Abstract
Free text fields are often used to store clinical drug data in electronic health records. The use of free text facilitates rapid data entry by the clinician. Errors in spelling, abbreviations, and jargon, however, limit the utility of these data. We designed and implemented an algorithm, using open source tools and RxNorm, to extract and normalize drug data stored in free text fields of an anesthesia electronic health record. The algorithm was developed using a training set containing drug data from 49,518 cases, and validated using a validation set containing data from 14,655 cases. Overall sensitivity and specificity for the validation set were 92.2% and 95.7% respectively. The mains sources of error were misspellings and unknown but valid drug names. These preliminary results demonstrate that free text clinical drug data can be efficiently extracted and mapped to a controlled drug nomenclature.
Introduction
The acquisition of accurate clinical drug data is an important problem in electronic health records1,2. While pick lists of pre-defined drug names and auto-completion are strategies that facilitate standardized data capture, the vast number of generic and brand name formulations available limit the utility of these approaches. One alternative, free text input, allows the user to record drug data quickly, but with poor data integrity. Errors in spelling, punctuation, and dosage complicate the utility of such data for medical, scientific, and administrative purposes.
The current preliminary report describes the design, development, and validation of a generic postprocessing algorithm to parse and normalize free text clinical drug data stored in an electronic health record into a standardized terminology. The aim of the system was to transform free text drug data into a form that can be used for export into other applications. We chose to create such a system using open source tools and RxNorm - the National Library of Medicine (NLM) repository of standard names for clinical drugs.
Methods
As a data source for development and testing, we used free text preoperative drug history data that had been typed into an anesthesia electronic health record (CompuRecord, Philips Medical Systems, Andover, MA) by attending/resident anesthesiologists, and subsequently imported into a relational database. The main software components used were as follows:
MySQL 5.0 (MySQL AB, Uppsala, Sweden). MySQL was used to store all configuration, reference information (drug vocabularies), input and output data.
RxNorm3,4. RxNorm is a cross-referenced lexicon of clinical drug nomenclature that is available at no cost. RxNorm was used as the reference source for drug name verification and for mapping trade (proprietary) names to their generic equivalents.
Metaphone5. The Metaphone algorithm6 is a variant of the Soundex algorithm (first used by census takers to classify similar sounding surnames by consonant groupings). Both work by mapping an input string to an encoding which is a rough approximation of the string’s English phonetic pronunciation. Strings that map to the same encoding are considered to have the same pronunciation. Practically, this has the effect of correcting for misspelling and typographical errors. Metaphone augments Soundex by additionally analyzing diphthongs, which makes it better suited to drug names.
We began by selecting a set of generic drugs that were appropriate to a specific medical domain–anesthesiology. We used a process of iterative frequency analysis of the training set. There was a clear cutoff point below which usage of any particular generic drug became uncommon in the electronic records.
Subsequently, the list of trade names was generated automatically by using the “tradename_of” relationship defined in RxNorm. Briefly, the RXNCONSO (“concept”) table was first queried to find the RxNorm Concept Unique Identifier (RXCUI) for each generic drug. The RXNREL (“relationship”) table was then queried to find all of the unique trade names associated with these generic drug RXCUI’s, as identified by the “tradename_of” relationship.
Hints List
The Hints List contained medical jargon, abbreviations (e.g., “dig” expands to “digoxin”), and common misspellings that were found to cause trouble for the Metaphone algorithm (i.e., truncated trailing consonants, swapped consonants, or missing syllables). It was developed via a process of iterative frequency analysis of the training data set. Additionally, several commonly used trade names that were not included in the version of RxNorm used for the project were placed in the Hints List. Two internet-based lists of common medical abbreviations7, 8 were also consulted during the training process to help the authors in expanding unrecognized/unfamiliar abbreviations.
Pre-processing and Ignore List Creation
The input strings were pre-processed in order to reduce the number of false positive matches. Clinicians often intermingled clinical drug names with commentary and other non-drug terms, and these non-drug terms interfered with the accuracy of the algorithm. These extraneous data were removed using regular expressions generated dynamically at runtime from a static list of terms and patterns to be removed. This Ignore List was manually compiled by the authors in a manner analogous to the creation of the Hints list.
Token generation
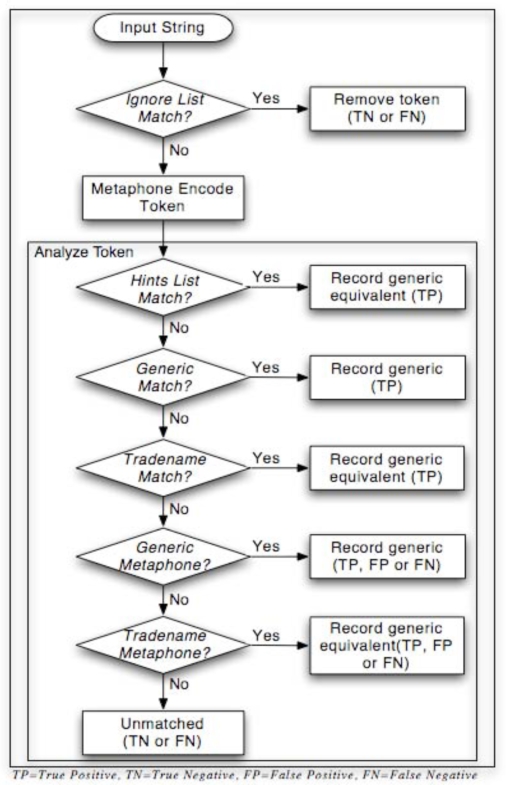
After normalizing all common input delimiters (e.g., tab, comma, semi-colon) to a single delimiter, Perl’s built-in split function was used to generate raw tokens. Both the Generic List and the Trade Name List were then encoded using Metaphone to create a Generic Metaphone List and a Trade Name Metaphone List. Tokens were then analyzed shown in Figure 1.
Figure 1.
Parsing Algorithm
Classification of Errors
As part of the validation of the algorithm, data on the type and number of errors were collected. There were several potential sources of error in the algorithm (see Figure 1):
False negatives were defined as: valid drug names that were inadvertently ignored and discarded during pre-processing; valid, known drug names that were misspelled, or inadvertently truncated during preprocessing and therefore not matched by Metaphone; or valid, unknown drug names that were spelled correctly, but were not matched because they were not among the commonly used generic or trade names included in our lists.
False positives included valid, unknown drug names that Metaphone mistakenly matched to a known generic or trade name; drug names misspelled such that they became a “sound-alike” to a known drug name and were mistakenly matched by Metaphone; or drug names that mapped to the same representation during Metaphone encoding (namespace collision). These false positives also generated a corresponding false negative (see example in Results).
Results
Using RxNorm (08/02/06 release), 6,931 trade names were found for 481 generic drugs. An additional 26 commonly used generic drugs had no trade name matches in RxNorm. These 507 generic drugs constituted the Generic List. The Ignore List contained 446 terms and patterns, and the Hints List contained 418 terms.
Two data sets were used during development, one for training and the other for validation of the algorithm. The training set consisted of 49,518 cases performed from 2002–2005. A total of 60,946 free text entries and 190,231 tokens were extracted. The validation set consisted of 14,655 cases performed in 2006. A total of 16,653 free text entries and 59,582 tokens were extracted.
The overall match rate, defined as the raw percentage of analyzed tokens (excluding ignored tokens) that registered as a match, is shown in Table 1.
Table 1.
Overall Match Rate
| Generic | Trade | Metaphone | Total | |
|---|---|---|---|---|
| Training | 35.6% | 49.5% | 7.7% | 92.9% |
| Validation | 36.5% | 48.3% | 7.3% | 92.3% |
Training Set Results
Of the 190,231 tokens in the set, 41,052(21.6%) were ignored and 149,179 tokens were further evaluated. There were 484 distinct trade names found. The Hints List matches accounted for 11.1% of the total. Of the 14.9% of tokens that did not match exactly, Metaphone matched an additional 3.1% to generic drugs and 4.6% to trade names. There were 10,580 (7.1%) terms that remained unmatched, 4,808 of which were unique.
Validation Set Results
Of the 59,582 tokens in the set, 11,341 tokens were ignored (19%) and 48,241 were further evaluated. The matching results are shown in Table 1. There were 460 distinct trade names found. The Hints List accounted for 11.2% of the total matches. Of the 15.2% of tokens that did not match exactly, Metaphone matched an additional 3.1% to generic drugs and 4.2% to trade names. There were 3,694 (7.7%) terms that remained unmatched, 1,954 of which were unique.
Classifiable Errors
Errors rates were estimated for the Validation Set as follows:
Pre-processing: Author AD reviewed a sample set of tokens from 1000 cases and the number of valid drug names that were inadvertently ignored was estimated to be 3.5%. For the Validation Set this corresponded to approximately 400 false negative tokens.
Namespace collisions: These could be calculated precisely. There was only one collision when Metaphone encoded the Generic List. The Trade Name List, in contrast, had 820 collisions, or close to 12% of total entries. These contributed to and were included in the misclassifications.
Misclassifications: The Metaphone matches were reviewed. Metaphone misclassified about 15% of the tokens that it matched (1.1% of all tokens). The error rate for trade names was approximately three times that of the generics. Of the generic name misclassifications, half could be accounted for by a single drug name mismatch (i.e., nystatin incorrectly mapped to nizatidine). As discussed in the Methods, this was logically a false negative for nystatin as well as a false positive for nizatidine. Other misclassifications were similar and also generated simultaneous false positive/false negative results.
Unmatched: The 3,694 tokens that did not match were reviewed and approximately 75% (2770) were found to be false negatives. These false negatives were equally distributed among valid generic or trade names not contained in any list, and misspellings that Metaphone did not classify correctly. The remaining 25% of unmatched tokens were true negatives.
Sensitivity and Specificity
Using the estimated error rates described above, sensitivity, specificity, positive predictive value and negative predictive value were calculated for the validation set. The results are shown in Table 2.
Table 2.
Validation Set Sensitivity & Specificity
| Sens. | Spec. | PPV | NPV | |
|---|---|---|---|---|
| Pre-processing | 99.2% | 92.2% | 98.1% | 96.5% |
| Metaphone | 61.1% | 76.9% | 85% | 48% |
| Overall | 92.2% | 95.7% | 98.8% | 76.2% |
Discussion
The reliable identification of medication data contained within free text fields has practical application. The current paper describes a generic post-processing algorithm that was developed and implemented to parse and normalize free text preoperative medication data stored in an anesthesia electronic health record into a form that can be used for export into other applications for multiple purposes. Particular attention was paid to the use of open source, freely available programming tools and resources, and in particular RxNorm. While several reports on the use of the UMLS Metathesaurus appear in the literature9–11, there are few, if any, documented uses of RxNorm. In our case, the use of RxNorm greatly enhanced the project since close to 50% of all identified tokens were trade names. The list of nearly 7,000 trade names found in RxNorm provided excellent coverage and nearly 500 distinct trade names were found in both the Training and Validation Sets.
The success of the algorithm was bolstered significantly by the use of the Hints List (more than 11% of the total matches in both sets). Metaphone contributed less than had been expected but still added more than 7% additional matches in both sets, although unfortunately many of these were not true matches. Additionally, the order in which tokens were analyzed was found to be important. Generic names had to be matched before trade names, and raw tokens before their Metaphone encodings. Matching against trade names or encodings too early resulted in excessive false positives because many trade names are phonetically similar to unrelated generic drug names.
Related work
As part of the preliminary work leading up to the creation of RxNorm, the NLM and the Veterans Administration (VA) first experimented with converting drugs listed in the VA National Drug File (VANDF) into a Semantic Normal Form (SNF). Of the 93,029 entries in the VANDF, they were able to algorithmically parse 70%.12 This project differed considerably from the current effort, which made no attempt to capture complete semantic information (dose, route, strength, etc). Nonetheless it is informative that even starting with a “clean, well-maintained file” (as the authors describe the VANDF), accurate automated drug name extraction was difficult.
More recently, Sirohi and Peissig13 published their work extracting medication names from the electronic medical record used by a single medical center. They used a commercially purchased natural language parser and focused their efforts on selecting the best drug lexicon to use with that product. As a data source they used the National Drug Data File (NDDF) (First DataBank, Inc., San Bruno, CA) — one of the seven source vocabularies incorporated into RxNorm. They also used classification information from the American Hospital Formulary Service (AHFS) (American Society of Health Pharmacists, Bethesda, MD). They experimented with three lexicons derived from the NDDF, ultimately settling on one which included short names (analogous to abbreviations), and excluded terms deemed not to be true drug names though a process of manual review and comparison with freely available English word lists. Their final medication list included 22,345 drug names, versus the 7,438 unique trade and generic names used in this project. Although the number of tokens in their training and test sets were of comparable magnitude to the current study, the number of occurrences of actual medication names was quite small, on the order of several hundred for each set. Thus they were able to fully verify the results of their extraction in contrast to the current study. It is notable that Sirohi and Peissig’s work and the current investigation achieved similar sensitivity and specificity, suggesting that these are reasonable results, given the available technology and source databases.
Limitations and Future Directions
There are several limitations to the algorithm as currently implemented. The false negative rate can be attributed primarily to the large number of unmatched terms that were valid drug names. This could be addressed by expanding the scope of the generic list for this domain. Updating RxNorm to the most current version would also help capture some of the trade names that were missed, as the NLM is continuously adding drug names to the RxNorm database. Expanded lists would also reduce the number of misclassification errors, by reducing the use of Metaphone. An alternate approach to addressing these unknown drug names would be to change the design of the algorithm so that RxNorm was directly queried for each term. This would in theory allow the capture of all valid drug names. However, preliminary experiments with such a method during the development process indicated that it might be unreasonably slow, because of the size of the RxNorm tables and the type of joins required. Further work would need to be done to optimize such a potential approach.
Another limitation was the disappointingly low sensitivity and specificity of the Metaphone algorithm. This is not entirely unsurprising given that Metaphone was designed for identifying surnames, not drug names. Extending and modifying the Metaphone algorithm could address this issue. An alternate strategy would be to modify the algorithm to look up any unmatched tokens directly in RxNorm before using Metaphone. This might represent an acceptable compromise between full ad-hoc querying of RxNorm and the current approach.
The terms and patterns in the Ignore List could also be refined. For example, some patterns were too broad or resulted in word fragments. The reported algorithm was also not designed to handle drugs where the name consists of more than one term (e.g., calcium chloride versus calcium gluconate). Moreover, the system does not construct a true Semantic Normal Form for the drug, as defined in RxNorm. Handling this type of information would add considerable complexity since it requires looking at each token’s surrounding context and building a true parse tree.
One additional achievable improvement might include more extensive use of RxNorm’s capabilities. The current implementation only exploits the RxNorm “tradename_of” relationship. There are other relationships defined in RxNorm, such as “consists_of”, which would allow for improved identification of individual drug components of compound formulations. (Currently, compound formulations were handled on a per-drug basis.) Drug classification data were also limited and could be made substantially more complete, for example by using the AHFS Classification schema.
Conclusion
Reliable identification and extraction of medication data from free text can be accomplished successfully using freely available tools and resources. In particular, RxNorm has proven to be a practical and convenient data source for trade name information. The current algorithm achieved an overall sensitivity and specificity of 92.3% and 95.7%, respectively. Continued refinement of the algorithm, as well as greater use of RxNorm features, should enable further increases in accuracy.
References
- 1.Cimino JJ, Patel VL, Kushniruk AW. Studying the human-computer-terminology interface. J Am Med Inform Assoc. 2001 Mar-Apr;8(2):163–73. doi: 10.1136/jamia.2001.0080163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sittig DF. Grand challenges in medical informatics. J Am Med Inform Assoc. 1994 Sep-Oct;1(5):412–3. doi: 10.1136/jamia.1994.95153429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RxNorm 2004[cited 2006 08/02]; Available from: http://www.nlm.nih.gov/research/umls/rxnorm/index.html
- 4.Simon L, Wei M, Robin M, Vikraman G, Stuart N. RxNorm: prescription for electronic drug information exchange. 2005. pp. 17–23.
- 5.Schwern M.Text-Metaphone1.96 ed; 1999. Available from: http://search.cpan.org/dist/Text-Metaphone/
- 6.Philips L. Hanging on the metaphone. Computer Language. 1990 Dec;7(12):39–43. [Google Scholar]
- 7.Berman J.Biomedical Abbreviations 2001[cited 2006; Available from: http://www.julesberman.info/abbtwo.htm
- 8.Goldberg H, Goldsmith D, Law V, Keck K, Tuttle M, Safran C. An evaluation of UMLS as a controlled terminology for the Problem List Toolkit. Medinfo. 1998;9 Pt 1:609–12. [PubMed] [Google Scholar]
- 9.Friedman C, Liu H, Shagina L, Johnson S, Hripcsak G. Evaluating the UMLS as a source of lexical knowledge for medical language processing. Proc AMIA Symp; 2001. pp. 189–93. [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg HS, Hsu C, Law V, Safran C. Validation of clinical problems using a UMLS-based semantic parser. Proc AMIA Symp; 1998. pp. 805–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc. 2001 Jan-Feb;8(1):80–91. doi: 10.1136/jamia.2001.0080080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson SJ, Brown SH, Erlbaum MS, Olson N, Powell T, Carlsen B, et al. A semantic normal form for clinical drugs in the UMLS: early experiences with the VANDF. Proc AMIA Symp; 2002. pp. 557–61. [PMC free article] [PubMed] [Google Scholar]
- 13.Sirohi E, Peissig P. Study of effect of drug lexicons on medication extraction from electronic medical records. Pac Symp Biocomput; 2005. pp. 308–18. [DOI] [PubMed] [Google Scholar]