Abstract
Obesity is a major public health problem in most developed countries and a major risk factor for diabetes and cardiovascular disease. Emerging evidence indicates that ciliary dysfunction can contribute to human obesity but the underlying molecular and cellular mechanisms are unknown. Bardet-Biedl syndrome (BBS) is a genetically heterogeneous human obesity syndrome associated with ciliary dysfunction. BBS proteins are thought to play a role in cilia function and intracellular protein/vesicle trafficking. Here, we show that BBS proteins are required for leptin receptor (LepR) signaling in the hypothalamus. We found that Bbs2−/−, Bbs4−/− and Bbs6−/− mice are resistant to the action of leptin to reduce body weight and food intake regardless of serum leptin levels and obesity. In addition, activation of hypothalamic STAT3 by leptin is significantly decreased in Bbs2−/−, Bbs4−/− and Bbs6−/− mice. In contrast, downstream melanocortin receptor signaling is unaffected, indicating that LepR signaling is specifically impaired in Bbs2−/−, Bbs4−/− and Bbs6−/− mice. Impaired LepR signaling in BBS mice was associated with decreased Pomc gene expression. Furthermore, we found that BBS1 protein physically interacts with the LepR and that loss of BBS proteins perturbs LepR trafficking. Our data indicate that BBS proteins mediate LepR trafficking and that impaired LepR signaling underlies energy imbalance in BBS. These findings represent a novel mechanism for leptin resistance and obesity.
INTRODUCTION
The increasing prevalence of obesity and its association with various human disorders, such as diabetes and hypertension has made studying the mechanisms that control energy homeostasis a high priority. The environmental and genetic factors contributing to the obesity ‘epidemic’ are currently under intense investigation but our understanding of the molecular, cellular and physiological processes that regulate energy homeostasis and the defects leading to energy imbalance and obesity is still far from complete. In mammals, body weight and energy homeostasis are controlled by the interplay between the peripheral signals and the brain, particularly the hypothalamus (1). Leptin is an adipocyte-derived hormone that circulates in proportion to the amount of body fat in order to inform the brain about the peripheral fat storage (2,3). Within the arcuate nucleus of the hypothalamus, a major site for leptin action, leptin is known to act on at least two neuronal populations: proopiomelanocortin (POMC) neurons, which are activated by leptin and neuropeptide Y (NPY) neurons [co-expressing agouti-related protein (AgRP)] and are inhibited by leptin (1–4). In rodents and humans, mutations in genes encoding leptin and the leptin receptor lead to severe obesity, infertility and endocrine dysfunctions (2,5).
Recent discoveries indicate that ciliary dysfunction is associated with human obesity. Bardet-Biedl syndrome (BBS) is the prototypical human genetic disorder associated with ciliary dysfunction and obesity (6,7). To date, more than 12 BBS genes have been identified and mutations in BBS genes are associated with obesity, polydactyly and retinal degeneration (6–8). Additional clinical features of BBS include diabetes, hypertension, cognitive impairment, renal anomalies and hypogenitalism (6–8). Although BBS proteins are involved in microtubule-based protein/vesicle trafficking (6,9,10), the precise molecular function of each BBS protein has not been characterized. Likewise, the pathophysiological mechanisms leading to each component of the BBS phenotype are unknown.
Previously, we demonstrated that obesity in BBS mice is associated with hyperleptinemia and leptin resistance (11). However, since leptin production is proportional to the amount of body fat and, hyperleptinemia and leptin resistance are frequently associated with obesity in animal models and humans (3,12–14), leptin resistance in BBS mice could be secondary to obesity, as opposed to being the cause of obesity. Here, we addressed this question and uncovered the molecular defects leading to energy imbalance in BBS. Using a BBS knockout mouse model (Bbs2−/−) (15), which recapitulate the major components of the human phenotype including obesity, we demonstrate that BBS proteins are required for LepR signaling. We confirmed this with additional BBS knockout mouse models (Bbs4−/− and Bbs6−/−) (16,17). We also demonstrate that BBS proteins physically interact with the LepR and mediate LepR trafficking. Our findings indicate that attenuated LepR signaling owing to altered trafficking of the LepR is the major cause of obesity in BBS.
RESULTS
Bardet-Biedl syndrome mice have defects in the hypothalamic leptin–melanocortin axis
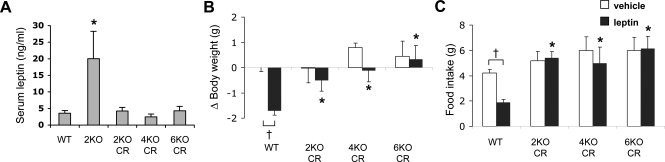
To test whether the leptin resistance in BBS mice is secondary to obesity, we examined the appetite-suppressing and weight-reducing effects of exogenous mouse leptin in BBS mice that had their circulating leptin levels normalized by calorie restriction. Bbs2−/−, Bbs4−/− and Bbs6−/− mice were given 70–80% of the chow pellets normally consumed daily by sex- and age-matched wild-type mice. This calorie-restriction protocol effectively prevented obesity and reduced serum leptin levels in BBS mice (Fig. 1A). While Bbs2−/−, Bbs4−/− and Bbs6−/− mice fed ad libitum showed 4–10-fold higher serum leptin levels [Fig. 1A and (11)], the leptin levels of calorie-restricted Bbs2−/−, Bbs4−/− and Bbs6−/− mice were comparable with wild-type animals. Intracerebroventricular (ICV) administration of leptin caused a significant reduction in food intake and body weight in wild-type mice. In contrast, ICV leptin failed to reduce food intake and body weight in Bbs2−/−, Bbs4−/− and Bbs6−/− mice despite the normal serum leptin levels (Fig. 1B and C). Food intake adjusted to body weight yielded similar results (data not shown). These results indicate that leptin resistance in BBS mice is not secondary to obesity. Rather, these results suggest leptin resistance as the cause of obesity in BBS mice.
Figure 1.
Bardet-Biedl syndrome mice are resistant to leptin. (A) Serum leptin levels of wild-type and calorie-restricted (CR) Bbs2−/− (2KO), Bbs4−/− (4KO) and Bbs6−/− (6KO) mice. Serum leptin level of normally fed Bbs2 null mice was shown for comparison (n = 4–16). (B, C) Bbs2, Bbs4 and Bbs6 null mice with normal circulating leptin levels are still resistant to the weight- and appetite-reducing action of leptin. Changes in body weight and food intake were measured 24 h after ICV leptin administration (n = 5–16). Data are mean ± SEM. *P < 0.05 compared with wild-type mice; †P < 0.05 versus vehicle.
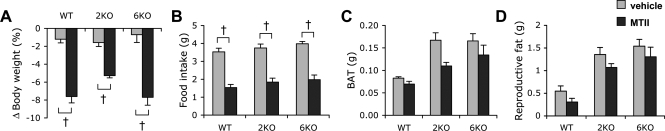
Melanocortin receptors, which are downstream to LepR signaling, are an important node for energy homeostasis (1,4). Therefore, we examined whether the ability of the melanocortin receptors to alter metabolism is also impaired in BBS mice. For this, we tested the effect of ICV administration of melanotan II (MTII), a melanocortin receptor agonist, on body weight and food intake (Fig. 2A and B). In contrast to leptin, ICV MTII reduced food intake and body weight in both wild-type and Bbs2−/− and Bbs6−/− mice. In addition, MTII administration reduced brown and white adipose tissues in both control and BBS animals (Fig. 2C and D). These data indicate that the ability of the melanocortin receptors to influence metabolism is intact in BBS mice. Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) results also demonstrated that the expression of melanocortin-3 and -4 receptors was normal in BBS mice (Supplementary Material, Fig. S1). Combined, these data indicate that obesity in BBS animals is owing to defect(s) downstream or at the level of LepR but upstream of melanocortin receptors in the hypothalamic leptin–melanocortin axis.
Figure 2.
Bardet-Biedl syndrome mice are responsive to melanocortin receptor agonist, MTII. (A–D) BBS mice are responsive to the weight-, appetite- and fat mass-reducing effect of MTII. Wild-type, Bbs2−/− and Bbs6−/− mice were injected with MTII or vehicle, and changes in body weight and food intake were measured 24 h later. Brown adipose tissue and reproductive fat were measured after death in vehicle- or MTII-injected mice (n = 4–7). Data are mean ± SEM. *P < 0.05 compared with wild-type mice; †P < 0.05 versus vehicle.
Attenuated leptin receptor signaling in Bardet-Biedl syndrome animals
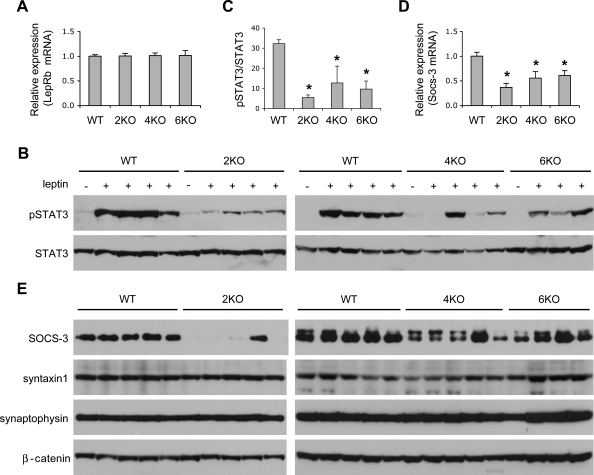
To determine the specific defect leading to leptin resistance in BBS mice, we examined the expression and signaling capability of the long isoform of the leptin receptor (LepRb), the isoform that has signaling capacity within the hypothalamus (18,19). The mRNA levels of LepRb were not altered in Bbs2−/−, Bbs4−/− and Bbs6−/− mice compared with wild-type controls (Fig. 3A). To examine whether the signaling capacity of the LepRb is normal in BBS mice, we compared the ability of leptin to activate hypothalamic signal transducer and activator of transcription-3 (STAT3) between wild-type and BBS mice. As above, leptin levels in BBS mice were normalized by calorie restriction to avoid leptin resistance secondary to obesity. In control mice, leptin caused a robust increase in STAT3 phosphorylation (Fig. 3B). However, leptin-induced STAT3 phosphorylation was greatly reduced in Bbs2−/−, Bbs4−/− and Bbs6−/− mice (to 17–39% of the control) albeit normal serum leptin levels in those mice (Fig. 3B and C). These data demonstrate that leptin resistance in BBS mice is caused by inability of the LepR to activate the downstream intracellular machinery associated with this receptor. In diet-induced obese mice, leptin resistance is thought to be owing to an increase in suppressor of cytokine signaling-3 (Socs-3), one of the targets induced by STAT3 and a negative feedback inhibitor of the LepR signaling (20,21). We found that both mRNA (Fig. 3D) and protein (Fig. 3E) levels of Socs-3 were decreased in Bbs2−/−, Bbs4−/− and Bbs6−/− animals, indicating that leptin resistance in BBS mice was not because of increased expression of Socs-3 in contrast to what is observed in diet-induced obesity model. The potential role of other mechanisms such as protein–tyrosine phosphatase 1B and the cyclic AMP responsive element-binding protein-1 (Creb1)-regulated transcription coactivator-1 in the leptin resistance associated with BBS remains to be determined (22,23). We also found that Bbs2−/−, Bbs4−/− and Bbs6−/− mice had normal expression of Syntaxin 1 (Fig. 3E), which was previously shown to be downregulated in mice deficient in leptin (ob/ob) or LepRb (db/db) (24). Synaptophysin is a pan-neuronal marker and its level was not changed in Bbs2−/−, Bbs4−/− and Bbs6−/− mice, indicating that there was no gross loss of neurons in BBS mice (Fig. 3E). In summary, our data indicate that leptin resistance in BBS mice is because of attenuated LepR signaling in the hypothalamus.
Figure 3.
Impaired LepRb signaling in Bardet-Biedl syndrome (BBS) mice. (A) Expression levels of the long form LepR (LepRb). Relative amounts of LepRb mRNAs were measured by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) after normalization with RPL19. Expression levels in wild-type were arbitrarily set at 1 (n = 6–9). (B) STAT3 phosphorylation upon leptin administration was reduced in BBS mice. BBS mice were calorie-restricted as above to normalize serum leptin levels and injected with leptin. Hypothalamic protein extracts were analyzed by standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blotting. (C) Quantification of phosphorylated STAT3 from data used in (B). (D) Socs-3 mRNA level was decreased in BBS mice. Socs-3 mRNA level was determined by qRT-PCR as above (n = 4–8). (E) Protein levels of Socs-3, Syntaxin1 and Synaptophysin. β-catenin was used as a loading control. Data are mean ± s.e.m. *P < 0.05 compared with wild-type mice.
Defective proopiomelanocortin neurons in Bardet-Biedl syndrome animals
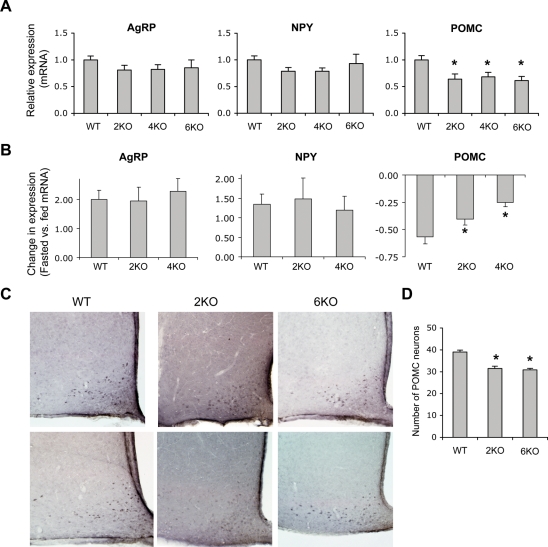
To gain further insight into the molecular and cellular mechanisms of obesity in BBS animals, we examined the expression levels of key hypothalamic genes involved in leptin action. Consistent with our previous findings (11), the expression levels of the orexigenic Agrp and Npy genes were normal, whereas the expression level of the anorexigenic Pomc gene was significantly lower in Bbs2−/−, Bbs4−/− and Bbs6−/− mice compared with wild-type controls (Fig. 4A). Next, we tested whether the hypothalamic orexigenic and anorexigenic genes responded properly to variations in the peripheral energy balance in BBS mice by comparing the mRNA levels of these neuropeptides in normally fed and 48 h fasted animals. In wild-type mice, AgRP and NPY mRNA levels increased and POMC mRNA levels decreased after fasting (Fig. 4B). In Bbs2−/− and Bbs4−/− mice, changes in AgRP and NPY mRNA levels in response to fasting were similar to that of wild-type controls. In contrast, fasting-induced decrease in POMC mRNA levels was significantly less in Bbs2−/− and Bbs4−/− mice when compared with wild-type controls (Fig. 4B). These data indicate that POMC neurons of BBS mice do not respond properly to changes in peripheral energy balance. Thus, POMC neurons appear to be selectively affected in BBS mice downstream of LepR signaling.
Figure 4.
Defective POMC neurons in Bardet-Biedl syndrome (BBS) mice. (A) Hypothalamic levels of agouti-related protein (AgRP), neuropeptide Y (NPY) and proopiomelanocortin (POMC) mRNA in normally fed Bbs2, Bbs4 and Bbs6 null mice when compared with wild-type controls (n = 6–9). (B) Effect of 48 h fasting (relative to the normally fed state) on hypothalamic AgRP, NPY and POMC mRNA levels in wild-type, Bbs2 and Bbs4 null mice (n = 4–5). (C) Immunohistochemical staining of POMC-positive neurons. Coronal sections of the wild-type, Bbs2−/− (2KO) and Bbs6−/− (6KO) brains were stained with anti-POMC antibodies and representative sections are shown. (D) Average number of POMC-positive neurons in the arcuate nucleus. POMC-positive cells in five sections within the similar position from each animal were counted and average number of POMC neurons per hemisphere is shown (n = 3–4). Data are mean ± SEM. *P < 0.05 compared with wild-type mice.
To test whether a reduction in the number of POMC neurons may be the main defect in BBS mice, we compared the number of these neurons in the hypothalamic arcuate nucleus between wild-type and BBS mice. Immunohistochemistry results revealed that there was a slight reduction (approximately 20%) in POMC-positive cells in Bbs2−/− and Bbs6−/− animals (Fig. 4C and D). However, this reduction in POMC neurons cannot explain the approximately 40% reduction in Pomc gene expression or the more than 60% decrease in STAT3 phosphorylation. In addition, immunostaining of POMC in BBS animals was often weak (which is consistent with the lower levels of POMC mRNA) suggesting that the number of POMC neurons might be underestimated in BBS mice. Combined with our biochemistry and gene expression data, these results indicate that leptin resistance in BBS mice is mostly because of attenuated LepR signaling.
Bardet-Biedl syndrome proteins interact with LepRb and mediate LepRb trafficking
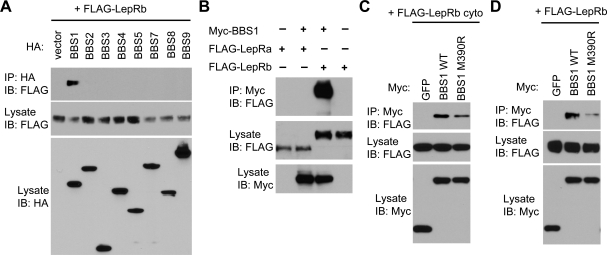
Recently, it was found that seven BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8 and BBS9) form a stable complex known as the BBSome (9). This complex is proposed as the functional unit that mediates protein/vesicle trafficking to cilia. We postulated that some of the BBSome subunits might interact with the LepRb and/or STAT3 to mediate their trafficking. Strikingly, we found that BBS1 protein specifically interacted with the LepRb in transient transfection and co-immunoprecipitation assays (Fig. 5A). No other BBSome subunits showed interaction with LepRb. In addition, the interaction between BBS1 and LepR was specific to the signaling isoform of the receptor, LepRb, because the non-signaling isoform, LepRa, did not interact with BBS1 (Fig. 5B). Consistent with this, we found that the C-terminal cytoplasmic domain of LepRb, which is unique to the LepRb, was sufficient to interact with BBS1 (Fig. 5C). We did not detect any interaction between STAT3 and BBS proteins (data not shown). The BBS1 M390R mutation, which is the most common mutation found in human BBS patients and sufficient to induce BBS phenotypes including obesity in a knock-in mouse model (25), greatly reduced the ability of BBS1 to interact with the LepRb (Fig. 5D). The BBS1 M390R mutant also showed decreased interaction with the LepRb cytoplasmic domain (Fig. 5C). To test whether the interaction between BBS1 and LepRb occurs in living cells, we transfected ARPE-19 cells with BBS1 either alone or with the LepRb. We found that although overexpressed BBS1 proteins were detected throughout the cytoplasm, co-transfection of LepRb with BBS1-restricted BBS1 localization to small punctuates in ARPE-19 cells (Supplementary Material, Fig. S2), supporting the physical interaction of LepR with BBS1. Altogether, these findings indicate that LepRb can be associated with the BBS1 subunit of the BBSome.
Figure 5.
Bardet-Biedl syndrome (BBS) proteins mediate LepRb trafficking. (A) Interaction of BBS1 with the LepRb. HA-tagged BBS proteins (BBS1–5 and 7–9) were transiently transfected with FLAG-tagged LepRb and interactions were examined by co-immunoprecipitation assays. Expression levels of each BBS protein and LepRb in the lysates were shown in the bottom and middle panels, respectively. Co-immunoprecipitated LepRb was shown in the top panel. IP, immunoprecipitation; IB, immunoblotting. (B) The signaling isoform of LepR, LepRb, but not the short isoform LepRa, interacts with BBS1. (C) C-terminal cytoplasmic domain of LepRb (LepRb cyto) is sufficient to interact with BBS1. Myc-GFP was used as a negative control. (D) BBS1 M390R mutation disrupts the BBS1–LepRb interaction. Others are the same as in (A).
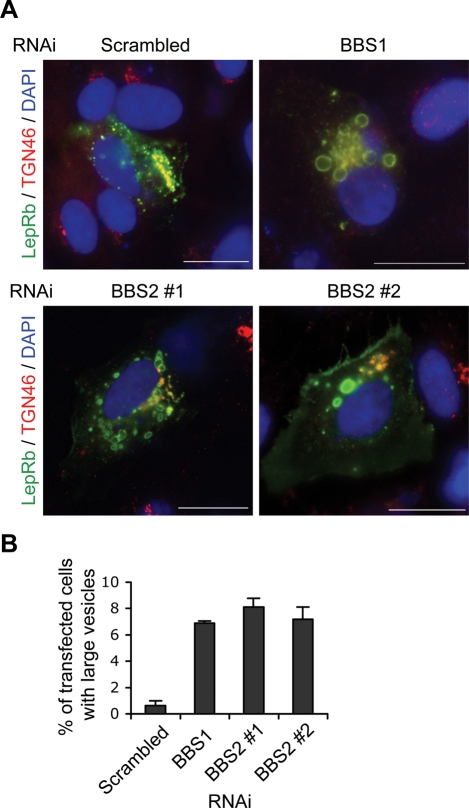
Finally, we tested whether LepRb trafficking was altered in the absence of BBSome proteins. Consistent with previous studies of the localization of the LepR (26,27), most LepRb immunoreactivity was found in the trans-Golgi network (TGN) and small vesicles, and only a very low level of the LepRb was detected in the plasma membrane (Fig. 6A and Supplementary Material, Fig. S3). Some of the small vesicles showing immunoreactivity for LepRb were positive for TGN46, a marker for post-Golgi vesicles as well as TGN (28), suggesting that these vesicles are in the secretory pathway or recycling endosomes. A fraction of the LepRb was also found in the early endosomes as marked by EEA1 (Supplementary Material, Fig. S3) (29). When BBS1 protein was depleted by shRNA-mediated RNA interference (Supplementary Material, Fig. S4), LepRb was found in abnormally large vesicles near the nucleus (approximately 7% of the transfected cells; Fig. 6). We found similar mistrafficking of LepRb when BBS2 protein was depleted (Fig. 6), indicating that the requirement of BBS proteins for LepRb trafficking is not limited to BBS1. These data suggest that BBS proteins are required for proper trafficking of the LepRb most likely between the Golgi and the ciliary and/or plasma membrane.
Figure 6.
Bardet-Biedl syndrome (BBS) proteins are required for LepRb trafficking. (A) Alteration of the LepRb trafficking in BBS1- and BBS2-depleted cells. ARPE-19 cells were transfected with FLAG–LepRb together with scrambled (control), BBS1 or BBS2 RNAi constructs. Localization of LepRb (green) and TGN46 (red) was probed by indirect immunofluorescence microscopy. Merged images with DAPI staining (blue; nuclei) are shown. (Scale bars: 10 µm). (B) Percentage of transfected cells (determined by FLAG immunofluorescence) with large vesicles. Values are average of three experiments and a minimum of 120 cells was counted in each experiment. Data are mean ± SEM.
DISCUSSION
BBS proteins have been implicated in protein/vesicle trafficking along the microtubule (9,10). However, how this general function is associated with individual components of the BBS phenotype was unknown. The present study provides the pathophysiological mechanism of obesity in BBS at the molecular level. Our study provides evidence that leptin resistance resulting from aberrant LepR trafficking and attenuated LepR signaling in the hypothalamus is a cause of obesity associated with BBS. We demonstrate that eliminating obesity and hyperleptinemia does not restore leptin sensitivity in BBS mice, indicating that leptin resistance is intrinsic rather than secondary to hyperleptinemia and obesity. We also demonstrate that the BBS1 protein physically interacts with the signaling isoform of LepR, LepRb and that BBS proteins are required for proper trafficking of the LepRb.
Our data suggest that BBS proteins mediate LepRb trafficking between the Golgi and specific areas of the plasma membrane (e.g. lipid raft, ciliary membrane). In the absence of BBS proteins, LepRb trafficking was perturbed and LepRb signaling was attenuated. This is consistent with the previous findings that the BBS proteins are necessary for the localization of G protein-coupled receptors (GPCRs) to neuronal cilia (30). Although we tested several antibodies against LepR, we were not able to detect LepRb in the mouse brain slices. This may be owing to the low level of surface expression and fast turnover of the LepRb (26,27). However, LepRb has been found in the cilium of the olfactory epithelial cells (31), suggesting that LepRb may localize to the cilium of hypothalamic neurons.
Of the BBSome subunits, only BBS1 showed physical interaction with LepRb. We previously demonstrated that BBS1 also interacted with Rabin8, a GDP/GTP exchange factor for Rab8 (9). These observations raise the possibility that BBS1 is the cargo-binding subunit of the BBSome. Thus, it is of interest to test whether the GPCRs mislocalized in BBS mutant neurons (e.g. Sstr3 and Mchr1) also interact with BBS1 (30). However, similar LepRb trafficking defects observed in BBS2 depleted cells and no obvious genotype–phenotype correlation among various BBS mutations indicate that integrity of the BBSome is important for its function in trafficking and that loss of any BBSome subunits could lead to the same phenotype (mistrafficking of the cargo).
Interestingly, in BBS animals loss of LepRb function appears to be selective. Indeed, although expression of Agrp, Npy and Pomc are all regulated by leptin (1), only Pomc gene expression was affected in BBS mice. Thus, the loss of leptin action appears to be restricted to the POMC neurons. Genetic ablation of LepRb specifically in POMC neurons leads to hyperphagia and increased fat mass in mice (32,33). In BBS mice, attenuated LepRb signaling in POMC neurons is likely the main cause of obesity. These results are consistent with the recent findings by Davenport et al. (34) showing that disruption of intraflagellar transport proteins, Tg737 or Kif3a, which are required for ciliogenesis, specifically in POMC neurons resulted in hyperphagia and obesity in mice. In addition, while BBS mice are resistant to leptin's action to decrease body weight and food intake, they have a preserved sympathetic/cardiovascular response to leptin (11), supporting the selective loss of LepRb function in BBS mice. In fact, leptin signaling is not completely abolished in BBS mice. A fraction of the LepRb signaling is present as shown by residual STAT3 activation following leptin treatment. Thus, BBS proteins appear to be required for ‘efficient’ LepRb signaling, which is necessary to control feeding behavior, while ‘low’ level of LepRb signaling is still transduced in the absence of BBS proteins and this level of signaling is sufficient to increase sympathetic nerve activity and blood pressure. Localization of the LepRb to a specific domain in the plasma membrane may be required for this ‘efficient’ signaling, while LepRb elsewhere in the plasma membrane may be sufficient to transduce the ‘low’ level of signal. Alternatively, the requirement for BBS proteins in the LepRb signaling may be specific to certain cell types and/or downstream effectors. These are interesting questions that warrant further studies.
In humans, variants of BBS2, BBS4 and BBS6 genes have been reported to be associated with obesity in non-BBS individuals (35). Indeed, a variant in the BBS2 gene was related to common adult obesity, and polymorphisms in BBS4 and BBS6 genes were associated with common early-onset childhood obesity and common morbid adult obesity. Whether the specific variants of the BBS genes associated with common obesity affect LepRb signaling and/or trafficking remains to be determined.
In summary, we have identified the specific defect that accounts for obesity in mouse models of BBS. Our findings indicate that mistrafficking and attenuated signaling of the LepRb is the main cause of energy imbalance leading to obesity in BBS. This defect represents a novel pathophysiological mechanism of leptin resistance with potential implication for common human obesity.
MATERIALS AND METHODS
For a detailed description of the Materials and Methods, please see Supplementary Material, which is published as supporting information on the Human Molecular Genetics website.
Animals
Generation of Bbs2−/−, Bbs4−/− and Bbs6−/− mice and genotyping was described previously (15–17). The University of Iowa Animal Research Committee approved all protocols.
Leptin normalization/leptin resistance study
Sex- and age-matched controls and BBS mice were housed in individual cages at 6–8 weeks of age. Bbs2−/−, Bbs4−/− and Bbs6−/− animals were given 70–80% of food consumed by wild-type controls everyday until the day before leptin treatment. Animals were equipped with ICV cannulae as previously described (36). Vehicle or recombinant mouse leptin (2 µg in 1 µl, R&D Systems, Minneapolis, MN) was injected ICV. Mice were sacrificed 24 h after ICV leptin administration, and food intake and body weight were measured. For STAT3 phosphorylation analysis, food was removed 18 h before ICV injection. Leptin or vehicle was injected ICV and animals sacrificed 2 h later by CO2 asphyxiation. Hypothalami were quickly dissected and homogenized in the lysis buffer. Protein extracts were loaded onto 4–12% NuPAGE Bis–Tris gels and analyzed by immunoblotting.
MTII study
Sex- and age-matched wild-type controls and Bbs2−/− and Bbs6−/− animals were housed in individual cages at 9–13 weeks of age with free access to food and water. Two weeks later, mice were implanted with ICV cannulae as described above. Body weight and food intake were measured daily for four consecutive days before ICV injection and used as baseline values. For each genotype, the mice treated with vehicle or MTII were weight-matched. Animals were sacrificed 24 h after MTII (1 µg in 1 µl; Phoenix Pharmaceuticals Inc., Burlingame, CA) or vehicle injection by CO2 asphyxiation, and body weight, food consumption and the weights of brown adipose tissue and reproductive fat were measured as described previously (11,25,36).
RNA extraction and quantitative reverse transcriptase-polymerase chain reaction
RNA was extracted from mouse hypothalami using TRIzol Reagent (Invitrogen, Carlsbad, CA). Complementary DNA was synthesized from 1 µg of total RNA and used for qPCR with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and Mx3000P QPCR System (Stratagene, La Jolla, CA). Relative gene expression was calculated by the ΔΔCt method following normalization with RPL19 (37). The PCR products were confirmed by the melt-curve analysis and sequencing. Primer sequences are shown in Supplementary Material text.
Immunohistochemistry
Mice were perfused with 4% paraformaldehyde/0.5% glutaraldehyde in phosphate-buffered saline (2.5 ml/min; 50 ml). Entire brain was excised and incubated in the same fixative overnight at 4°C then transferred to 30% sucrose solution. Brains were vibratome-sectioned with 45 µm thickness. Free-floating sections were permeabilized in phosphate-buffered saline with 0.25% Triton X-100 (0.25% Triton X-100), blocked by 3% normal goat serum and decorated with rabbit anti-POMC antibody (1:4000), biotinylated goat anti-rabbit IgG (1:200) and Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA).
Transfection and co-immunoprecipitation
HEK293T cells were transfected in six-well plates with 1 µg of CS2FLAG-LepR and 1 µg of indicated HA-tagged BBS proteins using FuGENE HD (Roche Applied Science, Indianapolis, IN). Cell lysates were immunoprecipitated with anti-Myc (9E10; SantaCruz, Santa Cruz, CA) or anti-HA (F-7; SantaCruz, Santa Cruz, CA) antibodies conjugated to agarose. Precipitated proteins were analyzed by standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting.
Immunofluorescence
ARPE-19 cells were seeded on glass cover slips in 24-well plates and transfected with total 1 µg of DNA using FuGENE HD (Roche, Indianapolis, IN). Cells were fixed with methanol for 6 min at −20°C, blocked with 5% BSA and 3% normal goat serum and decorated with indicated primary antibodies. Alexa Flour 488 goat anti-mouse IgG (Invitrogen, Carlsbad, CA) and Alexa Flour 568 goat anti-rabbit IgG (Invitrogen, Carlsbad, CA) were used to detect primary antibodies.
Statistical analysis
Results were expressed as mean ± SEM. Comparisons between groups were made by one-way or two-way analysis of variance (ANOVA) with Fisher LSD method post-test analysis for pairwise multiple comparisons, as appropriate. P < 0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
US National Institutes of Health (HL084207 to K.R. and V.C.S.) and (EY011298 and EY017168 to V.C.S.) and by the American Heart Association (0530274N to K.R.). V.C.S. is an investigator of the Howard Hughes Medical Institute. Funding to Pay the Open Access Charge was provided by Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Yves Rouillé for providing LepRa and LepRb expression plasmids.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Morton G.J., Cummings D.E., Baskin D.G., Barsh G.S., Schwartz M.W. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 2.Coll A.P., Farooqi I.S., O'Rahilly S. The hormonal control of food intake. Cell. 2007;129:251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahima R.S., Flier J.S. Adipose tissue as an endocrine organ. Trends Endocrinol. Metab. 2000;11:327–332. doi: 10.1016/s1043-2760(00)00301-5. [DOI] [PubMed] [Google Scholar]
- 4.Cone R.D. Anatomy and regulation of the central melanocortin system. Nat. Neurosci. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- 5.Friedman J.M., Halaas J.L. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 6.Blacque O.E., Leroux M.R. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell. Mol. Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tobin J.L., Beales P.L. Bardet-Biedl syndrome: beyond the cilium. Pediatr. Nephrol. 2007;22:926–936. doi: 10.1007/s00467-007-0435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore S.J., Green J.S., Fan Y., Bhogal A.K., Dicks E., Fernandez B.A., Stefanelli M., Murphy C., Cramer B.C., Dean J.C., et al. Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am. J. Med. Genet. A. 2005;132:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 10.Yen H.J., Tayeh M.K., Mullins R.F., Stone E.M., Sheffield V.C., Slusarski D.C. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 11.Rahmouni K., Fath M.A., Seo S., Thedens D.R., Berry C.J., Weiss R., Nishimura D.Y., Sheffield V.C. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J. Clin. Invest. 2008;118:1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frederich R.C., Hamann A., Anderson S., Lollmann B., Lowell B.B., Flier J.S. Leptin levels reflect body lipid content in mice: evidence for diet-induced resistance to leptin action. Nat. Med. 1995;1:1311–1314. doi: 10.1038/nm1295-1311. [DOI] [PubMed] [Google Scholar]
- 13.Maffei M., Halaas J., Ravussin E., Pratley R.E., Lee G.H., Zhang Y., Fei H., Kim S., Lallone R., Ranganathan S., et al. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat. Med. 1995;1:1155–1161. doi: 10.1038/nm1195-1155. [DOI] [PubMed] [Google Scholar]
- 14.Considine R.V., Sinha M.K., Heiman M.L., Kriauciunas A., Stephens T.W., Nyce M.R., Ohannesian J.P., Marco C.C., McKee L.J., Bauer T.L., et al. Serum immunoreactive-leptin concentrations in normal-weight and obese humans. N. Engl. J. Med. 1996;334:292–295. doi: 10.1056/NEJM199602013340503. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura D.Y., Fath M., Mullins R.F., Searby C., Andrews M., Davis R., Andorf J.L., Mykytyn K., Swiderski R.E., Yang B., et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl Acad. Sci. USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fath M.A., Mullins R.F., Searby C., Nishimura D.Y., Wei J., Rahmouni K., Davis R.E., Tayeh M.K., Andrews M., Yang B., et al. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum. Mol. Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 17.Mykytyn K., Mullins R.F., Andrews M., Chiang A.P., Swiderski R.E., Yang B., Braun T., Casavant T., Stone E.M., Sheffield V.C. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl Acad. Sci. USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee G.H., Proenca R., Montez J.M., Carroll K.M., Darvishzadeh J.G., Lee J.I., Friedman J.M. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen H., Charlat O., Tartaglia L.A., Woolf E.A., Weng X., Ellis S.J., Lakey N.D., Culpepper J., Moore K.J., Breitbart R.E., et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 20.Bjorbaek C., Elmquist J.K., Frantz J.D., Shoelson S.E., Flier J.S. Identification of SOCS-3 as a potential mediator of central leptin resistance. Mol. Cell. 1998;1:619–625. doi: 10.1016/s1097-2765(00)80062-3. [DOI] [PubMed] [Google Scholar]
- 21.Munzberg H., Myers M.G., Jr Molecular and anatomical determinants of central leptin resistance. Nat. Neurosci. 2005;8:566–570. doi: 10.1038/nn1454. [DOI] [PubMed] [Google Scholar]
- 22.Bence K.K., Delibegovic M., Xue B., Gorgun C.Z., Hotamisligil G.S., Neel B.G., Kahn B.B. Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat. Med. 2006;12:917–924. doi: 10.1038/nm1435. [DOI] [PubMed] [Google Scholar]
- 23.Altarejos J.Y., Goebel N., Conkright M.D., Inoue H., Xie J., Arias C.M., Sawchenko P.E., Montminy M. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat. Med. 2008;14:1112–1117. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahima R.S., Bjorbaek C., Osei S., Flier J.S. Regulation of neuronal and glial proteins by leptin: implications for brain development. Endocrinology. 1999;140:2755–2762. doi: 10.1210/endo.140.6.6774. [DOI] [PubMed] [Google Scholar]
- 25.Davis R.E., Swiderski R.E., Rahmouni K., Nishimura D.Y., Mullins R.F., Agassandian K., Philp A.R., Searby C.C., Andrews M.P., Thompson S., et al. A knocking mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc. Natl Acad. Sci. USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diano S., Kalra S.P., Horvath T.L. Leptin receptor immunoreactivity is associated with the Golgi apparatus of hypothalamic neurons and glial cells. J. Neuroendocrinol. 1998;10:647–650. doi: 10.1046/j.1365-2826.1998.00261.x. [DOI] [PubMed] [Google Scholar]
- 27.Belouzard S., Delcroix D., Rouille Y. Low levels of expression of leptin receptor at the cell surface result from constitutive endocytosis and intracellular retention in the biosynthetic pathway. J. Biol. Chem. 2004;279:28499–28508. doi: 10.1074/jbc.M400508200. [DOI] [PubMed] [Google Scholar]
- 28.Ponnambalam S., Girotti M., Yaspo M.L., Owen C.E., Perry A.C., Suganuma T., Nilsson T., Fried M., Banting G., Warren G. Primate homologues of rat TGN38: primary structure, expression and functional implications. J. Cell Sci. 1996;109(Pt 3):675–685. doi: 10.1242/jcs.109.3.675. [DOI] [PubMed] [Google Scholar]
- 29.Mu F.T., Callaghan J.M., Steele-Mortimer O., Stenmark H., Parton R.G., Campbell P.L., McCluskey J., Yeo J.P., Tock E.P., Toh B.H. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine ‘fingers’ and contains a calmodulin-binding IQ motif. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 30.Berbari N.F., Lewis J.S., Bishop G.A., Askwith C.C., Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl Acad. Sci. USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baly C., Aioun J., Badonnel K., Lacroix M.C., Durieux D., Schlegel C., Salesse R., Caillol M. Leptin and its receptors are present in the rat olfactory mucosa and modulated by the nutritional status. Brain Res. 2007;1129:130–141. doi: 10.1016/j.brainres.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 32.Balthasar N., Coppari R., McMinn J., Liu S.M., Lee C.E., Tang V., Kenny C.D., McGovern R.A., Chua S.C., Jr., Elmquist J.K., et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 33.van de Wall E., Leshan R., Xu A.W., Balthasar N., Coppari R., Liu S.M., Jo Y.H., MacKenzie R.G., Allison D.B., Dun N.J., et al. Collective and individual functions of leptin receptor modulated neurons controlling metabolism and ingestion. Endocrinology. 2008;149:1773–1785. doi: 10.1210/en.2007-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davenport J.R., Watts A.J., Roper V.C., Croyle M.J., van Groen T., Wyss J.M., Nagy T.R., Kesterson R.A., Yoder B.K. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr. Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benzinou M., Walley A., Lobbens S., Charles M.A., Jouret B., Fumeron F., Balkau B., Meyre D., Froguel P. Bardet-Biedl syndrome gene variants are associated with both childhood and adult common obesity in French Caucasians. Diabetes. 2006;55:2876–2882. doi: 10.2337/db06-0337. [DOI] [PubMed] [Google Scholar]
- 36.Rahmouni K., Haynes W.G., Morgan D.A., Mark A.L. Role of melanocortin-4 receptors in mediating renal sympathoactivation to leptin and insulin. J. Neurosci. 2003;23:5998–6004. doi: 10.1523/JNEUROSCI.23-14-05998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.