Abstract
The Gene Ontology (GO) is the most widely used ontology for creating biomedical annotations. GO annotations are statements associating a biological entity with a GO term. These statements comprise a large dataset of biological knowledge that is used widely in biomedical research. GO Annotations are available as “gene association files” from the GO website in a tab-delimited file format (GO Annotation File Format) composed of rows of 15 tab-delimited fields. This simple format lacks the knowledge representation (KR) capabilities to represent unambiguously semantic relationships between each field. This paper demonstrates that this KR shortcoming leads users to interpret the files in ways that can be erroneous. We propose a complementary format to represent GO annotation files as knowledge bases using the W3C recommended Web Ontology Language (OWL).
Introduction
With the advent of high-throughput technologies, biomedical research has seen an explosive growth in the type and amount of data that are generated during the course of research. Ontologies help researchers in managing the information explosion by providing explicit descriptions of biomedical entities and a means for annotating as well as analyzing the results of clinical and scientific research8.
Ontologies are currently viewed in biological sciences as a means 1) to achieve a high degree of interoperability among databases and 2) to enable conceptual integration of diverse datasets. In the former case, the Gene Ontology (GO)2 has shown demonstrable success in achieving interoperability among the Model Organism Databases (MODs), such as mice, rat, fruit fly and E. Coli, for describing the molecular functions, biological processes, and cellular locations of gene products. The GO is the most widely used biomedical ontology for annotation of genes and proteins as well as analysis of high throughput datasets – especially microarray data1.
In the context of gene and protein annotation, a GO annotation is a statement about a relationship between a biological entity and a concept represented by a GO term. For example, “[protein] DMP53 is associated_with cell death” is an annotation. Curators at MODs as well as institutes such as TIGR and EBI create these annotation statements in the course of careful review of the published literature. They comprise a large dataset of biological knowledge. Electronic methods, based primarily on sequence similarity, are also employed to create annotations in an automated manner. The GO annotations are made available as association files from the GO website. Those files, the primary format for the annotation statements, are an important resource for biomedical research.
As of this writing, there are 33,781 human gene products that are associated with 165,391 GO annotations. 116,620 of these annotations have been automatically inferred by electronic methods and 48,771 are the product of human curation. This means that there are about 48,000 statements about associations between a biological entity and the reference of a controlled GO term that have been created based on evidence reviewed by an expert. Creating these annotations is a time consuming process requiring skilled personnel. In total there are 14,654,921 annotations for more than 30 different organisms – 647,261 (4.42%) of which are expert curated.
The GO association files for storing annotations follow a tab-delimited file format with rows of 15 fields3. The semantics associated with each field in these files are declared in separate text descriptions (subject to human interpretation). This lack of formal semantics for knowledge representation can lead to errors because connections between fields in a tab delimited format – particularly between contextual fields that modify the meaning of other fields – are easily missed.
The goals of this paper are: (1) to document the shortcomings of the GO Annotation File Format, (2) to demonstrate that the shortcomings lead different users to different interpretations (and possible errors) and (3) to propose a format to represent annotations in the OWL language4 (a W3C recommendation), to deal with those problems.
The GO Annotation File Format
Curators at resources, such as UniProt and the various model-organism databases, create GO annotations after reading published literature. In the end, they export these annotations as a tab-delimited file, known as a gene association file that specifies links between gene products, and GO terms3. The gene or a gene product (transcript or protein) also has a database identifier. This flat file format, the GO Annotation File Format, is comprised of 15 tab-delimited fields, shown in Table 1.
Table 1.
The 15 tab delimited fields of the GO Annotation File Format.
| Column | Content | Required |
|---|---|---|
| 1 | DB | Y |
| 2 | DB_Object_ID | Y |
| 3 | DB_Object_Symbol | Y |
| 4 | Qualifier | N |
| 5 | GO ID | Y |
| 6 | DB:Reference (|DB:Reference) | Y |
| 7 | Evidence code | Y |
| 8 | With (or) From | N |
| 9 | Aspect | Y |
| 10 | DB_Object_Name | N |
| 11 | DB_Object_Synonym (|Synonym) | N |
| 12 | DB_Object_Type | Y |
| 13 | taxon(|taxon) | Y |
| 14 | Date | Y |
| 15 | Assigned_by | Y |
Each line in the file declares the gene product being annotated (fields 1-3 and 10-13) and the knowledge being asserted by a particular annotation (fields 4-9 and 14-15). Each gene product can have multiple annotations, needing multiple lines, and the fields specific to the gene product are repeated. The fields in each line can be grouped into identification fields (1, 2 and 3), lexical information fields (6, 10, 11, 14 and 15), and semantic fields (4, 5, 7, 8, 9, 12 and 13).
The semantic fields are the ones in which we are most interested, as they encode the knowledge represented about the gene product. Fields 4, 5, 9 and 12 determine the gene product associated with the GO term: DB_Object_Type (12) identifies the kind of gene product being annotated, GO ID (5) provides the GO term being used, Aspect (9) characterizes the term type (MF- Molecular Function, BP- Biological Process or CC- Cellular Component), and Qualifier (4) modifies the interpretation of an annotation, using the tags: NOT, contributes_to and colocalizes_with.
Fields 7 and 8 denote the kind of evidence that backs up the annotation: Evidence code (7) has a code specifying the evidence and With (or) from (8) has extra reference information for some evidence codes.
Field 13 (taxon) provides the ID, from the NCBI Taxonomy, for the species producing the gene product and the ID of the species interacting with or affected by the gene product; if the product (such as snake venom) is meant to interact with or affect other species.
Interpreting GO Annotation files
A particular gene product may participate in a number of annotations that assert its relationship with different biological concepts and the kind of evidence on which the assertion is based. The biological concepts come from four sources: the three Gene Ontologies (MF, BP and CC) and the NCBI Taxonomy. The restricted set of relation types that a canonical gene product can share with those concepts are shown in Table 2.
Table 2.
Relation types that a gene product can have and the biological concepts they can be related to.
| Relation Type | Term Type |
|---|---|
| associated_with | MF, BP or CC term |
| NOT associated_with | |
| contributes_to | MF term |
| NOT contributes_to | |
| colocalizes_with | CC term |
| NOT colocalizes_with | |
| produced_by | Taxon id |
| interacts_with | Taxon id |
For example, the annotations for gene FMN1 (riboflavin kinase) from the Saccharomyces Genome Database (SGD)5 tell us that FMN1:
is produced_by Saccharomyces cerevisiae,
is NOT associated_with function FMN adenylyltransferase activity,
is associated_with function riboflavin kinase activity,
is associated_with process FMN biosynthetic process,
is associated_with component microsome and
is associated_with component mitochondrial inner membrane.
Various programs and web sites read the GO Annotation files and extract the information to present a summary of the annotation for each gene product (for use in data analysis). Each tool or website can potentially interpret the format specifications differently however, because the semantics of the fields are declared in text form and are subject to human interpretation. The three main shortcomings of the GO Annotation File Format are:
It is a simple format of tab-delimited columns, which are not well suited to capture the semantics being represented.
It is an underspecified format, as syntactic (as well as semantic) information is subject to human interpretation.
It repeats information about each gene product multiple times leading to unnecessary redundancy.
Examples of misinterpretation of association files
These limitations lead to interpretation errors by users. The following are errors or omissions made by some important biomedical database resources when reading gene association files.
The Entrez Global Query Cross-Database Search System is a powerful federated search engine that allows users to search many discrete health sciences databases at the National Center for Biotechnology Information (NCBI) website. The Entrez Gene portal6 (gene-centered information) offers search using gene names. As an example, we searched two yeast (Saccharomyces cerevisiae) genes in the portal: FMN1 and DFM1.
FMN1 is annotated by SGD as NOT associated_with FMN adenylyltransferase activity (MF GO:0003919). DFM1 is annotated as NOT associated_with ER-associated protein catabolic process (BP GO:0030433). When we examine FMN1 and DFM1 on Entrez Gene, we find –under “General Gene Information - GeneOntology” (Figure 1) – the opposite information: Entrez Gene reports that the two genes are associated with these two GO terms. Because NCBI obtains this information from SGD5, we initially concluded that their program is not reading (or showing) the SGD GO associations file properly, most likely missing the Qualifier column. To test this hypothesis, we performed some additional test searches. We searched genes AHC1, with a contributes_to association with histone acetyltransferase activity (MF GO:0004402), and AGE2, with a collocated_with association with clathrin-coated vesicle (CC GO:0030136). These genes do not appear correctly on Entrez Gene. However, on all four genes pages, there are links to the SGD site where the genes FMN1, DFM1, AHC1 and AGE2 are correctly annotated – indicating a file-format interpretation error at Entrez Gene (we reported those problems to NCBI personnel and they acknowledged, identified and fixed them).
Figure 1.
Results of searches for the FMN1 and DFM1 genes in Entrez Gene. Arrows point to the misleading relationships.
We then searched other biomedical resources for the genes FMN1 and DFM1. Germonline {http://www.germonline.org} has the same problems as Entrez Gene. In their “Transcript-GO” section they show the same GO terms (in Figure 1) as been mapped to the two genes.
Ensembl {http://www.ensembl.org/index.html} (a joint project between EMBL - EBI and the Sanger Institute) and SwissProt {http://ca.expasy.org/sprot/} (Swiss Institute of Bioinformatics and the European Bioinformatics Institute) have a different behavior; they do not display GO annotations with the NOT qualifier. Hence, they are failing to display information that is available for those genes.
The examples we outline are not meant to be exhaustive, but to make the point that major biomedical resources are unable to interpret the GO association files in a uniform way (maybe even failing to read certain fields in the files). When professional users with access to documentation cannot properly interpret a format, it is a sure sign that this format needs improvement.
The two major weaknesses with the GO Annotation file format are (1) its lack of unambiguous syntactic information and (2) the fact that annotations are assertions about biological entities and a tab delimited format is not very suitable to represent such knowledge.
Currently, the number of errors resulting from the format limitations is not likely to be large because the use of qualifiers that modify the semantics of annotation statements is relatively new. In Table 3, we show the number of annotations that currently use qualifiers such as NOT.
Table 3.
Number of annotations with qualifiers and the percent they represent of the total and of the annotations manually curated.
| Organism | Annotations with qualifiers | Percent of the Total | Percent of the Human Curated |
|---|---|---|---|
| Human | 315 | 0.19% | 0.65% |
| Yeast | 559 | 1.59% | 1.59% |
| Mouse | 353 | 0.16% | 0.54% |
An OWL based solution
A proposed solution is to transform GO association files into a knowledge base in OWL. Using the Protégé tool7, this goal can be achieved using a Protégé Tab plugin, which we call the BioAnnotation Tab. Users can use the BioAnnotation Tab to read the GO annotation files into OWL. These GO Annotations in OWL will then generate an OWL ontology and a knowledge base, where the Gene Products are classes that have properties relating them to processes, functions, cellular locations, and species represented by GO terms and terms from the NCBI taxonomy. Curators at resources, such as the various model-organism databases (MODs), can still produce their annotations using the old format until they can migrate to a more expressive, knowledge representation scheme for the association files.
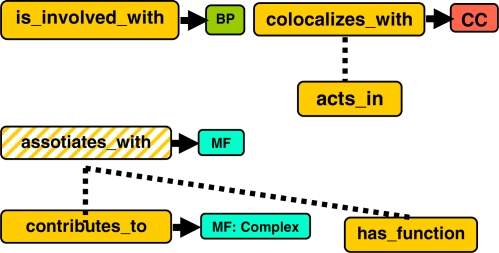
Embracing a format such as OWL also allows us to refine the relations described in Table 2 and to create the sub-properties shown in Figure 2.
Figure 2.
OWL properties that can be derived as specializations of the associated_with relation.
The associated_with GO relation (and its Qualifiers) can be partitioned into properties that are more specific for each kind of GO term with which it is used:
is_involved_with is used with BP terms.
colocalizes_with is used with CC terms, when the connection with the component is not very strong (example the resolution of an assay is not accurate).
acts_in is used with CC terms, it is a sub-property of colocalizes_with.
associates_with is a super-property for properties relating to MF terms (it isn’t actually used to annotate).
contributes_to is used with MF terms, when an individual gene product that is part of a complex can be annotated to terms that describe the function of the complex.
has_function is used with MF terms.
The relationships to the gene products are asserted by creating an OWL SomeValuesFrom restriction on the property relating a gene product to a GO term. If a relationship has a NOT qualifier, it adds the OWL ComplementClass over the restriction (actually saying that, apart from the GO term in the restriction, any value for that property is acceptable).
There are two more properties needed to represent relationships with species in the NCBI taxonomy: produced_by and interacts_with.
To produce the knowledge base/ontology in OWL, the BioAnnotation Tab reads each gene product from a GO Annotation file. In this example, we used the SGD gene_association.sgd file from the GO site. From the individual annotations for each gene product, the BioAnnotation Tab creates relationships to GO terms and to the NCBI taxonomy. Evidence codes are read as metadata for the created relationships.
Figure 3 shows the representation of the gene product FMN1 in yeast as part of the annotation knowledge base/ontology for Saccharomyces cerevisiae that we created with the BioAnnotation Tab. The screenshot shows the definition of the class FMN1 in OWL: It is a class derived from the Gene Product class with relations (OWL restrictions) to various GO terms and the taxon Saccharomyces cerevisiae. The NOT qualifier is appropriately represented as an OWL Complement class (not) to the relation has_function and the appropriate GO term FMN adenylyltransferase activity. The evidence code for each relationship is stored as an owl:annotationProperty of the relationship has_evidence. Currently there are no OWL reasoners that can take into account the certainty of restrictions, but this information is not lost.
Figure 3.
Section of the Protégé interface showing the FMN1 gene product representation.
Conclusion
We have shown the limitations of the GO Annotation File Format in terms of its ambiguity and inappropriateness for knowledge representation. We have shown how these limitations can lead to differing (and possibly erroneous) interpretations. We also propose a new format to represent GO Annotation statements. Being based on OWL (a W3C standard), this new format increases interoperability with other biological information sources in the Semantic Web. It is also an advance over the old tab-based format, as it represents the knowledge asserted in the annotations in the form of a knowledge base of assertions.
Curators, at biomedical resources (such the MODs), can still produce annotation files using the old simpler format. Interested users can use tools to convert those files. However, if curators, or other users, need to expand the scope of their annotations, including information such as phenotypes, an OWL based format offers them the expressiveness to do so.
Simple formats, such as tab-delimited files, can be a quick and easy initial solution to encode information, but as the content encoded gets more complex, they have to evolve into more expressive formats. We look forward to feedback from both the creators and users of the GO annotations.
We have also developed the BioAnnotation Tab, a Protégé plugin, to read association files in the GO Annotation File Format and populate an annotation knowledgebase. This plugin is available at http://bioontology.org/wiki/index.php/User:Dilvan
Acknowledgments
We would like to acknowledge Alex Morgan for useful suggestions about this work. This work was funded by NIH grant U54 HG004028 and a grant from CAPES-Brazil.
References
- 1.Khatri P, Draghici S.Ontological analysis of gene expression data: current tools, limitations, and open problems Bioinformatics 2005September1521183587–95.Epub, 2005 Jun 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GO Annotation Guide. http://geneontology.org/GO.annotation.shtml#file
- 4.W3C 2004OWL Web Ontology Language Reference, W3C Recommendation10 February 2004, http://www.w3.org/TR/owl-ref/
- 5.Saccharomyces Genome Database. http://www.yeastgenome.org/
- 6.NCBI Entrez Gene. http://ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
- 7.The Protégé Ontology Editor and Knowledge Acquisition System. http://protege.stanford.edu/
- 8.Rubin D, et al. National Center for Biomedical Ontology: advancing biomedicine through structured organization of scientific knowledge. OMICS. 2006 Summer;10(2):185–98. doi: 10.1089/omi.2006.10.185. [DOI] [PubMed] [Google Scholar]