Abstract
Proximal spinal muscular atrophy (SMA) is a neuromuscular disorder for which there is no available therapy. SMA is caused by loss or mutation of the survival motor neuron 1 gene, SMN1, with retention of a nearly identical copy gene, SMN2. In contrast to SMN1, most SMN2 transcripts lack exon 7. This alternatively spliced transcript, Δ7-SMN, encodes a truncated protein that is rapidly degraded. Inhibiting this degradation may be of therapeutic value for the treatment of SMA. Recently aminoglycosides, which decrease translational fidelity to promote readthrough of termination codons, were shown to increase SMN levels in patient cell lines. Amid uncertainty concerning the role of SMN's C-terminus, the potential of translational readthrough as a therapeutic mechanism for SMA is unclear. Here, we used stable cell lines to demonstrate the SMN C-terminus modulates protein stability in a sequence-independent manner that is reproducible by translational readthrough. Geneticin (G418) was then identified as a potent inducer of the Δ7-SMN target sequence in vitro through a novel quantitative assay amenable to high throughput screens. Subsequent treatment of patient cell lines demonstrated that G418 increases SMN levels and is a potential lead compound. Furthermore, treatment of SMA mice with G418 increased both SMN protein and mouse motor function. Chronic administration, however, was associated with toxicity that may have prevented the detection of a survival benefit. Collectively, these results substantiate a sequence independent role of SMN's C-terminus in protein stability and provide the first in vivo evidence supporting translational readthrough as a therapeutic strategy for the treatment of SMA.
INTRODUCTION
Proximal spinal muscular atrophy (SMA) is a neuromuscular disease and the leading heritable cause of infant mortality (1). It is linked at the molecular level to the survival motor neuron (SMN) genes. Humans contain two nearly identical copies of the SMN gene, SMN1 and SMN2, due to a recent inverted duplication at the 5q13 locus (2). However, only mutations in SMN1 are responsible for SMA (3,4). The critical difference between SMN1 and SMN2 is a silent, single-nucleotide transition within exon 7 that disrupts an exonic splicing enhancer in SMN2 (5–7). This results in production of small amounts of full-length transcripts from SMN2 and high levels of a differentially spliced form of SMN2 that lacks exon 7 (Δ7-SMN). The resulting transcripts are equivalent in stability (8). However, the Δ7-SMN protein is unstable and cannot oligomerize or self-associate as well as full-length protein (FL-SMN) (9,10). Consequently, the small amount of functional protein produced from SMN2 is not able to fully compensate for the loss of SMN1. Family studies and mouse models show that SMN2 enables survival and modifies disease severity (3,11–14). Thus, SMA is essentially a disorder that results from insufficient SMN dosage.
SMN2 is an ideal therapeutic target since it is capable of producing functional protein and it is ubiquitously present in the SMA patient population. Current approaches to elevate protein expression from this gene focus on upregulating transcription, altering splicing, increasing translation or delaying protein degradation [reviewed in (15)]. To date, there has been minimal investigation into the modulation of SMN stability as a therapeutic strategy. The disagreement concerning the molecular role of SMN exon 7 in protein function may be partially responsible for this. Some findings indicate that it plays a sequence independent role in protein function (16–18). Evidence supporting this includes a lack of exon 7 sequence conservation between species, an ability of non-specific sequences to restore Δ7-SMN localization and, most convincingly, that Δ7-SMN itself is capable of extending survival (19) and improving pathology of SMA model mice in vivo (16,17). It is hypothesized that Δ7-SMN produces this phenotypic improvement either through partial functionality or by ‘seeding’ oligomerization with and thereby increasing the amount of functional FL-SMN. Contrary to a sequence-independent role, some data suggest that exon 7 encodes a novel function. Such evidence includes the presence of two SMA point mutations (G279V and G279C) (20,21), and data suggesting exon 7 is important for axonal localization of specific mRNAs, loss of which leads to neuron-specific cell death (22–24). However, the SMA-essential function of SMN is still unclear, and the G279V point mutation encodes a protein less functional in oligomerization than even Δ7-SMN protein (10), suggesting such point mutations create a critical sequence-specific context that has a larger effect on SMN protein than the entire removal of exon 7 from SMN2 transcripts. Regardless of exon 7's function, the ability of elevated Δ7-SMN expression to extend survival of SMA model mice demonstrates that modulation of Δ7-SMN protein or its stability may be a viable therapeutic strategy for SMA.
Recently, the aminoglycoside antibiotics amikacin and tobramycin were shown to increase SMN within SMA patient fibroblasts (18). Aminoglycosides are an FDA-approved class of drug that acts within cells by binding to ribosomes to effect the translation of proteins from mRNA transcripts (25). Specific aminoglycosides differ in their effects on translation, some act on specific four base sequences (a stop codon plus the base immediately following it, i.e. UAG A) and cause ribosomes to misread stop codons (26). Instead of stopping, the ribosome will insert a residue and continue reading to translate sequences previously held silent by the stop codon. This mechanism, referred to as translational ‘readthrough’ or ‘termination suppression’, has attracted great interest due to its potential application to any genetic disease caused by nonsense mutations, such as Duchenne muscular dystrophy, cystic fibrosis and Hurler syndrome (26–33). This mechanism is especially intriguing for SMA because it would act on a monomorphic target that is present in all SMA patients: the Δ7-SMN stop codon sequence (UAG A).
Here, we demonstrate that SMN exon 7 plays a sequence-independent role in protein stability and that its absence can be partially compensated for by translational readthrough. We produce a novel assay to detect and quantify the induction of readthrough and, using this assay, we identify G418 as a compound acting upon the Δ7-SMN stop target sequence. This drug has previously been shown to be a strong inducer of readthrough at some disease-relevant stop codons and has been observed to produce a functional benefit in mouse models of genetic disease in vivo (34). Treatment with G418 resulted in the induction of SMN both in vitro and in vivo. Additionally, G418-treated SMA mice demonstrated a significant increase in motor function when compared with vehicle-treated SMA mice. Together, this work provides further support that SMN exon 7 acts as a structural tag important for SMN protein stability and provides the first in vivo evidence supporting the mechanism of translational readthrough as an SMA therapy.
RESULTS
Induction of SMN by drug treatment of patient fibroblasts
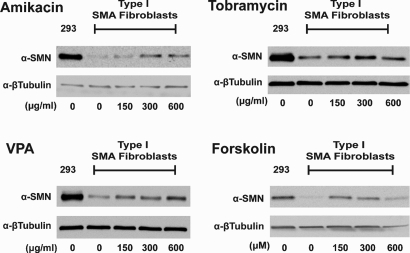
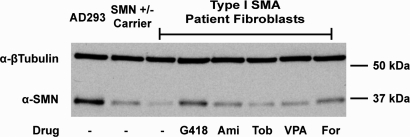
Initially, we compared and validated the effects of drugs reported to elevate SMN through distinct mechanisms in vitro. For this, primary cell lines derived from Type I SMA patients were treated with increasing doses of either amikacin, tobramycin, valproic acid (VPA) or forskolin. As aminoglycosides, amikacin and tobramycin were postulated to work by inducing translational readthrough (18). The histone deacetylase (HDAC) inhibitor VPA and the CRE-inducer forskolin were included as drugs that elevate SMN expression through effects on SMN2 transcription and/or splicing, respectively (35–37). Western blot analysis of treated cell lysates confirmed that all four of these drugs elevate SMN protein in patient fibroblasts (Fig. 1). Amikacin, tobramycin and VPA were found to increase SMN in a dose-dependent manner at concentrations of 150, 300 and 600 µg/ml. Forskolin treatment elevated SMN expression at concentrations of 20 and 40 µm but not at 80 µm. These data are in agreement with previously published reports on the ability of these drugs to increase SMN (18,35–37).
Figure 1.
Treatment of patient fibroblasts with SMN inducing drugs. Western blots of Type I SMA patient fibroblasts treated with various drugs working through discrete mechanisms. GM09677 cells containing 2 copies of the SMN2 gene and GM00232 cells containing one were treated with either amikacin, tobramycin, valproic acid (VPA) or forskolin for 48 h at the indicated concentration. Representative blots of treated GM09677 cells are shown. Amikacin and tobramycin are aminoglycosides, VPA is an HDAC inhibitor, and forskolin activates gene expression through cAMP response elements.
SMN exon 7 modulates protein stability in a sequence-independent manner
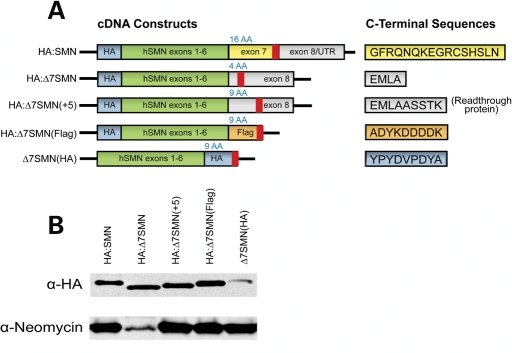
To determine if SMN exon 7 has sequence-independent effects, we generated a series of constructs that contained SMN isoforms with differing C-termini. These plasmids encoded either FL-SMN (HA:SMN), Δ7-SMN (HA:Δ7SMN), the protein that would be produced by translational readthrough of the Δ7-SMN mRNA target (HA:Δ7SMN[+5]) and hereafter referred to as ‘readthrough protein’, or proteins with generic C-termini that consist of either nine amino acid Flag (HA:Δ7SMN[Flag]) or HA (Δ7SMN[HA]) ectopic tags (Fig. 2A). Each of these isoforms also possess an HA tag to differentiate them from endogenous SMN protein. After confirming expression in transient transfection assays (data not shown), stable expressing cell lines of each construct were generated in AD293 cells (Fig. 2B). This allowed us to examine protein properties and eliminate artifacts of transient overexpression.
Figure 2.
Schematics and expression of SMN constructs with varying C-termini. (A) Schematic of constructs used to assay properties of SMN proteins with different C-termini. SMN exons 1–6 cDNA was fused to sequences encoding either SMN exon 7 or 8, or novel ectopic tags. HA tags were included in all constructs to differentiate them from endogenous SMN. The translational stop codon in each construct is denoted by a bold red line, whereas the number of C-terminal amino acids encoded by each construct is provided above in blue. Amino acid sequences of C-terminal peptides are provided to the right. (B) Western blot of stable cell lines verifies expression of each SMN construct and the drug-resistance marker.
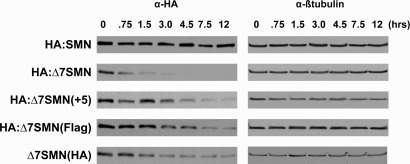
The role of SMN's C-terminus in protein stability was evaluated by treating the above-mentioned stable cell lines with the translation inhibitor cycloheximide. At 0, 0.75, 1.5, 3, 4.5, 7.5 and 12 h post-treatment, cells were lysed and qualitatively analyzed by western blot analysis to determine whether SMN protein was still present (Fig. 3). Data from this experiment revealed several new pieces of information. First, prior studies of FL-SMN and Δ7-SMN stability performed in transient transfection assays were extended from only 3 h to a final timepoint of 12 h (9). We found FL-SMN protein was stable and still appeared fully present at 12 h. In contrast, Δ7-SMN began diminishing almost immediately (0.75 h) and was almost undetectable at 3 h. Secondly, the readthrough protein was substantially more stable than Δ7-SMN protein. Notably, readthrough protein was still detected at 12 h after treatment with cycloheximide. Lastly, the stability of HA:Δ7SMN[Flag] and Δ7SMN[HA] matched that of the readthrough protein. Both were detectable at 12 h, the last timepoint assayed, which was four times longer than Δ7-SMN. Thus, the increased stability of readthrough protein was reproduced by addition of peptides that were different in sequence but equivalent in size. Furthermore, absence or presence of an N-terminal HA tag did not affect protein stability. Together, these findings demonstrate the ability of SMN's C-terminus to modulate protein stability in a manner not dictated by a specific sequence.
Figure 3.
Effect of C-termini on SMN protein stability. Stable cell lines expressing SMN constructs were treated with cycloheximide at 50 µg/ml and lysed after 0, 0.75, 1.5, 3.0, 7.5 and 12 h. To qualitatively assay stability, protein remaining at each timepoint was determined through western blot using an antibody recognizing the HA tag. β-Tubulin was included as a loading control. Blots are representative of three independent experiments.
G418 induces readthrough of the Δ7-SMN target stop codon and increases SMN
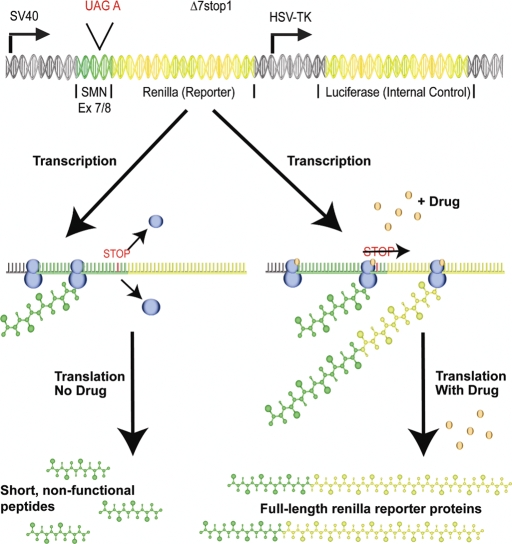
We next developed a novel, quantitative reporter assay to identify and compare efficiency of drugs that induce readthrough of the Δ7-SMN target stop codon sequence (UAG A) within human cells (Fig. 4). This assay is similar to a strategy used in creating inducible gene systems (38), and has been effective in vitro as well as in vivo with results in human cell lines successfully translated into mouse models. For our assay, a short sequence encoding the Δ7-SMN C-terminus was introduced upstream of a renilla reporter gene. The stop codon for this sequence (UAG A) was positioned close to the vector's start codon. Upon transfection of this reporter plasmid (referred to as Δ7stop), translation of its mRNA will normally generate only short, nonfunctional peptides. Treatment of transfected cells with drugs that effectively induce readthrough of Δ7-SMN transcripts will suppress translation termination at this target stop codon and thereby induce reporter expression for quantitative analysis. Normalization of the renilla reporter to an internal firefly luciferase control allowed for comparison between wells and transfections, and minimized any inaccuracy due to differences in transfection efficiency. Induction of a significant, dose-dependent fold increase in reporter activity was used as the criteria for successful induction of readthrough by a compound.
Figure 4.
A quantitative assay to detect translational read-through. Illustration of the read-through assay. A sequence encoding the C-terminus of SMN including its target stop codon was cloned into the 5′ end of a renilla reporter gene. Translation of the reporter transcript normally terminates at the Δ7-SMN stop codon to produce only short, nonfunctional peptides. Induction of translational readthrough produces a full-length reporter detectable through luciferase assay. The psi-CHECK 2 vector used for this assay independently coexpresses luciferase, allowing for normalization of the reporter signal to the luciferase internal control. Blue circles represent translational machinery and small brown circles represent read-through inducing drugs. Green DNA sequences represent those coding for SMN while yellow denote those coding for renilla or luciferase proteins.
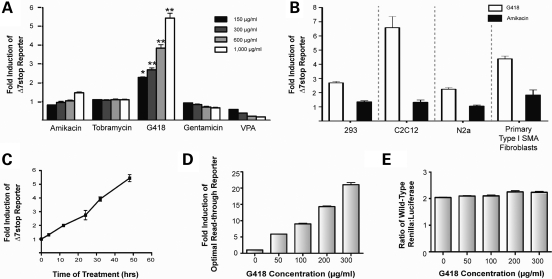
Several compounds applied at increasing concentrations were screened for their ability to induce readthrough of the Δ7stop reporter in AD293 cells (Fig. 5A). Included in this screen were the aminoglycosides amikacin, tobramycin, G418 and gentamicin as well as an HDAC inhibitor, VPA. Amikacin and tobramycin were the compounds initially reported to induce SMN protein in patient fibroblasts (18). Gentamicin and G418 were included because they are known to be efficient inductors of readthrough, have provoked clinical/therapeutic interest and represent discrete aminoglycoside families. VPA was included as a control that elevates SMN through a different mechanism.
Figure 5.
G418 induces readthrough of the therapeutic Δ7-SMN target. (A) Translational readthrough of the SMA therapeutic target stop codon (UAG A) detected by readthrough assay of AD293 cells treated with the indicated concentrations of amikacin, tobramycin, G418 or valproic acid (VPA). Values are expressed as fold induction of reporter activity over untreated cells and represent the mean ± SEM of three independent experiments, each performed in duplicate. (*P ≤ 0.005, **P ≤ 0.0005, Student's t-test, n = 6.) (B) Induction of readthrough in various cell lines. The Δ7stop reporter was transfected into AD293, HeLa, N2a, C2C12 and patient fibroblast cell types. Twenty-four hours later, cells were treated with G418 (150 µg/ml for fibroblasts, 300 µg/ml for others) for 2 days and assayed for reporter activity. Data for G418 treatment of fibroblasts is the mean ± SEM from a representative experiment performed in duplicate, all other values represent mean ± SEM from three independent experiments each performed in duplicate. (C) Reporter activity increases from 0 to 48 h of drug treatment. AD293 cells were transfected with the Δ7stop reporter, treated with G418 at 1000 µg/ml and assayed for reporter activity at 0, 4, 12, 24, 32 and 48 h after treatment. Data represent mean ± SEM from a representative experiment performed in triplicate. (D) Action of G418 upon a UGA C stop codon known to be highly susceptible to induction of translational readthrough. Here, cells were transfected with a reporter under translational control of a 5′ UGA C stop sequence and treated with increasing concentrations of G418. Values are mean ± SEM from a representative experiment performed in triplicate. (E) G418 treatment does not significantly alter normal ratios of renilla:luciferase activity. AD293 cells were transfected with a renilla control vector lacking a 5′ stop codon and treated with increasing concentrations of G418. Values represent mean ± SEM from a representative experiment performed in triplicate. Renilla activity is expressed as the ratio of renilla to luciferase.
G418 was identified by this assay as an agent capable of suppressing termination at the target Δ7-SMN stop codon in a dose-dependent manner. A significant increase in Δ7stop reporter activity was observed for all concentrations of G418 tested, peaking at 5.5 ± 0.3-fold at a concentration of 1000 µg/ml (t-test, P < 0.0005). Amikacin produced a small increase in reporter activity (1.5 ± 0.04) only at the highest concentration used; however, this did not reach statistical significance (t-test, P > 0.05). Tobramycin, gentamicin and VPA produced no increase in reporter activity. Thus, G418 was identified by this assay as a compound capable of inducing readthrough of the Δ7-SMN target in a dose-dependant manner.
To verify that our results were not cell type dependent and to demonstrate that translational readthrough could be induced in cell types relevant to an SMA therapy, we surveyed the ability of G418 and amikacin to induce readthrough of the Δ7stop reporter in other cell types. These included AD293 cells (a human embryonic kidney cell line), C2C12 cells (a muscle cell line derived from mouse myoblasts), N2a cells (a neuronal cell line derived from a murine neuroblastoma) and GM09677 cells (primary Type I SMA patient fibroblasts). G418 successfully induced readthrough in all cell types examined (Fig. 5B). Differences in readthrough efficiency or drug sensitivity between cell types were present, with fold increases of 2.7 ± 0.1 in AD293 cells, 8.8 ± 1.4 in HeLa cells, 6.6 ± 0.8 in C2C12 cells and 2.2 ± 0.1 in N2a cells treated with 300 µg/ml G418. At higher concentrations G418 induced reporter activity at levels >60-fold (corresponding to over 60% of WT renilla activity detected in control transfections), but these concentrations were associated with cytotoxicity (data not shown). Primary fibroblasts were particularly sensitive to aminoglycoside treatment. They displayed high levels of readthrough reporter activity at lower concentrations of G418 treatment (4.4 ± 0.2 at 150 µg/ml) than the other cell lines. They were also the only cell type that exhibited a >2-fold Δ7stop reporter induction in response to amikacin treatment, although this was at a higher concentration (2.1 ± 0.3 at 1000 µg/ml) than G418. In contrast to amikacin, which was only capable of inducing readthrough of the target Δ7-SMN stop codon in primary fibroblasts, G418 was capable of affecting a wide variety of cell lines—including cell types relevant to SMA.
To examine the sensitivity of this assay and validate its methodology, several additional experiments were performed. First, we determined the effects of drug exposure time on Δ7stop reporter activity. For this, AD293 cells were transfected with the reporter and treated with 1 mg/ml G418. Reporter activity was measured at 0, 4, 12, 24, 32 and 48 h (Fig. 5C). Results displayed a constant increase in renilla activity for all timepoints, indicating that readthrough-inducing drugs should produce a persistent increase in renilla activity through 48 h, the timepoint which we used in our dose and cell type-dependent assays. In order to determine our assay's sensitivity we evaluated the readthrough of the ‘UGA C’ stop codon, known to be highly affected by G418 (38). Treatment of cells expressing the UGA C reporter displayed a dose-dependent increase in renilla activity, with the lowest dose producing a 5.88-fold induction and the highest dose producing a 21.02-fold induction (Fig. 5D). Finally, we tested the effect of G418 treatment on the ratio of normal renilla:luciferase protein activity. This was done by transfecting AD293 cells with a reporter lacking an early translation termination signal, thus expressing wild-type renilla and luciferase proteins from the same vector used in our readthrough assays. Transfected cells treated with G418 at increasing concentrations displayed no change in renilla activity (Fig. 5E), demonstrating drug treatment does not effect activity of either renilla or the internal luciferase control. Overall, these experiments demonstrate G418 as a potent inducer of readthrough and confirm the ability of our methodology to accurately and efficiently identify compounds that induce readthrough of the Δ7-SMN target sequence.
Hypothesizing that induction of the Δ7stop reporter would correspond to an increase in SMN protein in SMA patient cells, we treated primary patient fibroblasts with G418. Amikacin, tobramycin, VPA and forskolin were included for comparison. Western blot analysis of treated GM09677 cells, derived from a Type I SMA patient, revealed an elevation of SMN protein by G418 treatment. Amikacin and tobramycin also increased SMN, though not to the degree induced by G418. VPA and forskolin increased SMN at levels higher than amikacin or tobramycin but lower than G418 (Fig. 6). Together, our data identify G418 not only as an inducer of translational readthrough at the Δ7-SMN stop codon within a variety of cell types, but also as an inducer of SMN protein within patient cells.
Figure 6.
G418 is a strong inducer of SMN protein in patient fibroblasts. Increase in SMN protein observed through western blot of Type I SMA patient fibroblasts treated with one of several drugs. GM09677 cells were treated for 48 h with 150 µg/ml amikacin, tobramycin, G418 or VPA, or with 20 µM forskolin. Wild-type AD293 cells and GMO8680 fibroblasts from an unaffected SMN carrier (SMN1 +/−) are included as controls.
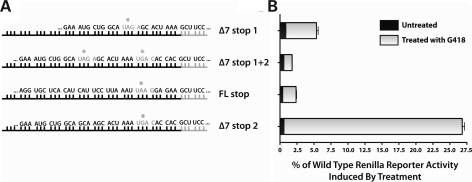
G418 induces readthrough of multiple SMN stop codons
To evaluate the effects of G418 on other stop codons that may be relevant to SMA therapy, we generated several other readthrough reporter plasmids in which renilla expression was repressed by one of several stop sequences found in the SMN genes (Fig. 7A). Here, the induction of readthrough was analyzed as a percentage of reporter activity relative to wild-type renilla. This allowed for comparison of one drug's effect on a variety of stop codons, independent of the sequence-specific effects on readthrough efficiency displayed by different aminoglycosides. This percentage is then directly relevant to how much overall reporter protein activity is being induced by treatment.
Figure 7.
G418 induces readthrough of multiple SMN stop codons relevant to SMA therapy. (A) Schematic of the constructs. Black bars represent SMN coding sequences, with stop codons highlighted in gray letters with asterisks. Gray bars represent renilla coding sequences. (B) Translational readthrough of each reporter induced by treatment of transfected AD293 cells with 1000 µg/ml G418. Values are expressed as the percentage of reporter activity compared with the activity of wild-type renilla protein. Data represent mean ± SEM of representative experiments.
Readthrough of the second stop codon sequence in Δ7-SMN transcripts (UGA C) is known to be highly inducible by G418. This indeed was the case in our study, as G418 treatment was able to induce Δ7stop2 readthrough reporter activity to levels >25% of wild-type renilla (Fig. 7B). To determine whether G418 is capable of inducing readthrough of multiple Δ7-SMN stop codons in the same transcript, the Δ7stop1 + 2 construct was used. Though magnitude of reporter induction was lower than for reporters containing one stop codon alone, G418 was able to induce readthrough of both stop codons to increase reporter activity. Readthrough of multiple stop codons in Δ7-SMN transcripts therefore is possible and would further lengthen the protein.
Ideally, a drug targeting the Δ7-SMN stop codon would act only on that codon to avoid off target effects. Since in actuality compounds will likely have effects on multiple stop codons we wished to observe off target effects on the FL-SMN stop codon, in particular to determine if readthrough of this codon may be prevalent. While readthrough of the FL-SMN stop codon would result in addition of a short peptide to the FL-SMN C-terminus, it is preferential not to interfere with the expression of FL-SMN already present in cells. We found that induction of the FLstop reporter, though present, occurred at a lower percentage than for the Δ7stop reporter (1.96 ± 0.22% versus 4.53 ± 0.61%, respectively). This stop codon also exhibited a lower level of ‘leakiness’ than that of Δ7-SMN, represented in untreated cells as lower levels of basal reporter activity. Hence, G418 has a greater effect on induction of the Δ7-SMN target than it does for off target effects on FL-SMN.
G418 treatment increases SMN and motor function in SMA mice
To examine the effects of G418 on SMN protein levels and the SMA phenotype in vivo, we administered the drug to a transgenic SMA mouse model (17). This mouse strain expresses transcripts from a Δ7-SMN cDNA transgene in high amounts. We hypothesized that the prevalence of this transcript would provide an abundant target for translational readthrough and improve the ability to detect a benefit in SMA mice (SMN2+/+;SMNΔ7+/+;Smn−/−) from treatment with drugs operating through this mechanism. Further, the utility of this model as a tool for preclinical drug studies has been previously demonstrated through treatment with the HDAC inhibitor trichostatin A and in studies assessing motor function (39–42). We administered G418 [14 mg/kg through intraperitoneal (IP) injection] to neonatal mice from postnatal day (PND) 5–13. Previous studies have identified this as the minimal window for drug treatment to produce a benefit in these SMA mice (42). This same dose and regimen of G418 treatment was previously found to be well tolerated by adult V2 vasopressin mutant mice and produced a functional benefit in vivo and correlated with a quantitative induction of readthrough in vitro (34) at concentrations roughly equivalent to those observed to induce readthrough in our experiments. Pharmacokinetic data of G418 concentrations in plasma are maximal <30 min after a single injection while substantial concentrations persist in tissues such as kidneys at extended timepoints, through 4 h after injection (34). Tolerance of the one week 14 mg/kg IP injection regimen in adult mice was confirmed in our lab, though 1 week treatment with higher doses (28–70 mg/kg) exhibited toxicity within a few days, while saline vehicle injections did not affect mouse weight, health or survival (data not shown).
Animals were monitored daily for health, weight and additionally evaluated every other day for motor function. At PND5, G418 and vehicle-injected SMA mice were equivalent in weight (2.71 ± 0.7 versus 2.61 ± 0.7 g, respectively) and litter size (6.9 ± 1.3 versus 6.8 ± 1.5, respectively).
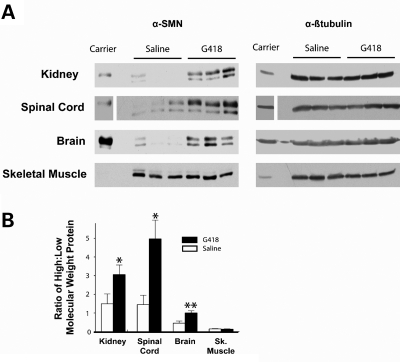
To assess the effects of G418 on SMN protein levels in vivo, tissue samples from treated and untreated SMA mice were analyzed by western blot. Western blot of untreated tissues from SMA mutant mice normally detects two SMN isoforms, a dark, low-molecular-weight protein corresponding to high levels of the Δ7-SMN protein and a faint upper protein corresponding to low levels of FL-SMN. Since readthrough should produce a protein higher in molecular weight than Δ7-SMN, we expected to visualize its induction as a shift towards a higher molecular weight that may or may not be discernable from FL-SMN. It is also possible that a more stable readthrough protein could ‘seed’ SMN complexes and thereby increase the levels of FL-SMN protein as well, which would also result in an increase in higher molecular weight protein. To account for conversion of Δ7-SMN into a readthrough protein and for a possible increase in FL-SMN induced by the seeding of complexes, we interpreted our protein data as a ratio of higher:lower molecular weight SMN as measured through densitometry. Tissues examined included the brain, kidney, spinal cord and skeletal muscle. Groups of mice were harvested at two timepoints: one group at PND10, during a mid-point of treatment, and the other at PND13, the final day of injection and when mice were at/near their functional death endpoint.
Western blot analyses of PND10 tissues revealed an increase of SMN for G418-treated brain, kidney and spinal cord tissues (Fig. 8A). This increase was confirmed using densitometry to detect a significant increase in the ratio of higher:lower molecular weight SMN in G418 treated as compared with untreated mice (P ≤ 0.05 kidney and spinal cord; P ≤ 0.001 brain) (Fig. 8B). No increase was detected in skeletal muscle. At PND13 (Supplementary Material) when mice were at/near their functional death endpoint, kidney continued to display an increase in SMN (P ≤ 0.05), while the spinal cord and brain exhibited a trend of increased SMN though no longer to a significant level as measured by densitometry, and the analysis of skeletal muscle was inconclusive due to lack of a distinct higher molecular weight protein. Very low levels of SMN within this tissue type are consistent with other reports (39). Thus, G418 was able to increase SMN in PND10 SMA mice in vivo within kidney and brain, as well as within spinal cord, a disease-relevant tissue.
Figure 8.
G418 treatment increases SMN in PND10 SMA mice. (A) Representative western blots of tissue from G418-treated and control mice. Tissues were harvested 2 h after their last injection on PND10. β-Tubulin is included as a loading control. A minimum of five samples were assayed for each treatment group. The kidney (100 µg), spinal cord (80 µg), brain (100 µg) and skeletal muscle (200 µg) were analyzed. Protein from an unaffected mouse was included to display the difference in band intensities for Smn +/− mice, with half of the protein amounts loaded for the kidney, spinal cord and skeletal muscle. (B) Graph of western blot densitometry used to qualitatively confirm G418 effects on SMN. Values represent mean ± SEM of the ratio of higher:lower molecular weight SMN. (*P ≤ 0.05, **P ≤ 0.001.)
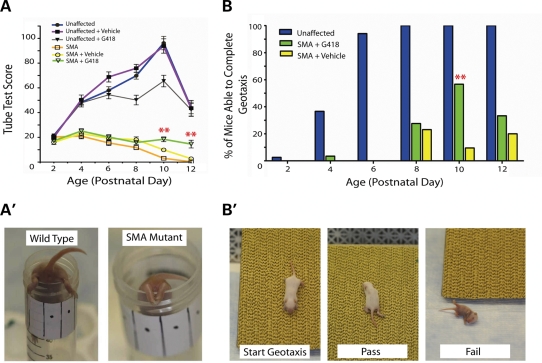
To analyze the effects of G418 on the SMA phenotype, we used the tube test as a functional motor scale starting at PND2. First, neonates receiving no injections were used to establish baseline data. No significant difference in Tube Test Score (TTS) was observed at any age between wild-type and unaffected carriers (data not shown), which reached a peak value for each at PND10. In contrast, SMA mice displayed scores significantly lower than unaffected mice beginning at PND4. From PND4 SMA mouse scores declined each day, from a score of 21.0 ± 7.8 at PND4 to a score of 0.24 ± 0.3 at PND12 (Fig. 9A).
Figure 9.
G418 treatment increases motor function of SMA mutant mice. (A) Graph of tube test score (TTS). Groups of untreated, vehicle-injected and G418-treated mice were administered the tube test from PND2-12. Treatment groups consisted of 29 G418-treated SMA mice, 35 G418-treated unaffected mice, 34 vehicle-injected unaffected mice and 20 vehicle-injected SMA mice. Values represent the mean ± SEM. (**P ≤ 0.01, Student's t-test) (A′) Photograph of mice performing the tube test at PND10. (B) Graph of the percentage of mice able to complete a negative geotaxis assay at each age. (**P ≤ 0.01, χ2.) (B′) Photographs illustrating the negative geotaxis assay.
After establishing baseline data, G418- and saline-injected mice were subjected to the tube test. G418-treated SMA mice displayed increased TTS scores over vehicle-injected SMA mice beginning at PND10 (Student's t-test, P ≤ 0.01) (Fig. 9A, videos available in Supplementary Material). Notably, the TTS of drug-treated SMA mice increased from PND8 to PND10, and maintained an elevated value at PND12 (15.8 ± 8.4 at PND8, to 18.4 ± 10.2 at PND10 and to 14.6 ± 12.2 at PND12). In contrast, vehicle-injected SMA mice had a decrease in TTS at each of these timepoints (18.1 ± 10.9 at PND8, to 9.9 ± 6.2 at PND10, to 2.7 ± 3.0 at PND12). A different effect of G418 treatment was observed in unaffected (WT and heterozygous carrier) mice. Upon G418 treatment, unaffected mice displayed reduced TTS values at PND8 and PD10 as compared with vehicle-injected controls. The increase in TTS value upon G418 injection indicated that treatment with this drug produced an increase in the motor function of SMA mice.
For a qualitative, all-or-nothing measure of mouse motor function we used the negative geotaxis assay. This test consisted of placing mice facing the bottom of an incline, a position from which they naturally try to re-orient themselves to face the top of the incline. Unaffected, healthy mice were typically able to do this beginning at PND4 or PND6. SMA mice that were untreated or injected with vehicle typically lack the strength and coordination needed to re-orient themselves 180º on the incline at any day. G418-treated SMA mice displayed an increased ability to complete geotaxis at PND10 over vehicle-injected SMA mice, with 17 of 30 (56.7%) completing geotaxis compared with 2 of 21 (9.5%), respectively (χ2, P ≤ 0.01). A representative video of this trend demonstrates a G418-injected SMA mouse that was able to complete geotaxis alongside a vehicle-injected SMA littermate that was unable to complete geotaxis (Supplementary Material). At PND12, 5 of 15 (33.3%) drug-treated mice were able to complete geotaxis as opposed to 2 of 10 (20.0%) vehicle injected. On PND8, the timepoint preceding increased motor function, 8 of 29 (27.6%) G418-treated mice were able to perform negative geotaxis compared with 6 of 26 (23.1%) vehicle-injected SMA mice. Through a qualitative improvement in the strength and coordination necessary to complete negative geotaxis in addition to a quantitative increase in TTS, SMA mice treated with G418 exhibited increased motor function.
Chronic administration of G418 is toxic to wild-type mice
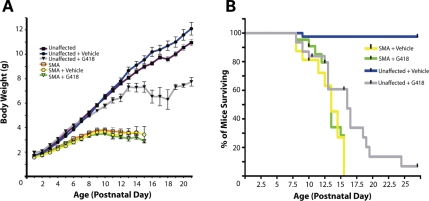
Although increased motor function was observed in SMA pups treated with G418, administration of this drug (14 mg/kg IP) for a period of 9 days was associated with toxicity, manifesting as effects on body weight and survival in unaffected mice. Drug toxicity in SMA mutants could not be distinguished from deterioration due to the SMA phenotype. In agreement with previous reports, SMA mice were significantly underweight at PND5 compared with unaffected mice (2.64 ± 0.5 versus 3.71 ± 0.4 g) (Fig. 10A). Cohorts of treated and untreated SMA pups displayed no significant difference in body weight at any age, though G418 pups were slightly smaller on average. However, upon administration of G418, a significant reduction in body weight was observed in unaffected mice beginning at PND9 (5.36 ± 0.6 versus 6.00 ± 0.8 g, Student's t-test, P < 0.005).
Figure 10.
Prolonged G418 treatment exhibits toxicity. (A) Body weight of treated and untreated mice from PND1-21. (B) Kaplan–Meier survival curve of treated and vehicle-injected groups of mutant and wild-type mice. Tick marks represent days at which mice were artificially removed for molecular analyses.
Ultimately, the majority of G418-treated wild-type and heterozygous mice died from drug toxicity caused by daily administration of the drug (Fig. 10B). The average survival for unaffected mice treated with G418 from PND5-13 and observed for a period of 1 month beginning at birth was 14.2 ± 4.5 with a median value of 16 days, while only 1 out of 42 vehicle-injected control mice were lost during this period. Vehicle-injected SMA pups displayed an average lifespan of 12.5 ± 3.0 days with a median value of 13.5 days and maximum value of 15.5 days. These numbers are in agreement with initial reports characterizing this model (17) but lower than those found in more recent studies (39,42). Upon G418 treatment, no significant difference was observed in survival of SMA pups. The average lifespan of drug-treated mice was 13.1 ± 2.0 days with a median of 13.5 days and a maximum of 15.5 days. Collectively, our data demonstrate an ability of G418 to increase motor function in SMA mice without significant effects on weight or survival of mutants; however, daily administration of this drug to young pups for a period of 9 days revealed toxicity in normally healthy genotypes of mice.
DISCUSSION
All SMA patients have at least one functional copy of SMN2. Hence strategies that increase or stabilize the transcripts and/or proteins produced from it are relevant towards developing a therapy for SMA. Here, we have demonstrated that the C-terminus of SMN is able to modulate protein stability in a manner independent of its specific sequence. Translational readthrough, by lengthening the Δ7-SMN protein to include a peptide normally held translationally silent, is a drug mechanism that is capable of increasing Δ7-SMN stability. Using this information, we developed an assay that is amenable to high throughput screens and capable of identifying compounds that act through this pathway to induce SMN in SMA cells. With this assay, G418 was identified as a target ‘hit’ and potential lead compound. Treatment with G418 was capable of inducing SMN in vitro, as well as increasing SMN in vivo and improving the motor function of SMA mice.
Currently, the general application of a readthrough mechanism towards genetic disease has attracted great attention but has been hindered by two issues: mutation heterogeneity and compound toxicity/bioavailability. Application of this mechanism to SMA is free of this first hurdle because it acts on a monormorphic, highly expressed target: Δ7-SMN transcripts. It is hypothesized that Δ7-SMN extends survival through either partial functionality (19) of the Δ7-SMN protein or by helping to ‘seed’ complexes containing FL-SMN (17); we hypothesize that the readthrough protein, being more stable, enhances the partial functionality and/or FL-SMN seeding capability of Δ7-SMN to further benefit the SMA phenotype. Other work indicates that the readthrough protein does indeed possess increased functionality over Δ7-SMN in snRNP assembly, and can both localize down the axon and increase neurite outgrowth in assays of SMN deficient cells (22,43). Through this increased functionality and/or by increasing pools of FL-SMN through the seeding of heteromeric SMN complexes, the readthrough-inducing drug G418 was observed here to produce a functional benefit to SMA mouse motor function. Improvement of motor function in SMA patients would be a significant step forward in improving quality of life, and thus translational readthrough is worth pursuing further as a therapeutic strategy whether as an independent mechanism or in combination with compounds working through different mechanisms.
Unfortunately, toxicity was encountered with G418 as our proof of concept drug. While G418 had no overt detrimental effect on body weight or survival of SMA mice, chronic administration produced lethality in normally healthy genotypes of neonate mice at a median of 16 days and may have prevented us from observing further benefits to SMA mouse health and survival. Mouse LD50 data indicate that G418 displays safety similar to other aminoglycosides in acute toxicity studies (34). However, the toxicity we observed through chronic administration together with toxicology studies in dogs (44) will likely prevent this drug from ever entering clinical use. Different compounds capable of inducing readthrough have entered clinical trials for other diseases, providing precedence that if a less toxic compound can be identified it could be moved further towards clinical application (33,45–47).
A limitation of aminoglycosides as a drug class in general is their limited penetrance of eukaryotic cells and inefficiency at crossing the blood–brain barrier. Here, we observed induction of SMN in the brain and spinal cord of young SMA mice. We believe this is due to the immature blood–brain barrier of neonate mice. Thus, this SMA model, with an early onset, may provide an advantage for studying drugs that have limited neuronal penetrance. Despite toxicity of chronic treatment, the ability of G418 to produce functional improvement in mouse models of SMA as well as other genetic diseases (34) is encouraging and provides proof-of-concept in support of translational readthrough as a therapy for genetic disease. Currently, there is a lack of G418 derivatives. With such evidence growing, there should be strong support for generating and assaying derivatives of G418 (or other readthrough compounds) which possess limited toxicity with optimal bioavailability and specificity profiles.
The increase in SMN stability provided by readthrough induction does not derive from the specific amino acid sequence produced but instead from the presence of a non-specific peptide at the C-terminus (Fig. 3). The ability of peptides to modulate SMN isoform stability to an intermediate between FL-SMN and Δ7-SMN could be explained by general effects on protein secondary structure or post-translational modification. To address these possibilities, we examined the amino acid sequence of the SMN C-terminus using a program that predicts secondary structure [JUFO (48)] and a program that predicts phosphorylation [GPS: Group-based Prediction System 2.0 (49)]. These programs reveal a likelihood that the FL-SMN C-terminus possesses a helix followed by loop secondary structure that contains two serine residues that are phosphorylated. Interestingly, the C-terminal end of Δ7-SMN is predicted to lack loop structure and contains no residues capable of being phosphorylated, while translational readthrough would produce a peptide predicted to restore loop structure and contain two serines predicted to be phosphorylated by kinases also predicted to phosphorylate FL-SMN. The ectopic tags we examined were also predicted to form secondary loop structure, and contain residues capable of either being phosphorylated or mimicking phosphorylation. In support of these computer predictions, recent evidence has emerged that FL-SMN is indeed phosphorylated at its C-terminus, that oligomerization stabilizes SMN protein and that these factors may play a role in its regulation through the ubiquitin proteasome system (B. Burnett and K. Fischbeck, personal communication). In regards to SMN's function and regulation, it will be interesting to ultimately determine the precise properties and pathways that convey full stability to the SMN protein.
Our quantitative assay to measure readthrough is capable of screening large numbers of drugs and can measure readthrough in a wide variety of cell types. Amikacin and tobramycin, previously identified as inducers of SMN in patient fibroblasts (18), did not induce readthrough in cell types other than fibroblasts and have been found by another report to not induce readthrough of the ‘UAG A’ stop codon in human cell lines efficiently (50). In contrast, G418, belonging to a separate structural class of aminoglycoside compounds, induced readthrough of the Δ7-SMN target stop codon in all cell types examined (Fig. 5). The observance of readthrough within human cells, and in multiple/target tissue types, as opposed to an extracellular transcription/translation systems, is important in order to account for the poor cellular penetration that many of these drugs exhibit. By ignoring the fact that many compounds in this drug class have poor cellular penetration, findings from cell-free assays could produce inconsistent results when translating their application in vivo to animal models and clinical trials. Our assay, performed within human cell lines and based on a strategy demonstrated by Murphy et al. (38) to correlate with the induction of readthrough in vivo within mice, should overcome these issues and be capable of identifying compounds relevant to an SMA therapy.
When we determined the ability of G418 to induce readthrough of other stop codons potentially relevant to SMA therapy, we observed several things: the amount of readthrough detected at the second stop codon in Δ7-SMN transcripts (UGA C) was particularly strong, G418 was capable of inducing readthrough of two stop codons in one transcript at the same time and readthrough of the FL-SMN stop codon was induced by G418, though at lower levels than the target Δ7-SMN stop codon. An ideal drug acting through this mechanism would be one that induces readthrough of the Δ7-SMN target alone. G418 does induce readthrough of the Δ7-SMN stop codon preferentially to that of FL-SMN, but also demonstrates high levels of termination suppression at the UGA C sequence and therefore could be having significant off-target effects.
In the future, less toxic and more specific readthrough drugs and a transgenic strategy should be employed to fully discern the therapeutic potential of translational readthrough as an SMA therapy. The identification of safer, more specific readthrough drugs would minimize toxicity interference issues encountered here and benefit studies of other genetic diseases that could potentially be treated by translational readthrough. A transgenic mouse expressing SMN readthrough protein on an SMA background would directly determine the therapeutic potential of translational readthrough for SMA without limitations imposed by drug toxicity or bioavailability. It would also address the possibilities that (i) efficient expression of SMN protein with a novel C-terminus could be detrimental and (ii) G418 may be inducing FL-SMN through off target effects on regulatory proteins.
In summary, it is clear that the drug mechanism of translational readthrough is capable of modulating SMN stability. The aminoglycoside G418 has demonstrated an ability to induce readthrough of the SMN target, to increase SMN protein and to increase SMA mouse motor function. Application of readthrough to SMA may be beneficial to the global SMA patient population as well as to the attempt of applying readthrough therapeutics to other, more heterogeneic, diseases. Together, this work provides a methodology for identifying readthrough-based therapeutics and provides the first in vivo evidence supporting their application towards SMA.
MATERIALS AND METHODS
Cell culture
AD293 cells, a human embryonic kidney cell line optimized for retention on tissue culture dishes (Stratagene), was cultured in Dulbecco's Modified Eagle's Medium (DMEM) with high glucose that had been supplemented with 10% fetal bovine serum and 2 mm l-glutamine. HeLa cells, a human cervical cancer cell line, and C2C12 cells, a mouse myoblast cell line, were also grown and maintained in this media. N2a cells, a mouse neuroblastoma cell line originating from brain neuroblast cells, were grown in EMEM supplemented with 10% fetal bovine serum, 1× non-essential amino acids, 2 mm l-glutamine and 100 U/ml penicillin and 100 µg/ml streptomycin. Low passage primary fibroblast cell lines, GM09677 and GM00232, were obtained from the Coriell Cell Repository (Camden, NJ, USA) and grown in alpha-minimal essential medium (α-MEM) supplemented with 15% fetal bovine serum, 1× nonessential amino acids, and 2 mm l-glutamine. The copy number of SMN1 and SMN2 was previously determined for the GM09677 and GM00232 primary cell lines (11,37). All cultures were maintained at ∼30–80% confluence, 37°C and 5% CO2 in a humidified atmosphere.
Drug treatment of patient fibroblasts
G418 sulfate (Gibco, Invitrogen) was reconstituted in sterile deionized water to a stock concentration of 80 mg/ml and stored at −20°C. Amikacin and Tobramycin (Fluka, Sigma) were reconstituted in sterile deionized water to stock concentrations of 50 mg/ml and stored at 4°C. VPA (Sigma) was also reconstituted in sterile deionized water at concentrations of 50 mg/ml, though due to its low stability this was done immediately prior to cell treatment. Twenty-four hours prior to drug treatment, GM09677 and GM00232 cells were seeded at a density of 1.5 × 105 cells/well in 6-well culture plates. At the time of treatment, cells were rinsed once with phosphate-buffered saline (PBS) and fed media containing drug diluted to the appropriate concentration. Media and drug were replaced every 24 h. Forty-eight hours after treatment, cells were harvested in radio immunoprecipitation assay (RIPA) buffer as described previously (51) and quantified by Lowry assay. Concentrations of 150 µg/ml amikacin, tobramycin, G418 and VPA correspond to 192, 321, 217 and 903 µm, respectively.
Production of plasmid constructs and stable cell lines
SMN cDNAs coding for varying C-terminal ends were generated through polymerase chain reaction (PCR) using high-fidelity Taq polymerase (Invitrogen) according to manufacturer's instructions, with SMN cDNA as a template and oligos (∼60mers) that had novel 5′ and 3′ ends containing homology to SMN (sequences available upon request). PCR products were ligated into the pCR 2.1 vector (Invitrogen). BglII and XhoI restriction sites, present in the oligos, were used to subclone inserts from the pCR 2.1 cloning vector directionally into the pCIneo mammalian expression vector (Promega). All plasmid constructs were verified through DNA sequencing.
Transfection of AD293 cells was performed using TransIT-293 Transfection Reagent (Mirus). Twenty-four hours prior to transfection, cells were seeded at a density of 1.5 × 105 cells/well in 6-well culture plates. Transfection was performed at ∼60% confluency with 2 µg DNA and 6 µl 293 transfection reagent in 200 µl OptiMEM. Forty-eight hours after transfection, cells were treated with drug to select for cells stably expressing the neomycin resistance gene. The HA:Δ7-SMN construct expresses a highly unstable isoform. Owing to difficulties in producing a stable cell line sufficiently expressing this protein, presumably due to this low stability, an alternate method was used. This was accomplished by co-transfection of AD293 cells with HA:Δ7-SMN in the pShuttle-IRES-hrGFP-1 vector (Stratagene) and an empty pCIneo vector. The stable cell line was then obtained by selecting for neomycin resistance through drug treatment in combination with enrichment for GFP expression via FACS sorting.
Cycloheximide assay for protein stability
One week prior to treatment of stable cell lines with cycloheximide, selective media were replaced with media containing no selective agent in order to prevent interference from antibiotic effect on protein stability. Forty-eight hours prior to treatment, cells were seeded at a density of 2.25 × 105 cells/well in 6-well culture plates. At the time of treatment, cells were rinsed once with PBS and re-fed with DMEM containing cycloheximide diluted to a concentration of 50 µg/ml. Cells were lysed in RIPA buffer at 0, 0.75, 1.5, 3, 4.5, 7.5 and 12 h after treatment. Protein was quantified by Lowry Assay (Bio-Rad). An amount of 120 µg of protein from each sample was assayed via SDS-PAGE followed by western blot analysis. Membranes were blocked in 5% milk and probed with HA polyclonal antibody (Santa Cruz) diluted at 1:500, followed by GAR (Bio-Rad) at 1:10 000. Protein was visualized through ECl chemiluminescence (Pierce).
Readthrough assay
Plasmid constructs for readthrough assays were produced by inserting an SMN sequence containing the desired stop codon into the 5′ end of a renilla luciferase reporter gene in the psiCHECK-2 vector (Promega). This was accomplished by PCR amplifying the renilla reporter with oligos encoding a novel 5′ end and the NheI and XhoI restriction sites (sequences available upon request). PCR products were initially inserted into the pCR 2.1 cloning vector, than subcloned into the psiCHECK-2 vector through restriction digest and re-ligation. Inserts were detected through EcoRI digest of the pCR 2.1 vector. SMN, renilla reporter and vector sequences were verified through DNA sequencing.
For the majority of experiments assaying readthrough, AD293 cells were seeded at 0.75 × 105 cells/well in 6-well culture plates and transfected 8 h later with the appropriate reporter plasmid. Transfections were performed using the 293 Trans-IT transfection kit with 1 µg DNA, 6 µl 293 reagent and 200 µl OptiMEM per well. Twenty-four hours after transfection, cells were rinsed once with PBS and re-fed with growth media containing the desired drug dilution with replacement every 24 h. Forty-eight hours after transfection, cells were trypsinized, redistributed in 96-well plates, and readthrough was analyzed through luciferase assay. Luciferase assays were performed using DualGlo Luciferase reagents (Promega) and read by a Veritas Microplate Luminometer (Turner BioSystems). Each drug treatment was performed at least three times, and each well from these experiments was analyzed by luciferase assay in duplicate.
For detecting readthrough in different cell types, HeLa, C2C12, N2a and GM09677 cells were transiently transfected with the Δ7stop readthrough reporter. HeLa cells were transfected using HeLa MONSTER transfection reagent (Mirus) with 1 µg DNA, 3 µl HeLa reagent and 0.5 µl MONSTER reagent. C2C12, N2a and GM09677 cells were all transfected using a Nucleofector II (Amaxa) electroporation device with appropriate reagents and electroporation protocols optimized for that cell type. Transfections of these cell lines were performed on 1 × 106 cells suspended in 100 µl of the appropriate buffer, followed by resuspension in a final volume of 800–1000 µl growth media and redistribution into 6-well plates.
SMA mouse model maintenance and drug treatment
Production and initial characterization of the intermediate SMA mouse model has been previously reported (17). Breeding pairs for SMA mice on an FVB background were purchased from Jackson Laboratories (strain # 005025). Animals were maintained in a controlled animal facility at 25°C, 60% humidity and fed ad libitum for water and food with a photoperiod of 12 h light/12 h dark, where they were monitored daily for health. All maintenance and procedures were approved and performed in accordance with the Children's Memorial Research Center's Institutional Animal Care and Use Committee regulations. The colony was maintained by mating SMN2+/+;SMNΔ7+/+;Smn +/− breeding mice. Mice were genotyped for the Smn knockout allele by PCR analysis of mouse tail DNA. Primers for PCR flanked the LacZ knockout cassette: 5′-GGT AAC GCC AGG GTT TTC C-3′/5′-CTC CGG GAT ATT GGG ATT G-3′/5′-CAA GGG AGT TGT GGC ATT CTT C-3′ (94°C 45 s/59°C for 45 s/72°C for 45 s, 35 cycles). Resulting PCR products were separated on 1.5% agarose gels.
G418 sulfate (GIBCO) was dissolved in sterile saline to a stock concentration of 10 mg/ml active solution and stored at 4°C. For injection into animals, G418 was diluted to 2.5 mg/ml. Mice were administered 14 mg/kg G418 or an equivalent volume of saline vehicle by intraperitoneal injection once daily from PND5-13. Treatment groups consisted of 29 drug-treated SMA mutants, 35 drug-treated unaffected mice, 20 vehicle-injected SMA mutants and 34 vehicle-injected unaffected mice.
For in vivo molecular studies, or when mice were considered to have met functional death endpoints (20% loss of body weight, inability to right and obvious state of distress), mice were euthanized with CO2 followed by cervical dislocation as a secondary measure. Upon sacrifice, tissues were harvested and flash frozen in liquid nitrogen. For protein analysis, tissues were lysed in RIPA, quantified by Lowry assay, then resolved on a 12% acrylamide gel and assayed by western blot. Densitometry was performed to confirm and analyze results using a Microtek 1000XL scanner and Openlab 5.0 software.
Functional motor scales
Functional motor scales used here were initially developed by Psychogenics, Inc. for characterizing motor function in SMA neonate mice. The tube test was administered every other day from PND2-12 by suspending a mouse by its hind limbs from the lip of a vertical 50 cc tube. Time spent hanging, number of pulls and hind limb strength (HLS) were measured and inserted into the following equation to obtain a quantitative score: TTS = [(time spent hanging) + 10(# of pulls)] × [(HLS score + 1)/4]. The TTS for a given mouse each day was recorded as an average of two consecutive trials. The negative geotaxis test was also administered every 2 days from PND2-12. This was performed by placing a mouse on a 30° incline so that it stood on all fours with its head facing the bottom of the incline. An inability of a mouse to reorient itself 180° within 30 s so that its head faced the top of the incline was recorded as a 0, or a failed attempt. If a mouse was able to reorient itself 180° to face the top of the incline it received a score of 1, corresponding to a successful completion of the task.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by the Medical Research Junior Board Foundation at Children's Memorial Hospital and also by Families of Spinal Muscular Atrophy. C.H. is supported by a National Institutes of Health [ST326M008061] training grant and additional support is provided by National Institutes of Health NINDS [1ROIN5060926] grant. During a portion of this work, C.J.D. was an American Academy of Neurology/SMA Foundation Young Investigator.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Barrington Burnett and Dr. Kenneth Fischbeck for discussion of data on the SMN C-terminus, Dr. Bassem El-Khodor, senior scientist at PsychoGenics Inc. who established the tube test and negative geotaxis test, and also Dr. Torsten Schoneberg for his insight into aminoglycoside pharmacokinetics.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Rochette C.F., Gilbert N., Simard L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 5.Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 7.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heier C.R., Gogliotti R.G., DiDonato C.J. SMN transcript stability: could modulation of messenger RNA degradation provide a novel therapy for spinal muscular atrophy? J. Child Neurol. 2007;22:1013–1018. doi: 10.1177/0883073807305669. [DOI] [PubMed] [Google Scholar]
- 9.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 10.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 11.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 12.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 14.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumner C.J. Therapeutics development for spinal muscular atrophy. NeuroRx. 2006;3:235–245. doi: 10.1016/j.nurx.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua Y., Zhou J. Modulation of SMN nuclear foci and cytoplasmic localization by its C-terminus. Cell. Mol. Life Sci. 2004;61:2658–2663. doi: 10.1007/s00018-004-4300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 18.Wolstencroft E.C., Mattis V., Bajer A.A., Young P.J., Lorson C.L. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 19.Frugier T., Tiziano F.D., Cifuentes-Diaz C., Miniou P., Roblot N., Dierich A., Le Meur M., Melki J. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 20.Talbot K., Ponting C.P., Theodosiou A.M., Rodrigues N.R., Surtees R., Mountford R., Davies K.E. Missense mutation clustering in the survival motor neuron gene: a role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum. Mol. Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 21.Wang C.H., Papendick B.D., Bruinsma P., Day J.K. Identification of a novel missense mutation of the SMN(T) gene in two siblings with spinal muscular atrophy. Neurogenetics. 1998;1:273–276. doi: 10.1007/s100480050040. [DOI] [PubMed] [Google Scholar]
- 22.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Bergeijk J., Rydel-Konecke K., Grothe C., Claus P. The spinal muscular atrophy gene product regulates neurite outgrowth: importance of the C terminus. FASEB J. 2007;21:1492–1502. doi: 10.1096/fj.06-7136com. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon K., Phillips I. Mechanisms of resistance to aminoglycosides in clinical isolates. J. Antimicrob. Chemother. 1982;9:91–102. doi: 10.1093/jac/9.2.91. [DOI] [PubMed] [Google Scholar]
- 26.Bidou L., Hatin I., Perez N., Allamand V., Panthier J.J., Rousset J.P. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 2004;11:619–627. doi: 10.1038/sj.gt.3302211. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson J. Antibiotics show promise as therapy for genetic disorders. J. Am. Med. Assoc. 2001;285:2067–2068. [PubMed] [Google Scholar]
- 28.Zingman L.V., Park S., Olson T.M., Alekseev A.E., Terzic A. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin. Pharmacol. Ther. 2007;81:99–103. doi: 10.1038/sj.clpt.6100012. [DOI] [PubMed] [Google Scholar]
- 29.Barton-Davis E.R., Cordier L., Shoturma D.I., Leland S.E., Sweeney H.L. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunant P., Walter M.C., Karpati G., Lochmuller H. Gentamicin fails to increase dystrophin expression in dystrophin-deficient muscle. Muscle Nerve. 2003;27:624–627. doi: 10.1002/mus.10341. [DOI] [PubMed] [Google Scholar]
- 31.Keeling K.M., Brooks D.A., Hopwood J.J., Li P., Thompson J.N., Bedwell D.M. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-l-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum. Mol. Genet. 2001;10:291–299. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- 32.Phillips-Jones M.K., Hill L.S., Atkinson J., Martin R. Context effects on misreading and suppression at UAG codons in human cells. Mol. Cell. Biol. 1995;15:6593–6600. doi: 10.1128/mcb.15.12.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilschanski M., Yahav Y., Yaacov Y., Blau H., Bentur L., Rivlin J., Aviram M., Bdolah-Abram T., Bebok Z., Shushi L., et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N. Engl. J. Med. 2003;349:1433–1441. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- 34.Sangkuhl K., Schulz A., Rompler H., Yun J., Wess J., Schoneberg T. Aminoglycoside-mediated rescue of a disease-causing nonsense mutation in the V2 vasopressin receptor gene in vitro and in vivo. Hum. Mol. Genet. 2004;13:893–903. doi: 10.1093/hmg/ddh105. [DOI] [PubMed] [Google Scholar]
- 35.Brichta L., Hofmann Y., Hahnen E., Siebzehnrubl F.A., Raschke H., Blumcke I., Eyupoglu I.Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 36.Majumder S., Varadharaj S., Ghoshal K., Monani U., Burghes A.H., Jacob S.T. Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene. J. Biol. Chem. 2004;279:14803–14811. doi: 10.1074/jbc.M308225200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Sumner C.J., Huynh T.N., Markowitz J.A., Perhac J.S., Hill B., Coovert D.D., Schussler K., Chen X., Jarecki J., Burghes A.H., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 38.Murphy G.J., Mostoslavsky G., Kotton D.N., Mulligan R.C. Exogenous control of mammalian gene expression via modulation of translational termination. Nat. Med. 2006;12:1093–1099. doi: 10.1038/nm1376. [DOI] [PubMed] [Google Scholar]
- 39.Avila A.M., Burnett B.G., Taye A.A., Gabanella F., Knight M.A., Hartenstein P., Cizman Z., Di Prospero N.A., Pellizzoni L., Fischbeck K.H., et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butchbach M.E., Edwards J.D., Burghes A.H. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol. Dis. 2007;27:207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Khodor B.F., Edgar N., Chen A., Winberg M.L., Joyce C., Brunner D., Suarez-Farinas M., Heyes M.P. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp. Neurol. 2008;212:29–43. doi: 10.1016/j.expneurol.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 42.Narver H.L., Kong L., Burnett B.G., Choe D.W., Bosch-Marce M., Taye A.A., Eckhaus M.A., Sumner C.J. Sustained improvement of spinal muscular atrophy mice treated with trichostatin a plus nutrition. Ann. Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattis V.B., Bowerman M., Kothary R., Lorson C.L. A SMNDelta7 read-through product confers functionality to the SMNDelta7 protein. Neurosci. Lett. 2008;442:54–58. doi: 10.1016/j.neulet.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Rocca P.T., Baker F., Frantz J.D., Szot R.J., Black H.E., Schwartz E. Skin and mucous membrane ulceration in beagle dogs following oral dosing with an experimental aminoglycoside antibiotic. Fundam. Appl. Toxicol. 1985;5:986–990. [PubMed] [Google Scholar]
- 45.Hirawat S., Welch E.M., Elfring G.L., Northcutt V.J., Paushkin S., Hwang S., Leonard E.M., Almstead N.G., Ju W., Peltz S.W., et al. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J. Clin. Pharmacol. 2007;47:430–444. doi: 10.1177/0091270006297140. [DOI] [PubMed] [Google Scholar]
- 46.Kerem E., Hirawat S., Armoni S., Yaakov Y., Shoseyov D., Cohen M., Nissim-Rafinia M., Blau H., Rivlin J., Aviram M., et al. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–727. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- 47.Politano L., Nigro G., Nigro V., Piluso G., Papparella S., Paciello O., Comi L.I. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol. 2003;22:15–21. [PubMed] [Google Scholar]
- 48.Meiler J., Baker D. Coupled prediction of protein secondary and tertiary structure. Proc. Natl Acad. Sci. USA. 2003;100:12105–12110. doi: 10.1073/pnas.1831973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howard M.T., Anderson C.B., Fass U., Khatri S., Gesteland R.F., Atkins J.F., Flanigan K.M. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann. Neurol. 2004;55:422–426. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- 51.DiDonato C.J., Parks R.J., Kothary R. Development of a gene therapy strategy for the restoration of survival motor neuron protein expression: implications for spinal muscular atrophy therapy. Hum. Gene Ther. 2003;14:179–188. doi: 10.1089/104303403321070874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.