Abstract
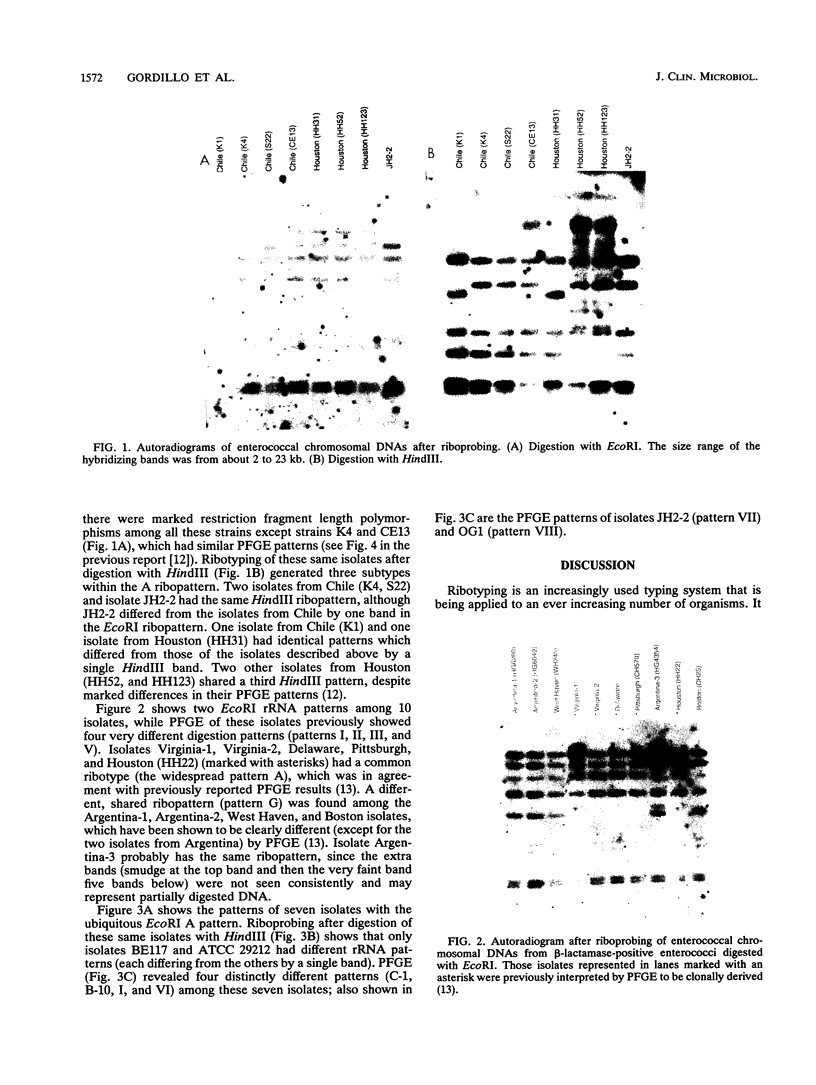
Hybridization of EcoRI- and HindIII-digested chromosomal DNAs from 41 isolates of Enterococcus faecalis with probes for rRNA genes was performed (ribotyping). The ability of ribotyping to distinguish strains at the subspecies level was compared with results previously determined by pulsed-field gel electrophoresis (PFGE). With EcoRI, seven ribopatterns (usually differing by only one band) were found, while PFGE had previously shown 25 clearly different patterns plus six related variants. Digestion with HindIII generated a few additional patterns but still failed to differentiate some strains that had very different PFGE patterns. Ribotyping with BscI has also been reported to be inadequate for subspecies strain differentiation (L. M. Hall, B. Duke, M. Guiney, and R. Williams, J. Clin. Microbiol. 30:915-919, 1992). Although ribotyping with other restriction endonucleases may perform better in distinguishing different strains, at present PFGE appears to be superior for strain differentiation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Derlot E., Courvalin P. Mechanisms and implications of glycopeptide resistance in enterococci. Am J Med. 1991 Sep 16;91(3B):82S–85S. doi: 10.1016/0002-9343(91)90348-2. [DOI] [PubMed] [Google Scholar]
- Hall L. M., Duke B., Guiney M., Williams R. Typing of Enterococcus species by DNA restriction fragment analysis. J Clin Microbiol. 1992 Apr;30(4):915–919. doi: 10.1128/jcm.30.4.915-919.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnen E., Richter F., Richter K., Andries L. Establishment of a typing system for group D streptococci. Zentralbl Bakteriol Mikrobiol Hyg A. 1988 Jan;267(3):322–330. doi: 10.1016/s0176-6724(88)80048-8. [DOI] [PubMed] [Google Scholar]
- Mederski-Samoraj B. D., Murray B. E. High-level resistance to gentamicin in clinical isolates of enterococci. J Infect Dis. 1983 Apr;147(4):751–757. doi: 10.1093/infdis/147.4.751. [DOI] [PubMed] [Google Scholar]
- Miranda A. G., Singh K. V., Murray B. E. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J Clin Microbiol. 1991 Dec;29(12):2752–2757. doi: 10.1128/jcm.29.12.2752-2757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A. G., Singh K. V., Murray B. E. Determination of the chromosomal size of three different strains of Enterococcus faecalis and one strain of Enterococcus faecium. DNA Cell Biol. 1992 May;11(4):331–335. doi: 10.1089/dna.1992.11.331. [DOI] [PubMed] [Google Scholar]
- Murray B. E. Beta-lactamase-producing enterococci. Antimicrob Agents Chemother. 1992 Nov;36(11):2355–2359. doi: 10.1128/aac.36.11.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Lopardo H. A., Rubeglio E. A., Frosolono M., Singh K. V. Intrahospital spread of a single gentamicin-resistant, beta-lactamase-producing strain of Enterococcus faecalis in Argentina. Antimicrob Agents Chemother. 1992 Jan;36(1):230–232. doi: 10.1128/aac.36.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Singh K. V., Heath J. D., Sharma B. R., Weinstock G. M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990 Sep;28(9):2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Singh K. V., Markowitz S. M., Lopardo H. A., Patterson J. E., Zervos M. J., Rubeglio E., Eliopoulos G. M., Rice L. B., Goldstein F. W. Evidence for clonal spread of a single strain of beta-lactamase-producing Enterococcus (Streptococcus) faecalis to six hospitals in five states. J Infect Dis. 1991 Apr;163(4):780–785. doi: 10.1093/infdis/163.4.780. [DOI] [PubMed] [Google Scholar]
- Murray B. E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990 Jan;3(1):46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J. E., Wanger A., Zscheck K. K., Zervos M. J., Murray B. E. Molecular epidemiology of beta-lactamase-producing enterococci. Antimicrob Agents Chemother. 1990 Feb;34(2):302–305. doi: 10.1128/aac.34.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preheim L., Pitcher D., Owen R., Cookson B. Typing of methicillin resistant and susceptible Staphylococcus aureus strains by ribosomal RNA gene restriction patterns using a biotinylated probe. Eur J Clin Microbiol Infect Dis. 1991 May;10(5):428–436. doi: 10.1007/BF01968023. [DOI] [PubMed] [Google Scholar]
- Prevost G., Jaulhac B., Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992 Apr;30(4):967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaberg D. R., Culver D. H., Gaynes R. P. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991 Sep 16;91(3B):72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Zervos M. J., Kauffman C. A., Therasse P. M., Bergman A. G., Mikesell T. S., Schaberg D. R. Nosocomial infection by gentamicin-resistant Streptococcus faecalis. An epidemiologic study. Ann Intern Med. 1987 May;106(5):687–691. doi: 10.7326/0003-4819-106-5-687. [DOI] [PubMed] [Google Scholar]