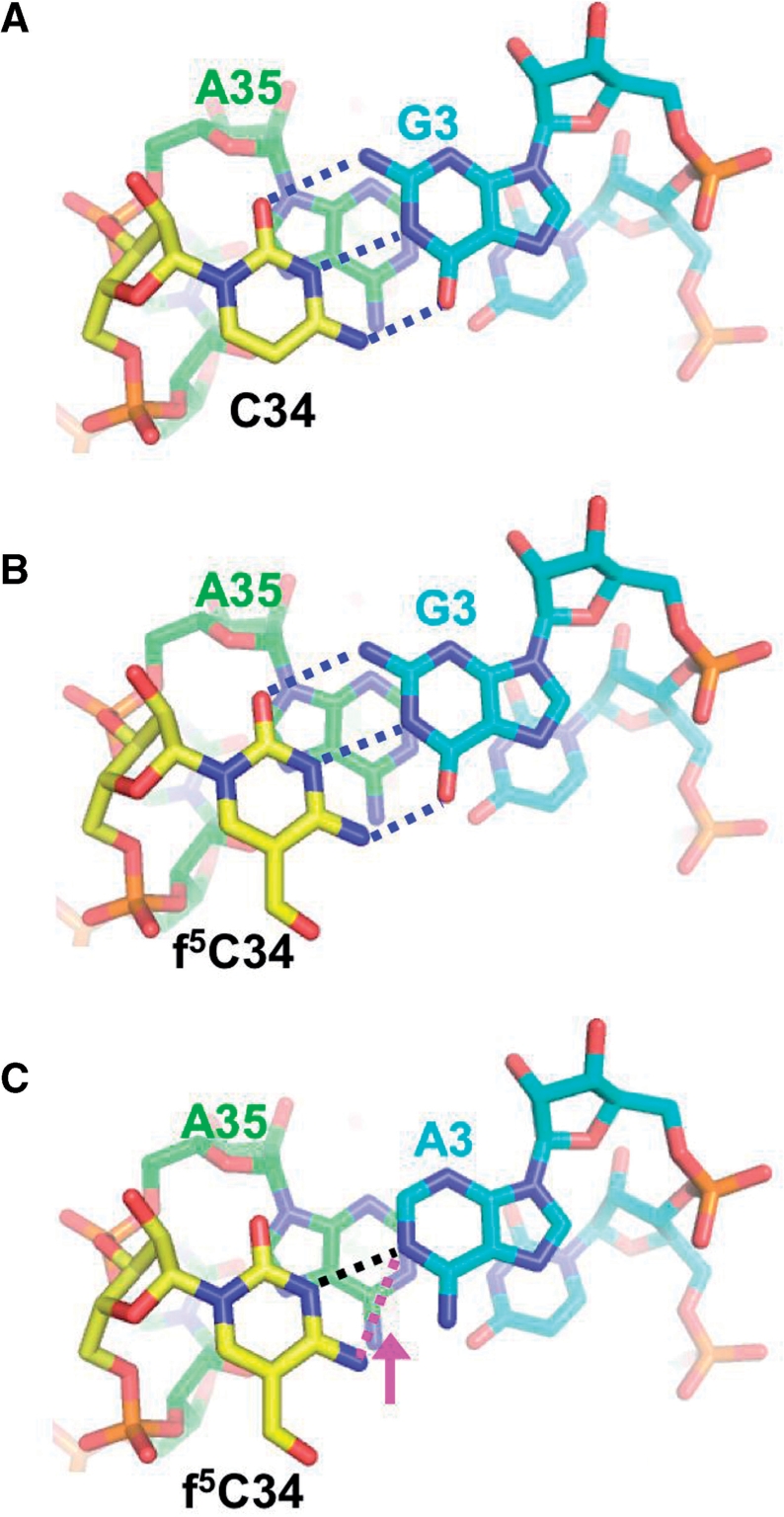
Base pairing model of f5C-G and f5C-A. (A) C34-G1 base pair of the initiator tRNAMet and the AUG codon (PDB: 2J00). The first letter of the anticodon C34 (yellow) base pairs with the third letter of the codon G3 (cyan) and stacks on the 3′-adjacent nucleotide A35 (green). Hydrogen bonds between C34 and G3 are shown as blue dotted lines. (B) f5C-G base pair model. The coordinates of f5C were generated with the PRODRG server (http://davapc1.bioch.dundee.ac.uk/prodrg) (72) and superposed to the C34 in (A). Putative hydrogen bonds between f5C34 and G3 are shown as blue dotted lines. (C) The f5C-A base pair model. The coordinates for adenine were also generated with the PRODRG server and superposed on G3 in (B). Putative hydrogen bonds between f5C34 and A3 are shown as dotted lines. The distance between N3 of f5C34 and N1 of A3 or N4 of f5C34 and N1 of A3 is 2.95 Å (black dotted line) or 3.44 Å (magenta dotted line), respectively. An arrow (magenta) represents an expected shift to form a hydrogen bond between N4 of f5C34 and N1 of A3. These figures were prepared with PyMOL, from DeLano Scientific (http://www.pymol.sourceforge.net/).
It is more difficult to visualize how f5C base pairs with A. Two views of an f5C-A base pair can be proposed. In the first view, a hydrogen bond forms between N1 of A and N3 of f5C (Figure 7C, black line). In the second view, N1 of A is hydrogen bonded to N4 of f5C (Figure 7C, magenta line). Although the distance between N1 of A and N3 of f5C is adequate to form a hydrogen bond (2.95 Å), N1 of A must be protonated for this interaction to occur. On the other hand, hydrogen bonding between N4 of f5C and N1 of A does not require protonation of the A residue.
There have been several observations of the C–A+ base pair (A+ means adenosine protonated at N-1) by NMR and crystallographic studies. The protonation of A (A+) in an A–C pair has been shown in double-stranded (ds) DNA (56) and in tandem C–A base pairs in dsRNA (57). NMR studies demonstrated that pKa values of A residues closed to the cleavage site of ribozymes were shifted to 6.2 and 6.5 (58,59). In the anticodon loop of E. coli tRNALys3 where an A residue is observed with a pKa of about 6 (60). In the case of mt tRNAMet, there is no direct evidence for the protonation of N1 of A3. However, our NMR analyses have revealed that the f5C nucleoside adopts the most rigid C3′-endo conformation in all of the modified cytidine nucleosides studied to date and that the pKa value of f5C is very low (61,62). It suggests that the approach of f5C could bring about the protonation of A to form the f5C-A+ base pair.
Alternatively, in the second model above f5C shifts slightly from the position taken in the base pair with G to form a hydrogen bond with A (magenta arrow in Figure 7C). In our previous NMR study, using the transferred nuclear Overhauser effect (TRNOE) method, we found that the conformation of the f5C-A pair was different from that of the f5C-G pair, and that the AUA codon adopted an extended A-form RNA conformation (63–65). During the decoding process the codon should be fixed in the A-form (66,67), so that the anticodon loop of mt tRNAMet would adapt its own conformation to keep the mRNA in the A-form in vivo. This conformational change in the sugar–phosphate backbone would be tolerant at the wobble position like the inosine–adenosine base pair (68). In addition, NMR studies of 2′-deoxycytosine derivatives, f5C and hm5C show that the 4-amino group (N4) and the oxygen group of 5-substituent (O5) are fixed in the planar configuration against the cytosine base through intrabase hydrogen-bonding between N4 and O5 (47). The reduction of f5C to hm5C maintains the expanded codon recognition ability (Figure 6C), suggesting that common characteristics between f5C and hm5C enable the modified cytidine to base pair with A3 (the third letter of the codon). The tautomeric form of these 5-substituents of C34 would have the advantage of enhanced stacking against A35, compensating for the single hydrogen bond. This idea is supported by recent crystallographic studies of an RNA anticodon stem-loop fragment with modified bases in the ribosomal A-site. This structure clearly shows that the anticodon nucleotides exhibit a C3′-endo conformation and stack against the 3′-adjacent base (46,68–70). Because of the 3′ stack of the anticodon, the modified U34 prefers to base pair with G3 in the standard Watson–Crick base pairing geometry, not in the G–U wobble geometry (46,69). Moreover, even a single hydrogen bonding in the wobble base pair is compensated through increased stacking of the modified base and additional hydrogen bonding among the modification moiety and proximate groups in the adjacent nucleotide (46,69). Considering these steric interactions, it seems more probable that f5C acquires an altered conformation, perhaps the tautomeric form, due to the presence of the 5-substituent group, and forms a 3′-stack with A35, which would compensate for a single hydrogen bonding in the f5C-A base pair. Further investigations are required to clarify which nitrogen group of f5C, N3 or N4, interacts with N1 of A3.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS Research Fellowship for Young Scientists (08004075 to C.T.); Grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (1130802 and 14035206 to K.W.); the Human Frontier Science Program Organization (to K. W.); National Institutes of Health (GM32734 to L.L.S.). Funding for open access charge: Human Frontier Science Program Young Investigator Grant (RGY0067/2007 to C.T.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Dr H. Takaku, of Chiba Institute of Technology, for his kind gift of the pseudouridine amidite, and Dr T. Ohtsuki, presently in Okayama University, for his technical assistance in the synthesis of oligoribonucleotides. We also thank Prof. Dr K. Miura, of Gakushuin University, for his encouragement during this study.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Himeno H, Masaki H, Kawai T, Ohta T, Kumagai I, Miura K, Watanabe K. Unusual genetic codes and a novel gene structure for tRNA(AGYSer) in starfish mitochondrial DNA. Gene. 1987;56:219–230. doi: 10.1016/0378-1119(87)90139-9. [DOI] [PubMed] [Google Scholar]
- 3.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 4.Xu X, Arnason U. The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteroplasmy of the control region. Gene. 1994;148:357–362. doi: 10.1016/0378-1119(94)90713-7. [DOI] [PubMed] [Google Scholar]
- 5.Krettek A, Gullberg A, Arnason U. Sequence analysis of the complete mitochondrial DNA molecule of the hedgehog, Ernaceus europaeus, and the phylogenic position of the Lipotyphla. J. Mol. Evol. 1995;41:952–957. doi: 10.1007/BF00173175. [DOI] [PubMed] [Google Scholar]
- 6.Osawa S, Juckes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe K, Osawa S. In: tRNA: Structure, Biosynthesis, and Function. Soll D, RajBhandary U, editors. Washington DC: American Society for Microbiology; 1995. pp. 225–250. [Google Scholar]
- 8.Roe BA, Wong JFH, Chen EY. In: Proceedings of the Third Cleveland Symposium on Macromolecules. Walton AG, editor. Amsterdam: Elsevier; 1981. pp. 167–176. [Google Scholar]
- 9.Crick FHC. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 10.Mikelsaar R. C-A base pairs in transfer ribonucleic acids and codon-anticodon complexes. J. Theoritical. Biol. 1981;92:163–180. doi: 10.1016/0022-5193(81)90390-8. [DOI] [PubMed] [Google Scholar]
- 11.Samuelsson T, Axberg T, Boren T, Lagerkvist U. Unconventional reading of glycine codons. J. Biol. Chem. 1983;258:13178–13184. [PubMed] [Google Scholar]
- 12.Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McClosky JA, Kawai G, et al. A novel modified nucleoside found at the first position of the anticodon of methionine trna from bovine liver mitochondria. Biochem. 1994;33:2234–2239. doi: 10.1021/bi00174a033. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe Y, Tsurui H, Ueda T, Furushima R, Takamiya S, Kita K, Nishikawa K, Watanabe K. Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem. 1994;269:22902–22906. [PubMed] [Google Scholar]
- 14.Tomita K, Ueda T, Watanabe K. 5-formylcytidine (f5C) found at the wobble position of the anticodon of squid mitochondrial tRNAMetCAU. Nucleic Acids Symp. Ser. 1997;37:197–198. [PubMed] [Google Scholar]
- 15.Tomita K, Ueda T, Ishiwa S, Crain PF, McClosky JA, Watanabe K. Codon reading patterns in Drosophila melanogaster mitochondria based on their tRNA sequences: a unique wobble rule in animal mitochondria. Nucleic Acids Res. 1999;27:4291–4297. doi: 10.1093/nar/27.21.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takemoto C, Ueda T, Miura K, Watanabe K. Nucleotide sequences of animal mitochondrial tRNAsMet possibly recognizing both AUG and AUA codons. Nucleic Acids Symp. Ser. 1999;42:77–78. doi: 10.1093/nass/42.1.77. [DOI] [PubMed] [Google Scholar]
- 17.Kumazawa Y, Schwartzbach CJ, Liao H-X, Mizumoto K, Kaziro Y, Miura K, Watanabe K, Spremulli LL. Interaction of bovine mitochondrial phenylalanyl-tRNA with ribosomes and elongation factors from mitochondria and bacteria. Biochem. Biophys. Acta. 1991;1090:167–172. doi: 10.1016/0167-4781(91)90097-6. [DOI] [PubMed] [Google Scholar]
- 18.Eberly SL, Locklear V, Supremulli LL. Bovine mitochondrial ribosomes, elongation factor specificity. J. Biol. Chem. 1985;260:8721–8725. [PubMed] [Google Scholar]
- 19.Takemoto C, Koike T, Yokogawa T, Benkowski L, Spremulli LL, Ueda T, Nishikawa K, Watanabe K. The ability of bovine mitochondrial transfer RNAMet to decode AUG and AUA codons. Biochimie. 1995;77:104–108. doi: 10.1016/0300-9084(96)88112-0. [DOI] [PubMed] [Google Scholar]
- 20.Scaringe SA, Franklyn C, Usman N. Chemical synthesis of biologically active oligoribonucleotides using beta-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990;18:5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews DE, Hessles RA, Denslow ND, Edwards JS, O’Brien TW. Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 1982;257:8788–8794. [PubMed] [Google Scholar]
- 22.Schwartzbach CJ, Farwell M, Liao H-X, Spremulli LL. Bovine mitochondrial initiation and elongation factors. Methods Enzymol. 1996;264:248–261. doi: 10.1016/s0076-6879(96)64025-7. [DOI] [PubMed] [Google Scholar]
- 23.Ravel J, Shorey R, Foehner S, Shive W. A study of the enzymatic transfer of aminoacyl-tRNA to E. coli ribosomes. Arch. Biochem. Biophys. 1986;125:514–526. doi: 10.1016/0003-9861(68)90609-7. [DOI] [PubMed] [Google Scholar]
- 24.Tsurui H, Kumazawa Y, Sanakawa R, Watanabe Y, Kuroda T, Wada A, Watanabe K, Shurai T. Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem. 1994;221:166–172. doi: 10.1006/abio.1994.1393. [DOI] [PubMed] [Google Scholar]
- 25.Wakita K, Watanabe Y, Yokogawa T, Kumazawa Y, Nakamura S, Ueda T, Watanabe K, Nishikawa K. Higher-order structure of bovine mitochondrial tRNAPhe lacking the ‘conserved’ GG and TYCG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res. 1994;22:347–353. doi: 10.1093/nar/22.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seno T, Kobayashi M, Nishimura S. Purification of Escherichia coli methionine tRNAf and tRNAm and studies on their biophysical and biochemical properties. Biochem. Biophys. Acta. 1968;169:80. doi: 10.1016/0005-2787(68)90010-5. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura S. Proceedings in Nucleic Acid Research. New York: Harper & Row; 1971. [Google Scholar]
- 28.Chomcznski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 29.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenosines, guanosines, pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donis-Keller H. PhyM: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980;8:3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuchino Y, Hanyu N, Nishimura S. Analysis of modified nucleosides and nucleotide sequence of tRNA. Methods Enzymol. 1987;155:379–396. doi: 10.1016/0076-6879(87)55026-1. [DOI] [PubMed] [Google Scholar]
- 32.Kuchino Y, Nishimura S. Enzymatic RNA sequencing. Methods Enzymol. 1989;180:154–163. doi: 10.1016/0076-6879(89)80099-0. [DOI] [PubMed] [Google Scholar]
- 33.Ohtsuki T, Kawai G, Watanabe Y, Kita K, Nishikawa K, Watanabe K. Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res. 1996;24:662–667. doi: 10.1093/nar/24.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokogawa T, Watanabe Y, Kumazawa Y, Ueda T, Hirao I, Miura K, Watanabe K. A novel cloverleaf structure found in mammalian mitochondrial tRNA(Ser) (UCN) Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nierhaus KH. In: Ribosomes and Protein Synthesis. Spedding G, editor. New York: Oxford University Press; 1990. pp. 161–189. [Google Scholar]
- 36.O'Brien TW, Denslow ND. Bovine mitochondrial ribosomes. Methods Enzymol. 1996;264:237–248. doi: 10.1016/s0076-6879(96)64024-5. [DOI] [PubMed] [Google Scholar]
- 37.Schwartzbach CJ, Spremulli LL. Bovine mitochondrial protein synthesis elongation factors identification and initial characterization of an elongation factor Tu- elongation factor Ts complex. J. Biol. Chem. 1989;264:19125–19131. [PubMed] [Google Scholar]
- 38.Spencer A, Heck A, Takeuchi N, Watanabe K, Spremulli L. Characterization of the human mitochondrial methionyl-tRNA synthetase. Biochem. 2004;43:9743–9754. doi: 10.1021/bi049639w. [DOI] [PubMed] [Google Scholar]
- 39.Lusic H, Gustilo EM, Vendeix FA, Kaiser R, Delaney MO, Graham WD, Moye VA, Cantara WA, Agris PF, Deiters A. Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res. 2008;36:6548–6557. doi: 10.1093/nar/gkn703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stern L, Schulman LH. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J. Biol. Chem. 1978;253:6132–6139. [PubMed] [Google Scholar]
- 41.Forster C, Chakraburtty K, Sprinzl M. Discrimination between initiation and elongation of protein biosynthesis in yeast: identity assured by a nucleotide modification in the initiator tRNA. Nucleic Acids Res. 1993;21:5679–5683. doi: 10.1093/nar/21.24.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Pawel-Rammingen U, Astrom S, Bystrom AS. Mutation analysis of conserved position potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1432–1442. doi: 10.1128/mcb.12.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 44.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 45.Rozenski J, Crain PF, McCloskey JA. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 47.LaFrancois CJ, Fujimoto J, Sowers LC. Synthesis and characterization of isotopically enriched pyrimidine deoxynucleoside oxidation damage products. Chem. Res. Toxicol. 1998;11:75–83. doi: 10.1021/tx970186o. [DOI] [PubMed] [Google Scholar]
- 48.Karino N, Ueno Y, Matsuda A. Synthesis and properties of oligonucleotides containing 5-formyl-2′-deoxycytidine: in vitro DNA polymerase reactions on DNA templates containing 5-formyl-2'-deoxycytidine. Nucleic Acids Res. 2001;29:2456–2463. doi: 10.1093/nar/29.12.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keith G. Mobilities of modified ribonucleotides on two-demensional cellulose thin-layer chromatography. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 50.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 51.Quigley GJ, Teeter MM, Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc. Natl Acad. Sci. USA. 1978;75:64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown RS, Dewan JC, Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985;24:4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- 53.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg(2+) in 15-year old crystals. J. Mol. Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 54.Woriax VL, Burkhart W, Spremulli LL. Cloning, sequence analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim. Biophys. Acta. 1995;1264:347–356. doi: 10.1016/0167-4781(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 55.Andersen G, Thirup S, Spremulli L, Nyborg J. High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP. J. Mol. Biol. 2000;297:421–436. doi: 10.1006/jmbi.2000.3564. [DOI] [PubMed] [Google Scholar]
- 56.Hunter W, Brown T, Anand N, Kennard O. Structure of an adenine-cytidine base pair in DNA and its implications for mismatch repair. Nature. 1986;320:552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- 57.Jang SB, Hung LW, Chi YI, Holbrook EL, Carter RJ, Holbrook SR. Structure of an RNA internal loop consisting of tandem C-A+ base pairs. Biochemistry. 1998;37:11726–11731. doi: 10.1021/bi980758j. [DOI] [PubMed] [Google Scholar]
- 58.Cai Z, Tinoco I. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 59.Legault P, Pardi A. In situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 1994;116:8390–8391. [Google Scholar]
- 60.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 61.Kawai G, Yokogawa T, Nishikawa K, Ueda T, Hashizume T, McCloskey JA, Yokoyama S, Watanabe K. Conformational properties of a novel modified nucleoside, 5-formylcytidine, found at the first position of the anticodon of bovine mitochondrial tRNAMet. Nucleosides Nucleotides. 1994;13:1189–1199. [Google Scholar]
- 62.Kawai G, Hashizume T, Yasuda M, Mayazawa T, McCloskey JA, Yokoyama S. Conformational rigidity of N4-acetyl-2′-O-metylcytidine found in tRNA of extremely thermophilic Archaebacteria (Archaea) Nucleosides Nucleotides. 1992;11:759–771. [Google Scholar]
- 63.Kashiwai T, Takemoto C, Ohtsuki T, Kawai G, Watanabe K. NMR study on non-universal codon recognition by mitochondrial tRNA possessing a modified nucleoside 5-formylcytidine. Nucleic Acids Symp. Ser. 1998;39:155–156. [Google Scholar]
- 64.Rosevear P, Mildvan A. Ligand conformations and ligand-enzyme interactions as studied by the nuclear Overhauser effect. Methods Enzymol. 1989;177:333–358. doi: 10.1016/0076-6879(89)77019-1. [DOI] [PubMed] [Google Scholar]
- 65.Clore G, Gronenborn A, Mclaughlin L. Structure of the ribonucleoside diphosphate codon UpUpC bound to tRNAPhe from yeast. J. Mol. Biol. 1984;174:163–173. doi: 10.1016/0022-2836(84)90370-x. [DOI] [PubMed] [Google Scholar]
- 66.Topal MD, Fresco JR. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976;263:289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- 67.Lim VI, Curran JF. Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA. 2001;7:942–957. doi: 10.1017/s135583820100214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy FV, Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 69.Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008;283:18801–18811. doi: 10.1074/jbc.M800233200. [DOI] [PubMed] [Google Scholar]
- 71.Sprinzl M, Hartmann T, Weber J, Blank J, Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17:r1–r171. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schüttelkopf AW, von Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.