Abstract
We recently reported that the intronic splice-site mutation IVS3-8G>A of CHRNA1 that encodes the muscle nicotinic acetylcholine receptor α subunit disrupts binding of a splicing repressor, hnRNP H. This, in turn, results in exclusive inclusion of the downstream exon P3A. The P3A(+) transcript encodes a non-functional α subunit that comprises 50% of the transcripts in normal human skeletal muscle, but its functional significance remains undetermined. In an effort to search for a potential therapy, we screened off-label effects of 960 bioactive chemical compounds and found that tannic acid ameliorates the aberrant splicing due to IVS3-8G>A but without altering the expression of hnRNP H. Therefore, we searched for another splicing trans-factor. We found that the polypyrimidine tract binding protein (PTB) binds close to the 3′ end of CHRNA1 intron 3, that PTB induces skipping of exon P3A and that tannic acid increases the expression of PTB in a dose-dependent manner. Deletion assays of the PTB promoter region revealed that the tannic acid-responsive element is between positions −232 and −74 from the translation initiation site. These observations open the door to the discovery of novel therapies based on PTB overexpression and to detecting possible untoward effects of the overexpression.
INTRODUCTION
Tannins are plant-derived polyphenols and are divided into two groups of hydrolyzable and condensed tannins (proanthocyanidins) (1). Hydrolyzable tannins are derivatives of gallic acid (3,4,5-trihydroxyl benzoic acid) in which a variable number of gallic acids are esterified to a core phenol. The simplest hydrolyzable tannins are gallotannins that are polygalloyl esters of glucose. The prototypical gallotannin is tannic acid or 1,2,3,4,6-penta-O-β-d-glucose (CAS 1401-55-4, C76H52O46) comprised five galloyl esters and a glucose. Commercially available tannic acid additionally includes gallotannins with a variable number of galloyl esters. Tannins are rich in nuts, red wine, tea and coffee, but hydrolyzable tannins including tannic acid are not rich in tea (2,3) or red wine (4).
Polypyrimidine tract binding protein (PTB), encoded by PTBP1, is multifunctional, which participates in pre-mRNA splicing (5,6), internal ribosome entry site (IRES)-mediated translation (7), mRNA polyadenylation (8), RNA localization (9) and mRNA stabilization (10). PTB carries four RNA recognition motifs and each motif binds to a CU-rich element (11). In terms of a splicing trans-factor, CU-rich splicing cis-elements are located upstream and/or downstream of alternatively spliced exons. PTB bound to the 3′ splice site directly competes with binding of U2AF65 that recognizes the polypyrimidine tract (12–15). Alternatively, PTBs bound to the flanking introns repress splicing of the exon (16–20).
We recently identified a pathogenic intronic mutation in a congenital myasthenic syndrome, IVS3-8G>A of CHRNA1 that encodes the muscle nicotinic acetylcholine receptor α subunit (21). We reported that IVS3-8G>A disrupts binding of a splicing repressor, hnRNP H, and causes exclusive inclusion of the downstream exon P3A. The P3A(+) transcript encodes a non-functional α subunit and comprises 50% of the transcripts in normal human skeletal muscle, but its functional significance remains unknown (22–24).
Here, we report that PTB binds to the polypyrimidine tract of CHRNA1 intron 3 immediately upstream of the hnRNP H-binding site, and silences recognition of exon P3A in cooperation with hnRNP H. IVS3-8G>A, however, has no effect on the binding of PTB. Screening of chemical compounds revealed that tannic acid mitigates exclusive inclusion of exon P3A due to IVS3-8G>A by activating the promoter region of PTBP1 and causes a dose-dependent increase of PTBP1 mRNA.
RESULTS
Screening of off-label effects of 960 chemical compounds
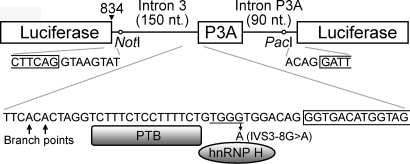
In an effort to search for a potential therapeutic modality for aberrant inclusion of CHRNA1 exon P3A due to IVS3-8G>A, we screened for off-label effects of 960 chemical compounds (GenPlus, MicroSource Discovery Systems). Most were FDA-approved drugs, and some were bioactive compounds or natural products. We constructed a chimeric minigene carrying the mutant exon P3A with its flanking introns in the middle of the firefly luciferase cDNA (pcDNA3.1-Luc-mtP3A) (Fig. 1). Skipping of exon P3A should generate luminescence, whereas its inclusion should not. After the first round of screening in duplicate, we narrowed the list to 80 compounds and performed the second round of screening in duplicate. Among the 80 compounds, 24 consistently exhibited beneficial effects (Table 1). We observed no shared feature among the 24 compounds. We hereafter focused on tannic acid, because tannic acid was expected to have less toxicity or untoward effects than the other compounds.
Figure 1.
Chimeric CHRNA1 exon P3A minigene for screening of chemical compounds. CHRNA1 exon P3A and its flanking introns are inserted after position 834 (arrowhead) of the firefly luciferase cDNA. Skipping of exon P3A should generate an intact luciferase molecule, whereas inclusion of exon P3A should abrogate it. The hnRNP H-binding ‘TGGG’ motif is underlined.
Table 1.
Twenty-four best compounds with averaged normalized relative luciferase activity ≥1.20 and CV% <0.20 (n = 4)
| Compounds | Mean | SD |
|---|---|---|
| Tomatine | 1.50 | 0.16 |
| Tannic acid | 1.47 | 0.20 |
| Troxerutin | 1.44 | 0.22 |
| Camptothecin | 1.43 | 0.12 |
| Mexiletine hydrochloride | 1.43 | 0.21 |
| Halcinonide | 1.42 | 0.07 |
| Clobetasol propionate | 1.40 | 0.21 |
| 6α-Methylprednisolone acetate | 1.40 | 0.27 |
| Flurandrenolide | 1.33 | 0.19 |
| Oxethazaine | 1.31 | 0.11 |
| Ketorolac tromethamine | 1.28 | 0.09 |
| Avocadynone acetate | 1.27 | 0.10 |
| Medrysone | 1.26 | 0.19 |
| Propafenone hydrochloride | 1.26 | 0.10 |
| Etodolac | 1.24 | 0.11 |
| Propylthiouracil | 1.23 | 0.06 |
| Pergolide mesylate | 1.23 | 0.03 |
| Hydrocortisone butyrate | 1.22 | 0.10 |
| 5-Azacytidine | 1.22 | 0.05 |
| Alexidine hydrochloride | 1.22 | 0.10 |
| Fluconazole | 1.21 | 0.10 |
| Methoxyamine hydrochloride | 1.21 | 0.09 |
| N-Formylmethionylalanine | 1.21 | 0.11 |
| Dinitolmide | 1.20 | 0.08 |
Among the 80 compounds that we employed for the second round of screening, 24 compounds consistently demonstrated beneficial responses. The values are the global mean and SD of the first (n = 2) and second (n = 2) rounds of screening.
Tannic acid induces skipping of CHRNA1 exon P3A in a dose-dependent manner
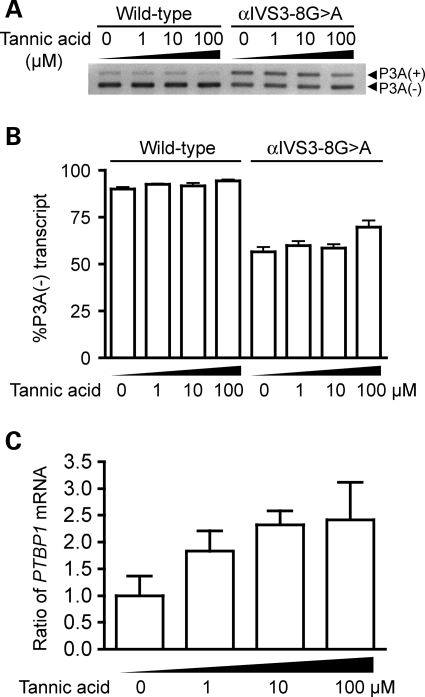
We next examined a dose-dependent effect of tannic acid. We added 0, 1, 10 and 100 µm of tannic acid to transfected HEK293T cells and examined its effect on splicing of exon P3A by real-time RT–PCR. Tannic acid exhibited no effect on splicing of wild-type exon P3A, whereas 100 µm tannic acid minimally but significantly increased the ratio of the P3A(−)-transcript (Fig. 2A and B). Lack of an effect on wild-type exon P3A may represent that the wild-type exon P3A is mostly spliced out even in the absence of tannic acid and that we cannot observe an effect of tannic acid in the wild-type construct.
Figure 2.
Tannic acid alleviates aberrant inclusion of CHRNA1 exon P3A due to IVS3-8G>A by facilitating the expression of PTBP1. (A) RT–PCR of wild-type and mutant CHRNA1 minigenes in HEK293T cells with increasing concentrations of tannic acid. (B) Real-time RT–PCR to quantify the ratio of P3A-skipped transcript arising from wild-type and mutant CHRNA1 minigenes in HEK293T cells with increasing concentrations of tannic acid. The ratios are represented by the mean and SD. For the mutant minigene, 100 µm tannic acid increases the ratio of the P3A(−)-transcript. (C) Real-time RT–PCR to demonstrate a dose-dependent increase of PTBP1 mRNA by tannic acid. The ratios are normalized to that in the absence of tannic acid. Means and SD are represented.
Tannic acid has no effect on the expression of hnRNP H
We next searched for the molecular basis of the tannic acid-mediated amelioration of the aberrant inclusion of exon P3A. We previously reported that recognition of exon P3A is down-regulated by binding of hnRNP H to ‘UGGG’ at positions −11 to −8 of intron 3 (Fig. 1), and IVS3-8G>A attenuates its bindings ∼100-fold (21). We hypothesized that tannic acid increases the expression of hnRNP H and subsequently silences the recognition of exon P3A. We therefore analyzed the effect of tannic acid on mRNA levels of hnRNP H in HEK293T cells by real-time RT–PCR, but found no effect (data not shown).
PTB binds to CHRNA1 intron 3 and silences splicing of exon P3A
We next looked for a new splicing trans-factor that regulates recognition of exon P3A. UV cross-linking of nuclear extracts of HeLa, HEK293T and C2C12 cells to wild-type CHRNA1 intron 3 disclosed a ∼60 kDa fragment (Fig. 3A). Its molecular weight prompted us to hypothesize that the bound molecule was PTB. Immunoprecipitation of the UV-cross-linked nuclear extract (Fig. 3B) and western blotting of affinity purified molecules (Fig. 3C) indeed identified that the bound molecule was PTB. Additionally, siRNA-mediated down-regulation of PTB enhanced recognition of exon P3A, whereas overexpression of PTB silenced it (Fig. 3D), which supports the notion that PTB binds to intron 3 and induces skipping of exon P3A.
Figure 3.
PTB binds close to the 3′ splice site of exon P3A and induces skipping of exon P3A. (A) 32P-labeled RNA probe is incubated with nuclear extracts of indicated cells, followed by UV cross-linking and SDS–PAGE. Autoradiographs demonstrate a ∼60-kDa molecule (arrowhead). (B) 32P-labeled RNA probe is incubated with nuclear extracts of HEK293T cells followed by UV cross-linking and RNase digestion. The bound molecules are immunoprecipitated with anti-PTB (At-PTB) or control antibody (cont Ab), and separated on SDS–PAGE. Only anti-PTB antibody precipitates a ∼60-kDa molecule. (C) Western blotting with anti-PTB or anti-SRp55 antibody of affinity purified HEK293T nuclear extract. Affinity purification also demonstrates the binding of PTB to the wild-type (WT) intron 3, but not to the scrambled RNA probe (Scr). NUC represents a lane loaded with nascent nuclear extract. (D) Real-time RT–PCR analysis of HeLa cells transfected with the wild-type minigene along with the indicated siRNA or cDNA. The %P3A values are normalized to those of control values. Down-regulation of PTBP1 enhances, whereas overexpression of PTBP1 silences the recognition of exon P3A. Bars represent the mean and SD of three experiments.
Surface plasmon resonance analysis demonstrated that IVS3-8G>A had no effect on the binding of PTB (data not shown). As we previously found that hnRNP H binds close to the 3′ end of intron 3 and silences splicing of exon P3A (21), we asked if hnRNP H and PTB interact with each other. Anti-hnRNP H antibody did not co-immunoprecipitate PTB in a nuclear extract of HEK293T cells in the absence of substrate mRNA (data not shown). To summarize, splicing of exon P3A is down-regulated by both hnRNP H and PTB, and IVS3-8G>A only compromises binding of hnRNP H but not of PTB.
Tannic acid induces the expression of PTB by activating its promoter region
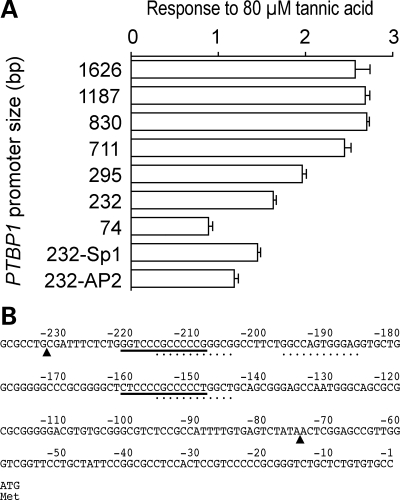
We next asked if tannic acid exerts its effect by inducing expression of PTB, and found that tannic acid increased the expression of PTBP1 mRNA in a dose-dependent manner (Fig. 2C). We further asked how PTBP1 is transcriptionally induced by tannic acid, and examined the effect of tannic acid on the PTBP1 promoter region using the luciferase reporter assay (Fig. 4). We measured responses to 80 µm of tannic acid in constructs harboring 1626, 1187, 830, 711, 295, 232 and 74 nucleotides upstream of the translation initiation site of the human PTBP1 gene. Tannic acid increases luciferase activity ∼2.5-fold when the promoter length is 711 nucleotides or longer (Fig. 4A). This degree of response is similar to that observed with the native PTBP1 gene (Fig. 2C). The 295- and 232-nucleotide constructs also respond to tannic acid but to a lesser extent. The 74-nucleotide construct exhibited no response to tannic acid. We then asked why the 232-nucleotide construct responds to tannic acid, whereas the 74-nucleotide construct does not. We deleted two putative Sp1 sites and three putative AP2 sites from the 232-nucleotide construct (Fig. 4B). Both deletion constructs minimally reduced the responses to tannic acid but not to the level of the 74-nucleotide construct. Real-time RT–PCR also revealed that 1, 10 and 100 µm of tannic acid did not increase the expression level of the SP1 gene encoding Sp1 or the TFAP2A gene encoding AP-2α (data not shown). To summarize, tannic acid-responsive cis-element likely resides between positions 232 and 74 of PTBP1, but the exact transcriptional cis-element remains unidentified.
Figure 4.
Scanning deletion analysis of the PTBP1 promoter region. (A) Luciferase promoter assays to demonstrate responses to 80 µm tannic acid. Relative luciferase activities are first calculated by dividing the firefly luciferase activity by the cotransfected Renilla luciferase activity. The calculated relative luciferase activity in the presence of 80 µm tannic acid is then divided by that in the absence of tannic acid to estimate the response. Means and SD (n = 4) are represented. The sizes of the PTBP1 promoter region before the translation initiation site are indicated. ‘232-Sp1’ and ‘232-AP2’ indicate a 232-nucleotide construct lacking two Sp1 elements [underlined in (B)] or three AP2 elements [indicated by dots in (B)], respectively. (B) PTBP1 promoter region from position –239 to +3. Arrowheads point to the 5′ ends of the 232-nucleotide and 74-nucleotide constructs in (A).
DISCUSSION
PTB silences splicing of exon P3A
We identified that PTB binds close to the 3′ end of intron 3 and silences splicing of the downstream exon P3A. In our previous studies, Syproruby staining of the affinity purified nuclear extract disclosed binding of hnRNP H but not of PTB (21). In our current studies, UV cross-linking of the nuclear extract to a radiolabeled probe detected binding of PTB but not of hnRNP H. Binding of hnRNP H and PTB, as well as their silencing effects on splicing of exon P3A, have been confirmed by several other methods for both factors. For RNA-binding molecules, we frequently observe this kind of inconsistency. This is likely because an RNA-binding molecule recognizes the tertiary structure of RNA (25,26), and the tertiary structure likely differs depending on the size, position and the attached molecule of the RNA probe employed for the binding assay.
Although both PTB and hnRNP H bind to the similar intronic region and both silence splicing of exon P3A, we failed to detect a direct interaction of the two molecules. Markovtsov et al. (27) similarly found that hnRNP H/F and PTB/nPTB cooperatively enhance binding to a splicing cis-element of the c-src N1 exon, but did not demonstrate a direct protein–protein interaction. Further analysis is required to prove if the two molecules silence splicing of exon P3A cooperatively or independently.
Off-label effects of FDA-approved drugs
FDA-approved drugs and bioactive compounds have been employed for screening of off-label effects for neurological diseases, because the identified compounds can be readily applied to clinical practice (28,29). The identified off-label compounds include digitoxin, nerifolin, peruvoside and suloctidil for spinobulbar muscular atrophy (30); β-lactam antibiotics for amyotrophic lateral sclerosis and brain ischemia (31); and dorsomorphin for fibrodysplasia ossificans progressiva (32). Additionally, kinetin alleviates aberrant splicing of IKBKAP in familial dysautonomia (33). (−)-Epigallocatechin gallate also down-regulates the expression of hnRNP A2/B1 and normalizes the aberrant splicing of mutant IKBKAP (34). Similarly, sodium valproate corrects splicing of SMN2 by increasing the expression of a splicing trans-factor, Htra2-β1, and also increases the expression of SMN2 (35,36). Sodium valproate is a histone deacetylase (HDAC) inhibitor and activates a variety of genes (37). We identified that tannic acid works on the promoter region of PTBP1 and facilitates its expression in a dose-dependent manner. Tannic acid, however, is unlikely to act directly on the promoter region of PTBP1, but rather modulates unidentified cellular processes that culminate in the activation of the PTBP1 promoter region. The exact activation mechanisms still remain to be elucidated.
Therapeutic effects of tannic acid in other diseases
Tannic acid has protein-aggregating, astringent, anti-microbial, anti-oxidant, anti-mutagenic and anti-proliferative properties (38). For example, the constringing action of tannic acid on mucous tissues is exploited to treat diarrhea in medical practice. Tannic acid inhibits the 3C-like protease encoded by the severe acute respiratory syndrome (SARS) coronavirus with IC50 of 3 µm (39). Tannic acid has anti-oxidant properties (40) and is protective against toxin-induced skin tumors (41). Tannic acid similarly suppresses skin tumor promotion induced by UV-B radiation by 70% in mice (42). Tannic acid is protective against spontaneous liver tumor development in mice by 87% (43). In addition, tannic acid raises the survival rate of mice bearing syngeneic tumors by 30% (44). Apoptosis-inducing activity of tannic acid is mediated by suppression of protease activity in tumor cells (45), reduction of ERK1, 2 phosphorylation by a cyclic adenosine 5′,3′-monophosphate-protein kinase A-dependent pathway (46) and also down-regulation of the superoxide dismutase (47). Up-regulation of PTB by tannic acid, however, has never been reported to our knowledge.
Potential clinical application of tannic acid
The LD50 of orally administered tannic acid is as high as 2260 mg/kg in rat (48). Rabbit has a higher LD50 of 5000 mg/kg (49). The human threshold, the tolerable daily intake (TDI), is extrapolated from the rodent threshold, the no observed adverse effect level (NOAEL), by dividing the NOAEL with the uncertainty factor (UF) (50). Assuming that the default UF representing the interspecies variation is 10 (51,52), the NOAELs of 2260 and 5000 mg/kg should give rise to the TDIs of 13.6 and 30.0 g of tannic acid for a 60 kg human, respectively. If these doses were completely absorbed and evenly dissolved in 60% body water, the tannic acid concentrations in body water would become 221 and 490 µm in human.
The adverse effects of tannic acid have not been thoroughly investigated in humans. Koide et al. (44) report that 8750 mg/kg/day of dietary tannic acid for 35 days reduces the weight gain in mice, but causes no pathological changes in kidney, liver or lung. This would be equivalent to 52.5 g/day for a 60 kg human according to the NOAEL approach described above. Afsana et al. (53) report that 20 g/kg/day of dietary tannic acid for 3 weeks has no effect on the body weight gain or food intake in rats, but 10 g/kg/day or more of tannic acid for 3 weeks induces anemia by reducing the iron absorption, which would be equivalent to 60 g/day for a 60 kg human according to the NOAEL approach.
Clinically available tannic acid is albumin tannate (CAS 9006-52-4) composed of 50% tannic acid and 50% albumin. In clinical practice, 1–2 g of albumin tannate is given for diarrhea which is equivalent to 0.5–1 g of tannic acid. If these doses were completely absorbed and evenly dissolved in 60% body water in a 60 kg human, the tannic acid concentrations in body water would become 8–16 µm. Our studies using HEK293T cells demonstrate that even 1 µm of tannic acid up-regulates the expression of PTBP1 but 100 µm of tannic acid is required to attenuate the aberrant splicing of exon P3A. As the absorption efficiency and elimination rate of dietary tannic acid has not been determined in humans, we cannot predict how much tannic acid would be required to treat patients, and whether it is toxic or not.
Diseases and physiological states in which the up-regulation of PTB is beneficial or detrimental
Raponi et al. (54) recently reported that PTB recognizes two splicing cis-elements in the pseudoexon of NF1 activated by an A-to-G mutation 270 nucleotides upstream of the nascent intron/exon boundary. Up-regulation of PTB ameliorated the aberrant inclusion of the pseudoexon. Fred and Welsh (10) found that PTB stabilizes the preproinsulin mRNA by binding to its 3′-UTR and participate in the glucose-induced increase of preproinsulin mRNA. In both of these conditions, up-regulation of PTB by tannic acid might be beneficial.
On the other hand, PTB is overexpressed in tumor cells and promotes cell growth (55,56), although PTB itself does not transform cells (57). Viruses exploit PTB to facilitate the IRES-mediated translation initiation (58,59). PTB also silences the splicing of a cassette exon of the neuronal PTB (nPTB) and down-regulates the expression of nPTB in non-neuronal cells. In neuronal cells, microRNA miR-124 down-regulates the translation of PTB and hence up-regulates the expression of nPTB (60). In these conditions, up-regulation of PTB by tannic acid could to be detrimental.
Although tannic acid in the form of albumin tannate has long been used as a safe anti-diarrhea medication, our observations that tannic acid causes overexpression of PTB now opens the door to the discovery of beneficial as well as possible detrimental effects of PTB overexpression.
MATERIALS AND METHODS
Firefly luciferase minigene carrying chimeric CHRNA1 exon P3A
We screened for off-label effects of 960 chemical compounds using chimeric minigenes carrying the firefly luciferase cDNA (Fig. 1). We first inserted the luciferase gene derived from the pGL3-Basic vector (Promega) into the CMV-based mammalian expression vector, pcDNA3.1 (Invitrogen), to make pcDNA3.1-Luc. Using the megaprimer-based site-directed mutagenesis method (61), we inserted a chimeric intron of 5′-GTAAGTATCAAGCGGCCGCNNNNNTTAATTAATCTTACTGACATCCACTTTGCCTTTCTCTCCACAG-3′ at position 834 of the luciferase cDNA, where position +1 represents the translation initiation site. The underlined are NotI and PacI restriction sites. We next inserted CHRNA1 exon P3A with its flanking introns of 150 and 90 nucleotides using the NotI and PacI sites to make pcDNA3.1-Luc-wtP3A. With this construct, the nonsense-mediated mRNA decay is not provoked even when exon P3A is spliced in. We then engineered IVS3-8G>A using the QuikChange site-directed mutagenesis kit (Stratagene), and made pcDNA3.1-Luc-mtP3A.
For all clones, the presence of the desired mutation and the absence of artifacts was determined by sequencing the entire insert with the CEQ8000 DNA analysis system (Beckman Coulter).
Screening of off-label effects of 960 chemical compounds
HEK293T cells were maintained in the Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (Sigma-Aldrich). At ∼50% confluency in a 75 cm2 flask, we transfected 6 µg of either pcDNA3.1-Luc or pcDNA3.1-Luc-mtP3A along with 2 µg of phRL-SV40 encoding the Renilla luciferase (Promega) using 24 µl of the FuGENE6 transfection reagent (Roche). Sixteen hours after transfection, we removed the transfection reagent and split the cells into three 96-well plates containing 10 µm of each chemical compound (GenPlus) and 2% DMSO. After growing the cells for 24 h, we measured the firefly and Renilla luciferase activities with the Dual luciferase reporter assay system (Promega) using the Luminoskan Ascent luminometer (Thermo Scientific).
We first assayed effects of each compound on the transcription activity and on stabilities of the luciferase mRNA and protein using pcDNA3.1-Luc that codes for the nascent firefly luciferase and harbors no chimeric exon. We normalized the firefly luciferase activities with the cotransfected Renilla luciferase activities. We repeated the experiments five times for each compound. The coefficients of variation (CV%) of five assays were 9.4 ± 4.0% (mean ± SD) for all the 960 compounds. We next performed duplicate assays to determine the effect of each compound on splicing of CHRNA1 exon P3A harboring IVS3-8G>A (pcDNA3.1-Luc-mtP3A) by measuring the relative luciferase activities in the presence of each compound. To estimate the effect of each compound on splicing of exon P3A, the relative luciferase activity of pcDNA3.1-Luc-mtP3A was normalized by dividing it by that of pcDNA3.1-Luc. The mean and SD of the normalized relative luciferase activities of 960 compounds was 1.05 ± 0.20, whereas the mean and SD of 192 negative controls without any compound was 1.00 ± 0.11. Eighty most effective compounds with normalized relative luciferase activities of 1.21 or higher (mean and SD, 1.36 ± 0.40) were subjected to a second round of screening in duplicate. The normalized relative luciferase activities of the 80 compounds in the second round of screening ranged from 0.63 to 1.92 with the mean and SD of 1.16 ± 0.25.
Analysis of effects of tannic acid on splicing of minigene carrying CHRNA1 exon P3A
In order to analyze the effect of tannic acid on splicing of exon P3A, we employed the previously reported pSPL3-wtP3A harboring a 288-bp genomic fragment spanning IVS3-123 to IVS3A+90 of CHRNA1 (21). We engineered IVS3-8G>A to make pSPL3-mtP3A. Transfection into HEK293T cells, RNA isolation and RT–PCR were performed as described (21). We also quantified the fraction of the P3A(−)-transcript by estimating the absolute copy numbers of P3A(+)- and P3A(−)-transcripts by real-time RT–PCR using Mx3000P (Stratagene) as previously described (21). Each real-time RT–PCR experiment was performed in triplicate. Patient cells were unavailable for the analysis of effects of tannic acid.
In vitro UV cross-linking and immunoprecipitation to isolate an RNA-binding protein
We synthesized [α-32P]-CTP labeled RNA using the Riboprobe System (Promega) from a PCR-amplified fragment according to the manufacturer’s instructions. The forward primer was 5′-TAATACGACTCACTATAGGGAGACAGG-3′, where the italicized is T7 promoter and the underlined is for annealing to the reverse primer. The reverse primer was 5′-CATGTCACCCTGTCCACCCACAGAAAAGGAGCCTGTCTC-3′. Nuclear extracts were prepared as previously described (62). The radioactively labeled RNA (1 × 104 cpm) was incubated at room temperature with 2 µl of nuclear extracts, 8 µg of yeast tRNA and 0.8 U of RNasin (Toyobo) in a final volume of 10 µl of the binding buffer (20 mm HEPES pH 7.8, 50 mm KCl, 3 mm MgCl2, 0.5 mm dithiothreitol, 0.5 mm EDTA and 10% glycerol). After 20 min, samples were exposed to UV light (254 nm) for 10 min, digested with 50 U of RNase T1, and subjected to SDS–PAGE (10% gel) and autoradiography. Immunoprecipitations were carried out under the same conditions but in 40 µl. After RNase T1 treatment, we added 360 µl of IP buffer (0.1% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris–HCl pH 8.1, and 167 mm NaCl) and 80 µl protein G-Sepharose (GE Healthcare) precoated with yeast tRNA. After eliminating the protein G-Sepharose, we incubated the mixture at 4°C for 30 min and added 5 mg of anti-PTB antibody (ZYMED Laboratory) or isotype-matched control antibody to the supernatant. Samples were mixed overnight at 4°C before the addition of 60 µl protein G-Sepharose. We mixed the sample for additional 30 min at 4°C. After the beads were washed five times with the IP buffer, bound proteins were eluted and subjected to SDS–PAGE (10% gel) and autoradiography.
Surface plasmon resonance analysis
We measured the binding affinities of PTB for the wild-type and mutant CHRNA1 ISS motifs by surface plasmon resonance using Biacore 3000 (GE healthcare). The synthesized RNA probe were 5′-UUUCUCCUUUUCUGUGGRUGGACAGGGUGACAUGGUA-3′ with a biotin tag at its 5′ end (Hokkaido System Science). The underlined R was G for the wild-type probe and A for the mutant probe carrying IVS3-8G>A. The probe was comprised 25 nucleotides in intron 3 and 12 nucleotides in exon P3. The probe was immobilized onto a streptavidin-coated sensor chip (GE healthcare) at a concentration of 200 resonance units by injecting 0.1 pmol/µl of the RNA probe in HBS-EP (0.01 m HEPES pH 7.4, 0.15 m NaCl, 3 mm EDTA and 0.005% P20).
The human PTBP1 cDNA was cloned into a pGEX6P1 vector (GE Healthcare) to make a GST fusion protein in bacteria. A fresh colony of Escherichia coli BL21 transformed with pGEX6P1-PTBP1 was grown in 5 ml of LB medium containing ampicillin at 37°C for 12 h. The culture was transferred to 200 ml of the LB plus ampicillin broth, and grown to OD600 of 0.1 at 37°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mm, and the culture was grown for another hour. The bacteria were harvested, resuspended in 10 ml of PBS and lysed by sonication. Bacterial debris was precipitated by centrifugation at 10 000g. We applied the supernatant onto a glutathione-Sepharose 4B column (GE Healthcare), washed the beads with PBS and eluted the bound protein with 10 mm glutathione.
Co-immunoprecipitation (Co-IP) to examine the binding of PTB and hnRNP H
We employed the Nuclear Complex Co-IP kit (Active Motif). We incubated 250 µg of a nuclear protein extract of HEK293T cells with 3 µg of anti-hnRNP F/H antibody (Santa Cruz) or isotype-matched control antibody in 500 µl of a low salt IP buffer (Active Motif) overnight at 4°C. We added 60 µl of protein G beads (GE Healthcare) and incubated for an additional hour on a rotary shaker. Beads were then washed six times with the low salt IP buffer. The immunoprecipitated proteins were dissolved in 20 µl of 1× SDS sample buffer, boiled for 5 min and analyzed by western blotting using anti-PTB antibody.
Real-time RT–PCR to quantify PTBP1 mRNA
For quantification of the expression level of PTBP1 mRNA in HEK293T cells, we employed Hs00259176_m1 for human PTBP1 (TaqMan Gene Expression Assays, Applied Biosystems) with the Premix Ex Taq (Takara Bio) in Mx3000P. We also measured the expression level of GAPDH with a pair of primers, 5′-GTCAAGGCTGAGAACGGGAA-3′ and 5′-GTGAAGACGCCAGTGGACTC-3′, using the SYBR Premix Ex Taq (Takara Bio). We then normalized the expression level of PTBP1 by that of GAPDH. Each experiment was performed in triplicate.
Gene silencing and overexpression of PTBP1 mRNA
For down-regulation of PTBP1 mRNA, we synthesized siRNA of 5′-GCCUCUUUAUUCUUUUCGGdTdT-3′. As a control, we employed the AllStar Negative Control siRNA 1027281 (Qiagen). HeLa cells were plated 24 h before transfection on 12-well plates. The transfection reagent included 300 ng of the minigene, 50 pmol of siRNA and 1 µl of Lipofectamine 2000 (Invitrogen) in 100 µl DMEM.
For overexpression of PTBP1 mRNA, we purchased an IMAGE clone 3863892 encoding the transcript variant 1 of human PTBP1 in a pCMV-SPORT6 vector from Invitrogen. As a negative control, we employed a CM-based pcDNA3.1(+) (Invitrogen). HeLa cells plated on 12-well plates as above was transfected with 100 ng of the minigene, 400 ng of a CMV-based vector and 1 µl of FuGENE6 (Roche) in 100 µl DMEM.
PTBP1 promoter assays
We introduced variable sizes of the human PTBP1 promoter region immediately upstream of the translational start site into pGL3-Basic vector encoding the firefly luciferase gene (Promega). To delete three AP2 sites and two Sp1 sites from the PTBP1 promoter region, we performed three and two rounds of the QuikChange site-directed mutagenesis kit, respectively. We transfected HEK293T cells in a 24-well plate with 0.3 µg of the pGL3 reporter vector and 0.1 µg of phRL-TK vector (Promega) using FuGENE6. Sixteen hours after transfection, we removed the transfection medium and added 0–80 µm of tannic acid (US Pharmacopeia and MicroSource Discovery Systems). We harvested cells 24 more hours and measured the firefly and Renilla luciferase activities. We performed all assays three or more times, and measured the luciferase activities in duplicate.
FUNDING
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology as well as the Ministry of Health, Labor, and Welfare of Japan, and by the National Institutes of Health grant NS6277 as well as by a Muscular Dystrophy Association research grant.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Keiko Itano for her technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hagerman A.E. 2002 Tannin Chemistry http://www.users.muohio.edu/hagermae/
- 2.Hamilton-Miller J.M. Antimicrobial properties of tea (Camellia sinensis L.) Antimicrob. Agents Chemother. 1995;39:2375–2377. doi: 10.1128/aac.39.11.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yam T.S., Hamilton-Miller J.M., Shah S. The effect of a component of tea (Camellia sinensis) on methicillin resistance, PBP2’ synthesis, and beta-lactamase production in Staphylococcus aureus. J. Antimicrob. Chemother. 1998;42:211–216. doi: 10.1093/jac/42.2.211. [DOI] [PubMed] [Google Scholar]
- 4.Cozzolino D., Cynkar W.U., Dambergs R.G., Mercurio M.D., Smith P.A. Measurement of condensed tannins and dry matter in red grape homogenates using near infrared spectroscopy and partial least squares. J. Agric. Food Chem. 2008;56:7631–7636. doi: 10.1021/jf801563z. [DOI] [PubMed] [Google Scholar]
- 5.Wagner E.J., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spellman R., Smith C.W. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y.K., Hahm B., Jang S.K. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J. Mol. Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- 8.Moreira A., Takagaki Y., Brackenridge S., Wollerton M., Manley J.L., Proudfoot N.J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote C.A., Gautreau D., Denegre J.M., Kress T.L., Terry N.A., Mowry K.L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 10.Fred R.G., Welsh N. The importance of RNA binding proteins in preproinsulin mRNA stability. Mol. Cell. Endocrinol. 2008 doi: 10.1016/j.mce.2008.06.007. [Epub (doi:10.1016/j.mce.2008.06.007)] [DOI] [PubMed] [Google Scholar]
- 11.Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 12.Lin C.H., Patton J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 13.Singh R., Valcarcel J., Green M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 14.Ashiya M., Grabowski P.J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 15.Sauliere J., Sureau A., Expert-Bezancon A., Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir-Ahmady B., Boutz P.L., Markovtsov V., Phillips M.L., Black D.L. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan R.C., Black D.L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlet B.N., Logan P., Singh G., Cooper T.A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 19.Southby J., Gooding C., Smith C.W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutually exclusive exons. Mol. Cell. Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma S., Kohlstaedt L.A., Damianov A., Rio D.C., Black D.L. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda A., Shen X.-M., Ito M., Matsuura T., Engel A.G., Ohno K. hnRNP H enhances skipping of a nonfunctional exon P3A in CHRNA1 and a mutation disrupting its binding causes congenital myasthenic syndrome. Hum. Mol. Genet. 2008;17:4022–4035. doi: 10.1093/hmg/ddn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beeson D., Morris A., Vincent A., Newsom-Davis J. The human muscle nicotinic acetylcholine receptor alpha-subunit exist as two isoforms: a novel exon. EMBO J. 1990;9:2101–2106. doi: 10.1002/j.1460-2075.1990.tb07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newland C.F., Beeson D., Vincent A., Newsom-Davis J. Functional and non-functional isoforms of the human muscle acetylcholine receptor. J. Physiol. 1995;489:767–778. doi: 10.1113/jphysiol.1995.sp021090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krull M., Petrusma M., Makalowski W., Brosius J., Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–1145. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielkopf C.L., Lucke S., Green M.R. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clery A., Blatter M., Allain F.H. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Markovtsov V., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heemskerk J., Tobin A.J., Ravina B. From chemical to drug: neurodegeneration drug screening and the ethics of clinical trials. Nat. Neurosci. 2002;5(suppl.):1027–1029. doi: 10.1038/nn931. [DOI] [PubMed] [Google Scholar]
- 29.Abbott A. Neurologists strike gold in drug screen effort. Nature. 2002;417:109. doi: 10.1038/417109a. [DOI] [PubMed] [Google Scholar]
- 30.Piccioni F., Roman B.R., Fischbeck K.H., Taylor J.P. A screen for drugs that protect against the cytotoxicity of polyglutamine-expanded androgen receptor. Hum. Mol. Genet. 2004;13:437–446. doi: 10.1093/hmg/ddh045. [DOI] [PubMed] [Google Scholar]
- 31.Rothstein J.D., Patel S., Regan M.R., Haenggeli C., Huang Y.H., Bergles D.E., Jin L., Dykes Hoberg M., Vidensky S., Chung D.S., et al. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- 32.Yu P.B., Hong C.C., Sachidanandan C., Babitt J.L., Deng D.Y., Hoyng S.A., Lin H.Y., Bloch K.D., Peterson R.T. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slaugenhaupt S.A., Mull J., Leyne M., Cuajungco M.P., Gill S.P., Hims M.M., Quintero F., Axelrod F.B., Gusella J.F. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004;13:429–436. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 34.Anderson S.L., Qiu J., Rubin B.Y. EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem. Biophys. Res. Commun. 2003;310:627–633. doi: 10.1016/j.bbrc.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Brichta L., Hofmann Y., Hahnen E., Siebzehnrubl F.A., Raschke H., Blumcke I., Eyupoglu I.Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 36.Brichta L., Holker I., Haug K., Klockgether T., Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann. Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 37.Gottlicher M., Minucci S., Zhu P., Kramer O.H., Schimpf A., Giavara S., Sleeman J.P., Lo Coco F., Nervi C., Pelicci P.G., et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung K.T., Wong T.Y., Wei C.I., Huang Y.W., Lin Y. Tannins and human health: a review. Crit. Rev. Food Sci. Nutr. 1998;38:421–464. doi: 10.1080/10408699891274273. [DOI] [PubMed] [Google Scholar]
- 39.Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3′-digallate (TF3) Evid. Based Complement. Alternat. Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sroka Z., Rzadkowska-Bodalska H., Mazol I. Antioxidative effect of extracts from Erodium cicutarium L. Z. Naturforsch. [C] 1994;49:881–884. [PubMed] [Google Scholar]
- 41.Gali H.U., Perchellet E.M., Klish D.S., Johnson J.M., Perchellet J.P. Hydrolyzable tannins: potent inhibitors of hydroperoxide production and tumor promotion in mouse skin treated with 12-O-tetradecanoylphorbol-13-acetate in vivo. Int. J. Cancer. 1992;51:425–432. doi: 10.1002/ijc.2910510315. [DOI] [PubMed] [Google Scholar]
- 42.Gali-Muhtasib H.U., Yamout S.Z., Sidani M.M. Tannins protect against skin tumor promotion induced by ultraviolet-B radiation in hairless mice. Nutr. Cancer. 2000;37:73–77. doi: 10.1207/S15327914NC3701_9. [DOI] [PubMed] [Google Scholar]
- 43.Nepka C., Sivridis E., Antonoglou O., Kortsaris A., Georgellis A., Taitzoglou I., Hytiroglou P., Papadimitriou C., Zintzaras I., Kouretas D. Chemopreventive activity of very low dose dietary tannic acid administration in hepatoma bearing C3H male mice. Cancer Lett. 1999;141:57–62. doi: 10.1016/s0304-3835(99)00145-7. [DOI] [PubMed] [Google Scholar]
- 44.Koide T., Kamei H., Hashimoto Y., Kojima T., Hasegawa M. Tannic acid raises survival rate of mice bearing syngeneic tumors. Cancer Biother. Radiopharm. 1999;14:231–234. doi: 10.1089/cbr.1999.14.231. [DOI] [PubMed] [Google Scholar]
- 45.Nam S., Smith D.M., Dou Q.P. Tannic acid potently inhibits tumor cell proteasome activity, increases p27 and Bax expression, and induces G1 arrest and apoptosis. Cancer Epidemiol. Biomarkers Prev. 2001;10:1083–1088. [PubMed] [Google Scholar]
- 46.Taffetani S., Ueno Y., Meng F., Venter J., Francis H., Glaser S., Alpini G., Patel T. Tannic acid inhibits cholangiocyte proliferation after bile duct ligation via a cyclic adenosine 5′,3′-monophosphate-dependent pathway. Am. J. Pathol. 2005;166:1671–1679. doi: 10.1016/S0002-9440(10)62477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen K.S., Hsiao Y.C., Kuo D.Y., Chou M.C., Chu S.C., Hsieh Y.S., Lin T.H. Tannic acid-induced apoptosis and -enhanced sensitivity to arsenic trioxide in human leukemia HL-60 cells. Leuk. Res. 2008 doi: 10.1016/j.leukres.2008.08.006. [Epub (doi:10.1016/j.leukres.2008.08.006)] [DOI] [PubMed] [Google Scholar]
- 48.Boyd E.M., Bereczky K., Godi I. The acute toxicity of tannic acid administered intragastrically. Can. Med. Assoc. J. 1965;92:1292–1297. [PMC free article] [PubMed] [Google Scholar]
- 49.Dollahite J.W., Pigeon R.F., Camp B.J. The toxicity of gallic acid, pyrogallol, tannic acid, and Quercus havardi in the rabbit. Am. J. Vet. Res. 1962;23:1264–1267. [PubMed] [Google Scholar]
- 50.Barnes D.G., Dourson M. Reference dose (RfD): description and use in health risk assessments. Regul. Toxicol. Pharmacol. 1988;8:471–486. doi: 10.1016/0273-2300(88)90047-5. [DOI] [PubMed] [Google Scholar]
- 51.Dourson M.L., Felter S.P., Robinson D. Evolution of science-based uncertainty factors in noncancer risk assessment. Regul. Toxicol. Pharmacol. 1996;24:108–120. doi: 10.1006/rtph.1996.0116. [DOI] [PubMed] [Google Scholar]
- 52.Foronda N.M., Fowles J., Smith N., Taylor M., Temple W., Darlington C. The use of myocardial and testicular end points as a basis for estimating a proposed tolerable daily intake for sodium monofluoroacetate (1080) Regul. Toxicol. Pharmacol. 2007;47:29–36. doi: 10.1016/j.yrtph.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Afsana K., Shiga K., Ishizuka S., Hara H. Reducing effect of ingesting tannic acid on the absorption of iron, but not of zinc, copper and manganese by rats. Biosci. Biotechnol. Biochem. 2004;68:584–592. doi: 10.1271/bbb.68.584. [DOI] [PubMed] [Google Scholar]
- 54.Raponi M., Buratti E., Llorian M., Stuani C., Smith C.W., Baralle D. Polypyrimidine tract binding protein regulates alternative splicing of an aberrant pseudoexon in NF1. FEBS J. 2008;275:6101–6108. doi: 10.1111/j.1742-4658.2008.06734.x. [DOI] [PubMed] [Google Scholar]
- 55.Venables J.P. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 56.He X., Pool M., Darcy K.M., Lim S.B., Auersperg N., Coon J.S., Beck W.T. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Norton J.T., Ghosh S., Kim J.J., Fushimi K., Wu J.Y., Stack M.S., Huang S. PTB differentially affects malignancy in a cell line dependent manner. J. Biol. Chem. 2008;283:20277–20287. doi: 10.1074/jbc.M803682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anwar A., Ali N., Tanveer R., Siddiqui A. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 2000;275:34231–34235. doi: 10.1074/jbc.M006343200. [DOI] [PubMed] [Google Scholar]
- 59.Song Y., Tzima E., Ochs K., Bassili G., Trusheim H., Linder M., Preissner K.T., Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohno K., Anlar B., Özdirim E., Brengman J.M., DeBleecker J.L., Engel A.G. Myasthenic syndromes in Turkish kinships due to mutations in the acetylcholine receptor. Ann. Neurol. 1998;44:234–241. doi: 10.1002/ana.410440214. [DOI] [PubMed] [Google Scholar]
- 62.Masuda A., Yoshikai Y., Kume H., Matsuguchi T. The interaction between GATA proteins and activator protein-1 promotes the transcription of IL-13 in mast cells. J. Immunol. 2004;173:5564–5573. doi: 10.4049/jimmunol.173.9.5564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.