Abstract
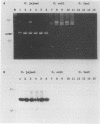
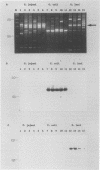
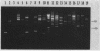
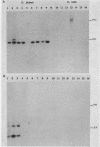
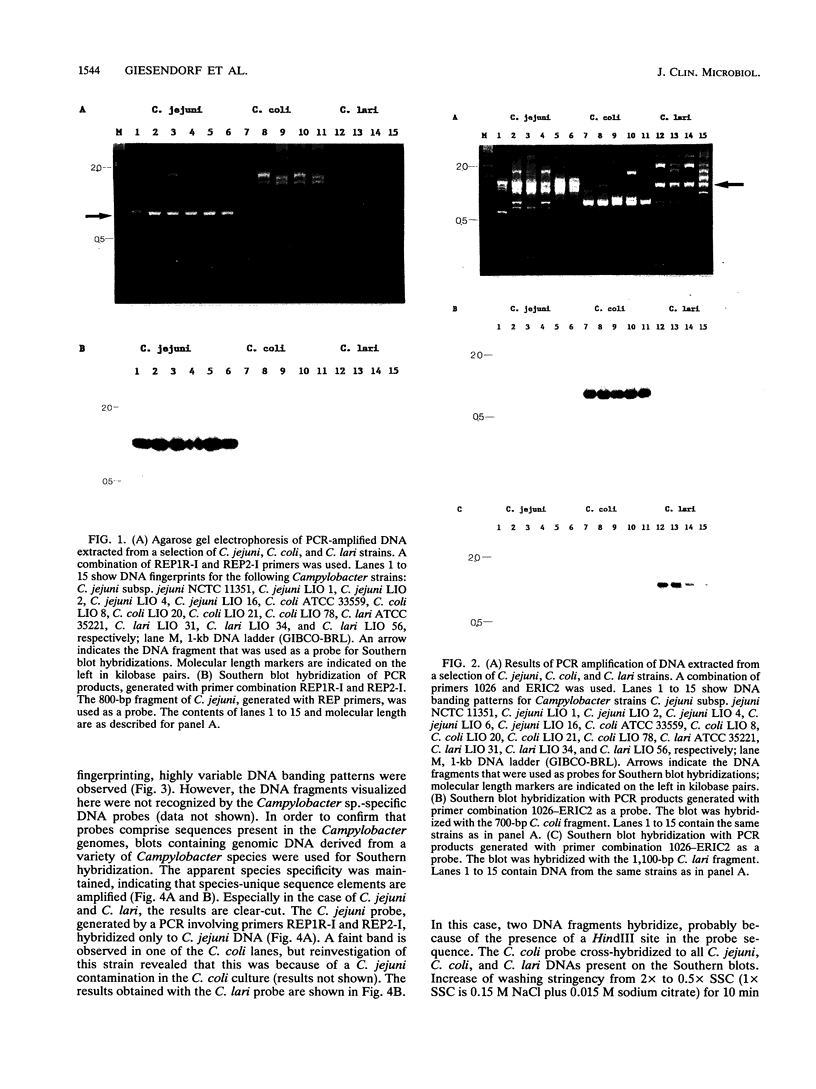
The application of polymerase chain reaction (PCR) fingerprinting assays enables discrimination between species and strains of microorganisms. PCR primers aiming at arbitrary sequences in combination with primers directed against the repetitive extragenic palindrome (REP) or enterobacterial repetitive intergenic consensus (ERIC) motifs generate isolate-specific DNA banding patterns. Analysis of these PCR fingerprints obtained for 33 isolates of Campylobacter jejuni, 30 isolates of Campylobacter coli, and 8 isolates of Campylobacter lari revealed that besides generation of isolate-specific fragments, species-specific DNA fragments of identical size were synthesized. It appeared that these DNA fragments could be used as species-specific probes, since they are unique for the pattern which they are deriving from. The probes do not cross-react with amplified DNA originating from a large panel of nonrelated microorganisms. Moreover, these probes displayed species specificity, as they reacted with a single restriction fragment on Southern blots containing DNA from C. jejuni, C. coli, and C. lari and other Campylobacter species. This combination of PCR fingerprinting and probe hybridization results in a highly specific identification assay and provides an example of specific test development without the prior need for DNA sequence information. The principle of the procedure holds great promise for the rapid isolation of DNA probes which, in combination with a general PCR assay, may lead to efficient typing and detection procedures for a multitude of medically important nonviral microorganisms.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Erlich H. A., Gelfand D., Sninsky J. J. Recent advances in the polymerase chain reaction. Science. 1991 Jun 21;252(5013):1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Giesendorf B. A., Quint W. G., Henkens M. H., Stegeman H., Huf F. A., Niesters H. G. Rapid and sensitive detection of Campylobacter spp. in chicken products by using the polymerase chain reaction. Appl Environ Microbiol. 1992 Dec;58(12):3804–3808. doi: 10.1128/aem.58.12.3804-3808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatus hernia. Lancet. 1982 Jan 16;1(8264):158–159. [PubMed] [Google Scholar]
- Jayarao B. M., Bassam B. J., Caetano-Anollés G., Gresshoff P. M., Oliver S. P. Subtyping of Streptococcus uberis by DNA amplification fingerprinting. J Clin Microbiol. 1992 May;30(5):1347–1350. doi: 10.1128/jcm.30.5.1347-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior H. New, extended biotyping scheme for Campylobacter jejuni, Campylobacter coli, and "Campylobacter laridis". J Clin Microbiol. 1984 Oct;20(4):636–640. doi: 10.1128/jcm.20.4.636-640.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior H., Woodward D. L., Edgar J. A., Laroche L. J., Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982 May;15(5):761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier S., van de Giessen A., Heuvelman K., Wernars K. RAPD analysis of Campylobacter isolates: DNA fingerprinting without the need to purify DNA. Lett Appl Microbiol. 1992 Jun;14(6):260–262. doi: 10.1111/j.1472-765x.1992.tb00700.x. [DOI] [PubMed] [Google Scholar]
- Medlin L., Elwood H. J., Stickel S., Sogin M. L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988 Nov 30;71(2):491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Penner J. L., Hennessy J. N., Congi R. V. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur J Clin Microbiol. 1983 Aug;2(4):378–383. doi: 10.1007/BF02019474. [DOI] [PubMed] [Google Scholar]
- Penner J. L. The genus Campylobacter: a decade of progress. Clin Microbiol Rev. 1988 Apr;1(2):157–172. doi: 10.1128/cmr.1.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucleic Acids Res. 1991 Oct 11;19(19):5275–5279. doi: 10.1093/nar/19.19.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C. R., Jr, Versalovic J., Koeuth T., Lupski J. R. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J Clin Microbiol. 1992 Nov;30(11):2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Bax R., Peerbooms P., Goessens W. H., van Leeuwen N., Quint W. G. Comparison of phage typing and DNA fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1993 Apr;31(4):798–803. doi: 10.1128/jcm.31.4.798-803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., De Jonckheere J., Quint W. G. Genotyping Naegleria spp. and Naegleria fowleri isolates by interrepeat polymerase chain reaction. J Clin Microbiol. 1992 Oct;30(10):2595–2598. doi: 10.1128/jcm.30.10.2595-2598.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]