Abstract
Integrating clinical research data entry with patient care data entry is a challenging issue. At the G. Pompidou European Hospital (HEGP), cardiovascular radiology reports are captured twice, first in the Electronic Health Record (EHR) and then in a national clinical research server. Informatics standards are different for EHR (HL7 CDA) and clinical research (CDISC ODM). The objective of this work is to feed both the EHR and a Clinical Research Data Management System (CDMS) from a single multipurpose form. We adopted and compared two approaches. First approach consists in implementing the single “care-research” form within the EHR and aligning XML structures of HL7 CDA document and CDISC ODM message to export relevant data from EHR to CDMS. Second approach consists in displaying a single “care-research” XForms form within the EHR and generating both HL7 CDA document and CDISC message to feed both EHR and CDMS. The solution based on XForms avoids overloading both EHR and CDMS with irrelevant information. Beyond syntactic interoperability, a perspective is to address the issue of semantic interoperability between both domains.
Keywords: HL7, CDA, CDISC, ODM, Medical Records Systems, Computerized, Biomedical Research, Clinical trial, Radiology, Interventional, Cardiovascular systems
Introduction
The computerization of medical data (observations, prescriptions, biological and imaging results) is a major issue for both patient care and biomedical research.
Considerable efforts were realized in these two areas for standardization and communication of medical information but in independent ways. In the healthcare domain, the HL7 organization creates standards for the exchange, management and integration of electronic clinical information. HL7 has developed since about ten years a version 3 of its messages, usually implemented in XML, which coherence is guaranteed by the Reference Information Model (RIM) adopted by the ISO. The first real use of this HL7 RIM was developed within a standard format for clinical documents (Clinical Document Architecture (CDA)) [1].
In the biomedical research domain, the CDISC (Clinical Data Interchange Standards Consortium) organization defines plateform-independent standards that support the electronic acquisition, exchange, submission and archiving of study data and metadata for pharmaceutical companies and Food and Drug Administration (FDA) [2]. CDISC Operational Data Model (ODM) was developed especially for data exchange in clinical trials between different study software [3].
In 2004, as part of the Cancer Biomedical Informatics Grid (caBIG) project, CDISC initiated the Biomedical Research Integrated Domain Group (BRIDG) to harmonize the semantics from available clinical trials information models into a shared model [4,5]. CDISC, HL7 and National Cancer Institute (NCI) are the three major stakeholders on the BRIDG project but it also includes regular representatives and content from other organizations including Food and Drug Administration (FDA), WHO, FAET (Federal Adverse Events Taskforce), PhRMA (Pharmaceutical Research and Manufacturers of America), and the Clinical Trials.gov [1,5,6]. The BRIDG model designed with the HL7 Development Framework defines standard entities, roles, attributes, and activities for the business process in standard clinical trials. It could be used as a core data standard for managing the workflow in clinical trials and for generating clinical trial software applications that share the same semantics and thus can exchange data more readily.
CDISC is also involved in projects promoting integration of clinical trials data entry with patient care data entry. The aim of the Oracle Clinical Project, in collaboration with NCICB (National Cancer Institute Center for Bioinformatics), HL7, and Oracle, is to propose an HL7 version 3 message to allow transmitting the definition of a form (DCI, Data Collection Instrument) between Electronic Health Record (EHR) and Clinical Research Data Management System (CDMS).
Another project aims at connecting healthcare and research within the framework of the Integrating Healthcare Enterprise (IHE) initiative. The goal of IHE initiative is to specify how data standards should be implemented to meet specific healthcare needs and to make systems integration more efficient and less expensive [7]. Based on working groups including users and manufacturers, IHE defines “Integration Profiles”, that are real-world situations describing exchange of information called “Transactions”, from various functional components of a distributed healthcare environment, called “Actors”. IHE provides implementation guides for “Transactions”, using established standards. The objective of the Retrieve Form for Data-capture Integration Profile (RFD) is to provide a method for gathering data within a user’s current application to meet the requirements of an external system. RFD aims at supporting the retrieval of forms from a form repository, display and completion of a form, and return of instance data from the display application to the source application. If an external agency, through some contractual agreement, requires data at the point of care, RFD will enable the EHR user to retrieve a data capture form from the external agency, to fill out the form, and to return the data to the external agency without leaving the EHR.
Although there is an issue to connect patient care and biomedical research, nowadays, in practice, biomedical research applications often require specific manual re-entry of data, some of which already reside in the EHR [8–10].
Our objective consists in implementing a solution to share data between EHR and CDMS:
Providing a single data capture form (avoiding double data capture that is time consuming and error prone);
Avoiding to overload either EHR or CDMS with useless information;
Guaranteeing the coherence of the information common to both contexts.
This article presents first the comparative analysis of the information required in both contexts – patient care and research in order to propose a unique multipurpose “care-research” form for data capture. Then we compared the HL7/CDA and CDISC/ODM models and proposed solutions for integrating data capture for both patient care and research. For this last step, we describe two approaches:
Implementing the single “care-research” form within the EHR and aligning XML structures of HL7 CDA document and CDISC ODM message to export relevant data from EHR to CDMS.
Displaying a single “care-research” XForms form within the EHR and generating both HL7 CDA document and CDISC message to feed both EHR and CDMS [11].
Finally we discuss the advantages and disadvantages of each approach and stress out the important issue of semantic interoperability.
Material and methods
Material
The EHR of the G. Pompidou European Hospital (HEGP) is administered by DxCare®, an application of the Medasys© company [12]. Structured documentation in DxCare is entered using standardized “questionnaires”. Clinical items are questions (attributes) for which answers (values) are expected and can be of different types: binary, text, numeric, date/time, lists, etc. Each attribute and each attribute value which data type is “list” is linked to a medical concept of a local dictionary of concepts that plays the role of a local reference terminology. For example, a DxCare questionnaire, called «cardiovascular (CV) radiology report», is dedicated to the cardiovascular radiology domain.
Air On Line (AOL)® (Kika Medical©) is a national repository of data for clinical research in the cardiovascular radiology domain [13].
Two types of forms are developed in AOL®: the General form which contains demographic data and past medical history of the patients and the Procedure form for data related to specific cardiovascular radiology procedures. Several Procedure forms were defined according to the processed anatomical site. Nowadays, radiologists enter clinical data in the «CV radiology report» of DxCare and then fill the «General form» and the corresponding «Procedure form» in AOL®.
Methods
Comparative analysis of the information required in both contexts – healthcare and research
The first step consisted in comparing items of «CV radiology report» of DxCare® to those of the General and Procedure forms in AOL®. All patient care and research items were defined and the common items were identified.
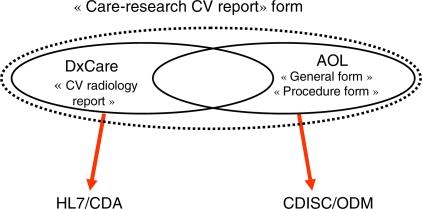
A multipurpose «care-research» data capture form for cardiovascular radiology, called «Care-Research CV report» was designed and implemented on one hand in DxCare and on the other hand separately by using the standard XForms (figure 1).
Figure 1.
The single multipurpose «Care-Research CV report» is the union of the DxCare «CV radiology report» and the AOL «General form» and «Procedure form»
In DxCare, the initial «CV radiology report» was completed to include the additional items required by AOL®. To evade semantic interoperability issues, the common items between patient care and research were reset in DxCare taking into account the data types, the values sets and the data type controls of the AOL® forms.
HL7/CDA and CDISC/ODM Comparison
Next step was to propose a CDA for daily practice in cardiovascular radiology based on HL7/CDA release 2 [14–17]. We also represented data for clinical research according to the ODM data model of CDISC version 1.2.
We analyzed the differences between the HL7/CDA and CDISC/ODM models on both a structural and organizational point of view (with regard to the context of data capture).
Integrating data capture for both patient care and research
We compared two approaches for integrating data capture for both patient care and research.
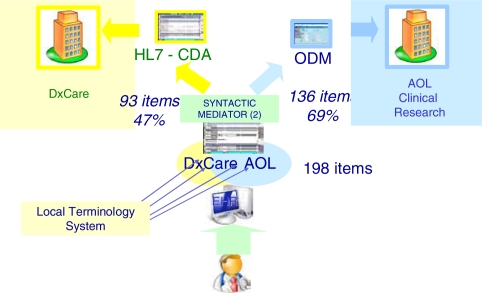
The first approach consisted in using a java DOM to develop a parser – also called “syntactic mediator”. This parser operates items selection from HL7 CDA documents, according to their relevance for clinical research, and syntactic transformation from HL7 CDA to CDISC ODM. A XML file includes selection criteria and mapping rules between HL7 CDA/CDISC ODM. Each instance of the “Care-Research CV report” available in DxCare is exported as an HL7 CDA document and translated into a CDISC ODM message (figure 3).
Figure 3.
Production of a CDISC ODM message from a HL7 CDA document, using a mapping file.
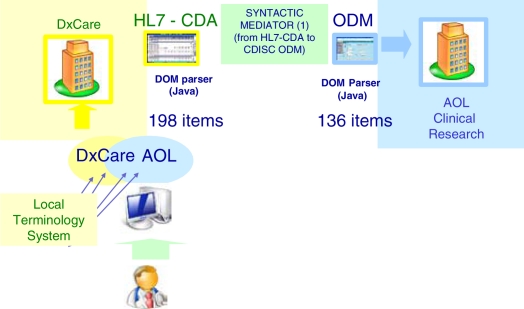
The second approach consisted in implementing the “Care-Research CV report” in XForms and generating, from this XForms form, both the HL7/CDA document and the CDISC/ODM message (figure 4).
Figure 4.
Production of a HL7 CDA document and a CDISC ODM message from a Xforms form
Results
Comparative analysis of the required information
The DxCare «CV radiology report» form includes 93 items. The AOL « General form » and « Procedure form» contain 136 items as a whole. The single multi-purpose “Care-Research CV report” form contains 198 items. There are 31 shared items in both contexts – patient care and clinical research.
HL7/CDA and CDISC/ODM models and comparative analysis
The HL7 CDA format for cardiovascular radiology includes the heading (meta-data) and the body that allows structuring the data of a cardiovascular radiology procedure. The proposed HL7 CDA contains one heading and the body where clinical data are organized into a hierarchy of sections.
The heading (meta-data) is structured according to the RIM and includes demographic data, information about healthcare provider(s) and encounter. The body contains both a non structured (text in natural language, legible by the user) and a structured part (hierarchy of sections containing coded items or “entries” without any depth limitation).
With regard to the items belonging to the list data type, we chose to use temporarily the HEGP local reference terminology.
The CDISC/ODM message format proposed for biomedical research in the cardiovascular radiology domain consists of three parts.
The first part, “Study”, contains general information on clinical trial (GlobalVariables), the units of measure of the clinical trial data (BasicDefinitions) and the data definitions (example: wording, types, constraints, coding, etc.). The second part, “AdminData”, contains information about the investigators. The third part, “ClinicalData”, contains the anonymous patient data and the clinical data organized into groups of items.
Some differences between CDISC/ODM and HL7/CDA are due to differences in the context of data capture between the healthcare and the clinical research domains. These differences intervene primarily in representation of the patient’s administrative data (table 1). A patient record concerns only one patient, whereas the clinical trial can concern several patients. Moreover, a CDA document concerns one visit, but there are several visits in one clinical study document.
Table 1.
Characteristics of both models for administrative data
| Patient record : HL7/CDA | Clinical study : CDISC/ODM |
|---|---|
| Only one patient | Several patients |
| One visit per document | Several visits for the same patient per message |
| Document identifier + patient identifier | Clinical trial identifier + document identifier |
| Detailed and complete information on the patient | Absence of administrative information on patients. |
| Links between the documents of the same patient | References to other clinical trials |
We decided to generate an ODM model for each patient and to group patients concerned by the same clinical study.
With regards to the structure of the models, the main differences between CDISC/ODM and HL7/CDA concern the representation of clinical data (table 2).
Table 2.
Characteristics of both models for clinical data representation
| Patient record: HL7/CDA | Clinical study: CDISC/ODM |
|---|---|
| Integration of definitions, references and data. | Separation of definitions, references and clinical data. |
| Infinite levels of depth | Three levels of depth available to define sections |
| RIM model | Absence of underlying reference model |
| Data entry control at the software level | Data entry control and generation of error messages |
| Coding at the software level | Possibility of coding elements |
The HL7/CDA model allows representing clinical data according to an infinite depth of sections.
For example let us take the case of the following sections:
<Section1>: CDA General form
<Section2>: Procedure
<Section3>: Description of the procedure
<Section4>: Medical device
<Section5>: Stents
<Section6>: Stent number 1
< Stent Indication >
< Stent Diameter >
CDISC/ODM proposes only three levels of depth to represent clinical study data.
CDISC_ODM 1.2
Structure: ClinicalData
SubjectData : Patient
StudyEventData: Visit
FormData : Form
ItemGroupData : Item grouping
ItemData : Data
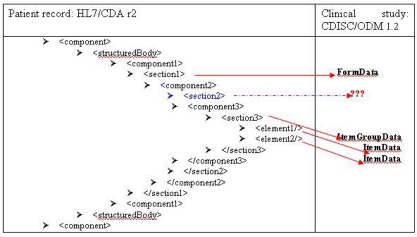
The following figure 2 shows that alignment is straightforward only in case of a two levels of depth section.
Figure 2.
Alignment between HL7/CDA sections and CDISC/ODM forms structures
For other cases, we decided to concatenate sections starting from three levels of depth, and to group them under only one item group.
The generic solution to generate an ODM from the example of HL7/CDA document is:
<FormData> : ODM General form
<ItemGroupData> : Procedure_Description of the procedure_Medical device_Stents_Stent1
<ItemData> : Stent Indication
<ItemData> : Stent Diameter
«Section» in the CDA document is aligned with the ODM «FormData» tag. Items in the CDA document « section 6 » are copied into the ODM «ItemData» tag. Wordings of «ItemGroupData» result from concatenation of wordings of the «section 2» (Procedure), «section 3» (Description of procedure), «section 4» (Medical device), «section 5» (Stents) and «section 6» (Stent1) tags.
Integration of healthcare and research data capture
Tools implementing both approaches have been implemented. The tool developed according to the first approach uses the mixed “Care-Research CV report” form implemented in DxCare (198 items) and produces a CDISC/ODM message including 136 items for biomedical research.
The tool developed according to the second approach uses the mixed “Care-Research CV report” form implemented in XForms (198 items) and produces an HL7/CDA document including 93 items dedicated to patient care and a CDISC/ODM message including 136 items dedicated to biomedical research.
Discussion
We showed that it was possible to exploit medical information of the EHR within the framework of clinical research. We used a method of alignment between XML formats of the clinical documents (HL7/CDA) model and the clinical research (CDISC/ODM) model. We also tested an alternative approach based on generation of both HL7/CDA documents and CDISC/ODM messages from a single XForms form.
Tools implementing these two approaches were developed and tested. For the implementation of the first approach, we had the choice between XSLT and the DOM parser. We used the Java DOM parser for the functional richness of its API (Application Programs Interface) and to facilitate the production of an intermediate step (XML mapping file) that can be very useful when one intends to make the alignment of CDISC towards HL7 (i.e., to import data resulting from clinical trials into the EHR). In contrast to the XSLT technology that requires a program for each direction of transformation, our mapping file is conceived for a bidirectional use.
The two approaches make it possible to produce clinical data using a single multipurpose form and to exploit data for both patient care and biomedical research. The benefit for healthcare providers consists in avoiding time consuming and error prone double data entry and allowing automatic transfer of administrative and medical data relevant for biomedical research towards a national server. Moreover, since relevant clinical data are stored in a structured manner in the EHR, they remain available for patient care and are both readable by healthcare professionals and exploitable by computers for decision support (e.g., alerts).
Limits and perspectives
Our work only makes it possible to ensure syntactic interoperability between two applications (DxCare® and AOL®). Indeed during the constitution of our material («care-research cardiovascular report» form), we ensured that the clinical items (including wording(s) and data types) were rigorously identical to those of AOL®. The HEGP local dictionary of concepts was supplemented and/or modified in order to take into account the medical concepts corresponding to items useful for biomedical research according to constraints of the AOL® application. Indeed, depending on the context (healthcare or biomedical research), similar items are often expressed by using different vocabularies.
Generalization to domains other than cardiovascular radiology one must be tested. In addition, our work must be supplemented by taking into account semantic interoperability issues in order to ensure that the DxCare® and AOL® applications have a common understanding of the exchanged data even when these data are expressed differently in both applications.
This future experiment will be conducted making use of the results of the BRIDG efforts [4]. We will focus on implementing solutions for semantic harmonization between various source models and/or reference terminologies such as MedDRA used for the designation of adverse drug reactions, LOINC used for the laboratory tests, ICD-10 or SNOMED-CT used for the procedures and diagnoses. Comparing manually semantically connected domain models is a tedious task. Defining the efficient use of assistance for model alignment such as receiving recommendations of merging options is a challenging issue [18].
Acknowledgments
Adeline MAGUERES, Nicolas DE SAINT JORRE, David OUAGNE, Olivier STEICHEN
References
- [1].HL7 : http://www.hl7.org
- [2].CDISC : http://www.cdisc.org
- [3].Kuchinke W, Wiegelmann S, Verplancke P, Ohmann C. Extended cooperation in clinical studies through exchange of CDISC metadata between different study software solutions. Methods Inf Med. 2006;45(4):441–6. [PubMed] [Google Scholar]
- [4].Weng C, Gennari JH, Fridsma DB. User-centered semantic harmonization: a case study. J Biomed Inform. 2007;40(3):353–64. doi: 10.1016/j.jbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- [5].NCI-caBIG : http://www.cabig.nci.nih.gov
- [6].Food and Drug Administration : http://www.fda.com
- [7].IHE : http://www.ihe.net/
- [8].Murray MD, Smith FE, Fox J, et al. Structure, Functions, and Activities of a Research Support Informatics Section. J Am Med Inform Assoc. 2003;10:389–398. doi: 10.1197/jamia.M1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].De Saint Jorre N. The eClinical Forum and PhRMA EDC Task Group. Mar 3, 2006. The Future Vision of Electronic Health Records as eSource for Clinical Research. [Google Scholar]
- [10].Gerdsen F, Muller S, Jablonski S, Prokosch HU. Standardized exchange of clinical documents--towards a shared care paradigm in glaucoma treatment. Methods Inf Med. 2006;45(4):359–66. [PubMed] [Google Scholar]
- [11].Xforms : http://www.w3.org/MarkUp/Forms/
- [12].MedaSys : http://www.medasys.com
- [13].Air on Line : http://www.air-online.org
- [14].CDA Implementation guide : www.hl7.org
- [15].HL7 France Groupe C/YK . Clinical Document Architecture release 2, Spécifications françaises, Définition des métadonnées (Volet Médical) Sep, 2005. [Google Scholar]
- [16].Dolin RH, Alschuler L. HL7 Clinical Document Architecture, Release 2. J Am Med Inform Assoc. 2006;13(1):30–9. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ferranti JM, Musser RC, Kawamoto K, Hammond WE. The Clinical Document Architecture and the Continuity of Care Record: A Critical Analysis. J Am Med Inform Assoc. 2006;13(3):245–52. doi: 10.1197/jamia.M1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Noy N, Musen M. PROMPT: algorithm and tool for automated ontology merging and alignment. In: Proceedings of the seventeenth national conference on artificial intelligence (AAAI-2000) Austin, TX; 2000. pp. 450–55. [Google Scholar]