Abstract
The National Library of Medicine (NLM) and the National Cancer Institute (NCI) are creating a digital archive of 100,000 cervicographic images and clinical and diagnostic data obtained through two major longitudinal studies. In addition to developing tools for Web access to these data, we are conducting research in Content-Based Image Retrieval (CBIR) techniques for retrieving visually similar and pathologically relevant images. The resulting system of tools is expected to greatly benefit medical education and research into uterine cervical cancer which is the second most common cancer affecting women worldwide. Our current prototype system with fundamental CBIR functions operates on a small test subset of images and retrieves relevant cervix images containing tissue regions similar in color, texture, size, and/or location to a query image region marked by the user. Initial average precision result for retrieval by color of acetowhite lesions is 52%, and for the columnar epithelium is 64.2%, respectively.
1. Introduction
Cervical cancer is the second most common cancer affecting women worldwide and the most common in developing countries. Cervicography is an inexpensive method for cervical cancer screening which sometimes allows physicians to visually detect abnormal tissues based on the appearance of the cervix after treatment with acetic acid (3%–5% concentration). The National Library of Medicine (NLM) and the National Cancer Institute (NCI) are creating a digital archive of 100,000 cervicographic images (cervigrams) and correlated, longitudinal clinical data, and supporting access and study tools. The data was obtained through two major studies in Costa Rica and the United States, the Guanacaste and ALTS1 projects, respectively. These projects enrolled a total of 15,060 volunteers and were designed to study the natural history of Human Papillomavirus (HPV) infection and cervical neoplasia. This neoplasia is closely related to anogenital types of HPV, which are sexually transmitted. There are more than 40 different types of HPV, but only about 15 of them are considered oncogenic because of their relation with pre-invasive lesions and cancer [1,2].
Access to this cervicographic image database by visual characteristics, such as color and texture of particular regions on the cervix, is expected to greatly benefit clinical teaching and research by providing a new and powerful capability to query this data. Our long-term goal is to create a content-based image retrieval (CBIR) system for this data.
This article discusses a prototype of this system with fundamental CBIR functions that operate on a small expert-annotated subset of the images. The prototype incorporates the following functions: feature selection and extraction, feature normalization, feature combination and weighting, feature dimension reduction, similarity measures, and a graphical user interface (GUI) for query and result. It does not include the functions of automatic image segmentation and recognition of the region of interest, which are initial steps necessary for CBIR. Our ongoing research on these steps has been reported in [3,4]. Regions of interest on images used in the CBIR tool database have been manually segmented and labeled by NCI medical experts, using our Boundary Marking Tool [5].
In using our system, the user creates a query by marking a region of interest on an image. The system calculates the feature vector of the query region for the specified features and compares it with the precomputed feature vectors of regions stored in the database. User-specified feature weights and feature combination strategy are applied in the similarity computation stage. The result images are displayed in order of decreasing similarity.
The prototype system discussed in this article is designed to help evaluate and identify key techniques among those found in the technical literature [6] by comparing their performance for the retrieval of these images. For example, for the color feature, the user may choose among a variety of representations, such as color histograms, color moments, color coherence vector, and dominant colors, in different color spaces. Currently, to the best of our knowledge, no study exists that provides a complete analysis of the content-retrieval task for cervigrams. Other studies only analyze individual uterine cervix images for pathology segmentation and classification [7].
2. Methods
2.1. Feature Selection and Extraction
We focus on extracting suitable features and corresponding feature representation for the retrieval of important tissues in cervigrams which are similar to the tissue region defined in the query image. We have selected color, texture, size, and location to represent the characteristics of the lesion region-of-interest and have implemented several possible representation options for each feature. Initial results obtained through various combinations of these features are presented. We expect to use this system to determine the most effective representation for each feature, based on retrieval results evaluated by medical experts which will include the senior author of the article.
2.1.1. Color
Color is one of the most dominant and distinguishing visual features used by physicians to identify lesions on the uterine cervix. For example, the acetowhite (AW) lesions appear opaque white, blood appears red, and the columnar epithelium (CE) tissue appears red or orange. In this CBIR tool, we have implemented multiple color descriptors that have been proposed for content-based retrieval tasks in the literature. These color descriptors are color histogram, color moments, color coherence vector, and dominant colors.
The color histogram is the most often used global color feature. It represents the joint probability of the intensities of the three color channels. The color moments descriptor is a compact representation that includes the first moment (mean), the second central moment (standard deviation) and the third moment of each color channel. The color coherence vectors descriptor [8] is similar to the color histogram, but, additionally incorporates spatial information. It classifies each pixel as either coherent or incoherent based on whether it belongs to a “large region” of similar color. The dominant colors descriptor [9] captures the representative colors in a region. Those representative colors are obtained by performing color clustering.
To calculate the above color descriptors, a color space must first be selected. The CBIR tool provides eight choices for color spaces: RGB, HSV, HSI, YUV, YIQ, YCbCr, Luv, and Lab. When calculating the color histogram and color coherence vector, the color needs to be quantized first in order to reduce the dimensionality of the feature vectors (and corresponding computation cost). Scalar quantization is used in the current cervigram CBIR tool.
2.1.2. Texture
Texture is another important feature used to differentiate different regions on the cervix. For example, the squamous epithelium (SE) tissue is relatively smooth and non-textured, while columnar epithelium (CE) tissue is relatively rough and textured. To extract texture features, we implemented five methods. These methods were based on the Gabor filter, the Log-Gabor filter, the gray-level co-occurrence matrix, the discrete wavelet transform, and the histogram of edge directions.
The Gabor filter has been widely used for extracting texture features since it is optimal in terms of minimizing the joint uncertainty in space and frequency [10]. In our CBIR tool, we use a Gabor filter bank with K =6 orientations and S =4 scales for texture analysis. The mean μmn and standard deviation σmn (m =1,...,S ; n =1,..., K) of the magnitude of the transform coefficients in the region are used to represent the region. The Log-Gabor filter was proposed by Field [11] for overcoming the limitations of Gabor filters. Similar to the Gabor filter, the mean μmn and standard deviation σ mn of the magnitude of the transform coefficients in the region are calculated. The gray level co-occurrence matrix (GLCM) features capture the spatial dependence of gray-level values within an image by using second-order statistics. We choose four of the statistical features proposed by Haralick [12] in our application: contrast, correlation, homogeneity, and energy. Discrete wavelet transform features are one of the multiscale features used to analyze the image texture [13]. We use a 4-level wavelet transform, which results in 13 sub-images. For each sub-image, the mean and standard deviation of the magnitude of its wavelet coefficients in the region are calculated as the features. A histogram of edge directions (HED) captures the spatial distribution of edges in the image [14]. In the CBIR tool, a 36-bin HED is used to represent the edge strength in the directions: (0, π/18,…., 35π/18).
2.1.3. Size
The size of the lesion tissues in the uterine cervix is one indicator of the degree of the lesion’s severity. The size feature is defined as the relative size of the region to that of the cervix.
2.1.4. Location
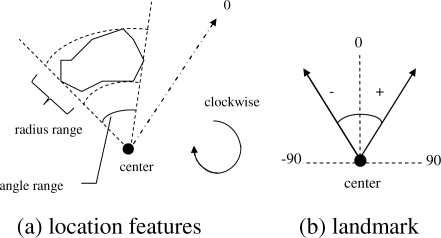
The spatial location of the region is characterized by its angle and distance with respect to the landmark angle and center in a clockwise polar coordinate system, as shown in Figure 1. The landmark center is the center location of the cervical os. The landmark angle is the orientation angle of the cervix in the image, with the reference axis (0 degree direction) lying in the “12 o’clock” direction.
Figure 1.
The definition of location features
The following location features are extracted for the region,
-
normalized angle range of the region
(3) where ρmax and ρmin are the maximal and minimal angles of the region.
normalized radius range of the region(4) where rmax and rmin are the maximal and minimal distance from the region to the landmark center; rcervix is the maximal distance from the cervix boundary to the landmark center.
-
normalized center of the region
(5) where N is the total number of pixels in the region
- extent of the region
(6)
2.2. Feature normalization
Normalization is required to compensate for the scale disparity between the feature components that are defined in different domains. The Gaussian normalization method is applied for both intra-feature normalization and inter-feature normalization.
2.3. Feature combination and weighting
The content of a region can be represented by the feature vectors in different feature classes, viz. color, texture, size and location. These features need to be combined together to decide the final retrieval results. Our CBIR tool provides two options for combining these feature classes. The first approach is to combine the feature classes at the feature level. That is, the feature vectors in different feature classes are combined into one overall feature vector. The system compares this overall feature vector of the query image to those of database images, using a predetermined similarity measurement. The second approach is to combine the feature classes at the rank level. That is, for each feature class, the system compares the feature vector of the query image to those of database images, using a pre-determined similarity measure, and ranks the retrieved images by their relevance score. Then the ranks in different feature classes are combined into one overall rank, and the overall similarity of database images are determined by the overall rank.
The degree of importance of each feature class can be specified by a weight. A weighted linear method is then applied for combining the feature classes or the ranked results.
2.4. Feature Dimension Reduction
When combining the feature classes at the feature level, the dimension of the overall feature vector may be high. Therefore, the system provides an option for performing a dimensionality reduction by principal components analysis (PCA).
2.5. Similarity measures
The similarity between two images is based on the distance between the feature vectors of the images in feature space. The similarity measures that we have implemented in our CBIR tool are: Minkowski-form distance, histogram intersection, Jeffrey divergence, quadratic-form distance [15], match distance, earth mover’s distance [16], weighted-mean-variance [13], and modified MDM [9]. Different feature representations may have different similarity measures. For example, histogram intersection is used for calculating the distance between two color histograms, while modified MDM is used for calculating the distance between two sets of dominant colors.
2.6. User Interface
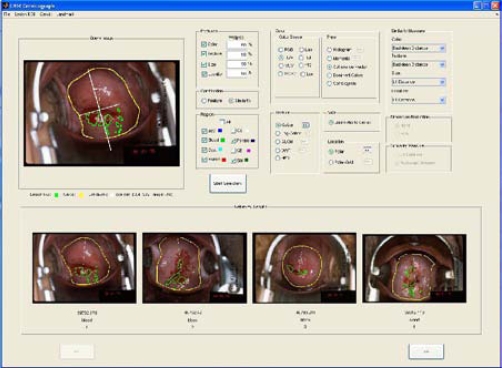
The system is implemented in MATLAB, and the GUI is shown in Figure 2. The three parts of the GUI are for (1) creating the query, (2) specifying system parameters, (3) displaying the retrieved results.
Figure 2.
User interface of CBIR system
For creating the query, the user may draw the region-of-interest (ROI) on the image, or load a binary mask of the ROI by using the Lesion ROI menu. To use the feature class Size, the cervix size information is required. For the feature class Location, the landmark’s center and orientation angle as well as the size of the cervix are required. The system provides a way to draw the cervix boundary or load a binary mask of the cervix and a method for drawing the landmark or inputting the landmark center and orientation angle values with the Cervix and Landmark menus, if the Size or Location features are selected.
The system parameters part of the GUI consists of panels that allow specification of features, feature class weights, feature combination, tissue types, color feature descriptors, texture descriptors, size feature descriptor, location feature descriptors, similarity measure per feature class (when rank level combination is selected), feature dimension reduction, and similarity measure for an overall feature vector (when feature level combination is selected).
The third part of the GUI presents the retrieved regions with the corresponding image tag, region label, and similarity rank. For convenience, a magnified version of any retrieved image is displayed by clicking on that image.
3. Discussion
One objective of the CBIR system in the medical domain is to assist doctors in diagnosis. For our application, given a query image with a certain region type (such as acetowhite), the system should retrieve images with the same type of region as the query image. In order to evaluate our CBIR system from this aspect, we presented each of the expert-marked regions in turn to the system as a query and calculated the fraction of the top (five) returned images that had the same region type as the query (the region used as query was not counted). We use the precision criterion to measure retrieval effectiveness.
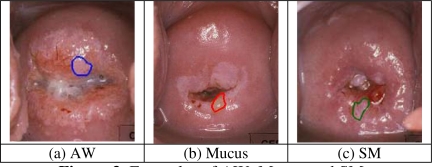
The system provides multiple choices for feature descriptors and similarity measures. As a starting point, we selected one set of system parameters and tested the system’s retrieval performance under this set. A comprehensive comparison will be conducted as future work. The ground truth data set we used for test has 120 images with overall 422 significant tissue regions marked by medical experts. There are six types of tissues, acetowhite (AW), blood, mucus, os, columnar epithelium (CE), and squamous metaplasia (SM). Table 1 lists the average precision for each region type for each feature class. The first column of Table 1 shows the number of cases for each tissue type. For color feature class, the descriptor used is color coherence vector, the color space selected is HSV color space, and the color quantization level is [8, 8, 4]. The texture descriptor is a Gabor filter (with 4 scales and 6 orientations) descriptor. The size feature descriptor is the size ratio explained in Section 2.1.3. The features used for representing the location property are those described in Section 2.1.4. The similarity measure for each feature class is Euclidean distance. Table 1 indicates that the importance of each feature class for differentiation is different for each tissue type. For example, location is the most essential property to recognize the os region, while color is crucial to identify the blood region. Therefore, it is advantageous that our system can provide users feature selection options. In Table 1, the precisions for tissue types Mucus and SM are relatively low. This can be explained by the close visual similarity between them and AW as shown in Figure 3 and the large appearance variations across images due to irregular illumination and different acquisition conditions. We plan continuing interactions with medical experts for refining our understanding of effective features for tissue classification.
Table 1.
The average precision for each feature class(Quantity is also number of queries)
| Type | Quantity | Color | Texture | Size | Location |
|---|---|---|---|---|---|
| AW | 76 | 52.1% | 44.2% | 41.8% | 41.6% |
| Blood | 51 | 62.0% | 14.5% | 13.7% | 22.4% |
| Mucus | 40 | 25.5% | 7.5% | 7.0% | 14.0% |
| Os | 120 | 52.5% | 41.2% | 55.8% | 72.0% |
| CE | 77 | 64.2% | 49.4% | 40.3% | 63.4% |
| SM | 58 | 29.7% | 19.0% | 19.3% | 29.0% |
| Total | 422 | 50.0% | 33.8% | 35.7% | 47.6% |
Figure 3.
Examples of AW, Mucus and SM
In addition to quantitative analysis of the retrieval performance of the system, we also evaluated the system using the judgment of one medical expert. Twelve queries and their corresponding top five images returned by the CBIR program were presented to the expert. The expert was asked to answer whether each returned region was a good match to the query region based on his own opinion and to provide his reasoning, as well as to provide his own similarity ranking if he disagreed with the similarity order given by the program. The expert concurred with our approach in using agreement between query region type and returned region type as one component of precision evaluation. We plan to use this measure to carry out a more comprehensive evaluation and comparison of different feature descriptors and similarity measures. For most cases, the similarity order given by the expert was different from the order given by the system. One reason for this disagreement may be due to the gap between the definition and weighted combination of the significant visual features used by the expert, as compared to the program. Another observation from this experiment is that, for a few returned regions, their types labeled by the evaluation expert were different from the labels in the database. If the difference between tissue types is subtle, on the other hand, suggesting a second opinion by presenting visually similar regions but different types is also a benefit of using CBIR system.
4. Conclusions and Future Work
In this paper, we have described work in progress toward building a CBIR system for a cervicographic image database. Basic CBIR functions included in the system are user options for feature selection and extraction, feature combination and weighting, and similarity measures, all available through a GUI. The intended system use is for investigation of feature and similarity measure choices to optimize retrieval of uterine cervix images by tissue characteristics. Preliminary objective and subjective assessment of this tool was described. Future goals include further retrieval evaluation in coordination with medical experts from NCI.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine, and Lister Hill National Center for Biomedical Communications.
Footnotes
References
- 1.Herrero R, Schiffman MH, Bratti C, et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa-Rica: the Guanacaste Project. Rev Panam Salud Publica. 1997;1(5):362–75. doi: 10.1590/s1020-49891997000500005. [DOI] [PubMed] [Google Scholar]
- 2.Schiffman M, Adrianza ME. ASCUS-LSIL Triage Study. Design, methods and characteristics of trial participants. Acta Cytol. 2000;44(5):726–42. doi: 10.1159/000328554. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S, Zimmerman G, Long R, Antani S, Jeronimo J, Greenspan H. Content analysis of uterine cervix images: initial steps towards content based indexing and retrieval of cervigrams. Proc SPIE Medical Imaging. 2006;6144:1549–56. [Google Scholar]
- 4.Srinivasan Y, Hernes D, Tulpule B, Yang S, Guo J, Mitra S, et al. A probabilistic approach to segmentation and classification of neoplasia in uterine cervix images using color and geometric features. Proc SPIE Medical Imaging. 2005;5747:995–1003. [Google Scholar]
- 5.Jeronimo J, Long R, Neve L, Michael B, Antani S, Schiffman M. Digital tools for collecting data from cervigrams for research and training in colposcopy. J Lower Genital Tract Disease. 2006 Jan;10(1):16–25. doi: 10.1097/01.lgt.0000194057.20485.5a. [DOI] [PubMed] [Google Scholar]
- 6.Müller H, Michoux H, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medicine - clinical benefits and future directions. International J Medical Informatics. 2004;73:1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Raad VV, Xue Z, Lange H. Lesion margin analysis for automated classification of cervical cancer lesions. Proc SPIE Medical Imaging. 2006;6144:1647–59. [Google Scholar]
- 8.Pass G, Zabih R, Miller J. Comparing images using color coherence vectors. Proc ACM Int Conf on Multimedia; Boston, MA. 1996. Nov, pp. 65–73. [Google Scholar]
- 9.Mojsilovic A, Hu J, Soljanin E. Extraction of perceptually important colors and similarity measurement for image matching, retrieval, and analysis. 11. Vol. 11. IEEE; Trans Image Processing: 2002. Nov, pp. 1238–48. [DOI] [PubMed] [Google Scholar]
- 10.Manjunath BS, Ma WY. Texture features for browsing and retrieval of image data. IEEE. Trans Pattern Analysis and Machine Intelligence. 1996 Aug;18(8):837–42. [Google Scholar]
- 11.Field DJ. J. Optical Society of America. 1987. Relations between the statistics of natural images and the response properties of cortical cells; pp. 2379–94. [DOI] [PubMed] [Google Scholar]
- 12.Haralick R, Shanmugam K, Dinstein I. Texture features for image classification. IEEE Trans Systems, Man and Cybernetics. 1973;3:610–21. [Google Scholar]
- 13.Rui Y, Huang T, Chang S. Image retrieval: current techniques, promising directions and open issues. J Visual Communication and Image Representation. 1999;10(4):39–62. [Google Scholar]
- 14.Jain AK, Vailaya A. Image retrieval using color and shape. Pattern Recognition. 1996;29(8):1233–44. [Google Scholar]
- 15.Hafner J, Sawhney HS, Equitz W, Flickner M, Niblack W. Efficient color histogram indexing for quadratic form distance functions. IEEE. Trans Pattern Analysis and Machine Intelligence. 1995 Jul;17(7):729–36. [Google Scholar]
- 16.Rubner Y, Tomasi C, Guibas LJ. The earth movers distance as a metric for image retrieval. Int J Computer Vision. 2000;40(2):99–121. [Google Scholar]