Abstract
Brain tumors’ treatment and prognosis depend to a large extent on their grades. Grading tumors follows a set of rules that refers to domain knowledge. Developing an automatic grading system requires explicit and formal representation of the domain. The NCI Thesaurus is the major ontological resource in the cancer domain. However, the description of brain tumors and grades in the NCI Thesaurus does not enable automatic grading. We have developed an ontology based on the NCI Thesaurus for automatic classification of glioma tumors based on a reference grading system. Two sets of tests have been done. The first one has been automatically generated and the second one consists of eleven pathology reports. The resulting ontology contains 243 classes, among which 234 correspond to NCI Thesaurus classes. Because all of the generated tests were correctly classified, we believe our system to be correct. Ten clinical reports are correctly graded and one is graded incompletely.
Introduction
Brain tumors represent 2.4 percent of all cancer deaths. Among tumor variables, tumor grade and histology appear to have the greatest effect on survival. Glioblastoma, with median survival less than twelve months, is a highly malignant (grade IV) glioma, which has the propensity to infiltrate throughout the brain in contrast to pilocytic astrocytoma of the posterior fossa, which does not spread and can be cured by surgery [1]. Traditionally, the grading (classification) of a tumor is determined by the evaluation of tumor characteristics by a pathologist.
The process of determining the grade of a tumor consists in checking if it meets a set of requirements. It is typically a classification task. The grading system requires domain knowledge in order to fill the granularity gap between the tumor descriptions and the grade descriptions. However, having formal representations of the grade definitions and the background knowledge is necessary for applications such as decision support for pathologists or integration of data graded using different systems. This is typically done by ontologies.
For the past years, a lot of biomedical ontologies have been developed including NCI thesaurus (NCIT) [2] a major resource in the cancer research domain. The NCIT provides descriptions for the brain tumors. It also has classes for the grades. However, those classes have neither descriptions nor definitions. Therefore, they can not be used for the automatic grading of tumors, which requires an explicit and formal representation.
The goal of this article is to show how the version of the NCIT in OWL (Web Ontology Language) can be extended to automatically perform classification of glioma using histological descriptions. We have focused our study on the malignant grade. For that, we have developed an ontology of the glioma tumors based on the World Health Organization grading system [3]. In this study, we focus on the reasoning tasks. Section 2 is an overview of this reference grading system. Section 3 is an overview of the NCIT, in which we conclude that it has to be extended in order to perform automatic tumor grading. Section 4 describes the method that we used and Section 5 presents the results obtained during the classification of a set of tests generated and the classification of eleven reports provided by a pathology department.
Reference Grading System
There are numerous grading systems for the grading of the glioma tumors. The reference grading system is the World Health Organization (WHO) grading system [3].
The WHO grading system assigns a grade from 1 to 4 to glioma, grade 1 being the least aggressive and grade 4 being the most aggressive. This classification is based on five histopathology criteria that are related to the degree of anaplasia: cellular density, nuclear atypia, mitosis, endothelial proliferation and necrosis. The WHO malignant grades are described as follows:
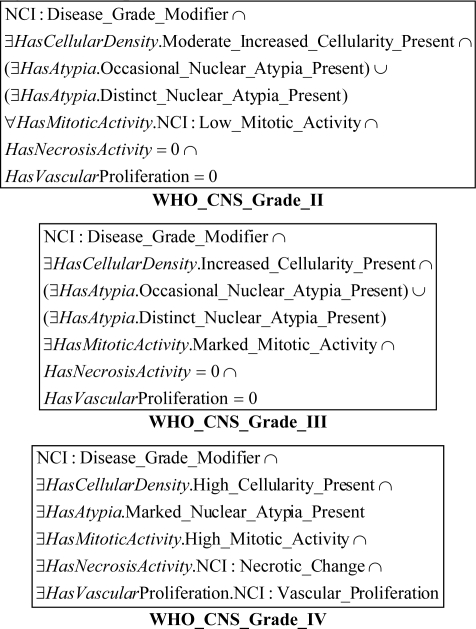
WHO Grade II: cellular density moderately increased, occasional nuclear atypia, mitotic activity absent or 1 mitosis, necrosis absent, endothelial proliferation absent
WHO Grade III: cellular density increased, distinct nuclear atypia, mitotic activity marked, necrosis absent, endothelial proliferation absent
WHO Grade IV: cellular density high, nuclear atypia marked, high mitotic activity, necrosis present, endothelial proliferation present
Overview (of grading) of brain tumor in the NCI Thesaurus
The NCI Thesaurus is a public domain description logic-based terminology to meet the needs of the cancer research community [2]. Its goal is to provide unambiguous codes and definitions for concepts used in cancer research. The NCIT has been converted into OWL-Lite [4]. The current version (07_01d) is composed of 55,458 named classes and 113 OWL Properties. Among these classes, 18% are defined classes, i.e. they have at least one necessary and sufficient constraint, and 82% are primitive classes, i.e. they can have constraints, but do not have any necessary and sufficient definitions. In the NCIT, glioma tumors and grades have been represented by defined and primitive classes.
The glioma tumors have been described as Central Nervous System Neoplasms. Each kind of tumors has been defined by necessary and sufficient conditions. For example, glioblastoma has been defined by the intersection of 17 restrictions. In Figure 1 we present some conditions used to define the glioblastoma class.
Figure 1.
Portion of the necessary and sufficient restrictions for the glioblastoma NCIT class
There are three main limitations preventing the NCIT from grading tumors or identifying tumors.
First, the classes representing the grades according to the WHO system have no restriction and are not semantically defined (Figure 2). Therefore, they are just placeholders as nothing can be inferred to be a subclass or an instance of these classes.
Figure 2.
WHO Central Nervous System Grade in the NCIT: The WHO grades have no definition
Second, the definitions for tumors involve the grade of the tumor (e.g. Figure 1). Since nothing can be classified as a grade (here, grade 4), the definitions for classes like glioblastoma cannot be used to infer the kind of tumor, from its histologic features. The structure of the NCIT imposes that the only way to determine the grade of a tumor is to determine which kind of tumor it is (e.g. by knowing that a tumor is a glioblastoma, we can deduce that its grade is 4). However, we have seen that for inferring the kind of tumor, we need to know its grade. Therefore, the definitions for the various kind of tumors cannot be used for any inference.
Third, some of the existing subclasses of the Morphologic_Finding class are too general with respect to the descriptions of the grades provided by the WHO. For example, Nuclear_Atypia has just one subclass which is Marked_Nuclear_Atypia.
Because of the open-world-assumption underlying the OWL semantics, if the grade of a tumor cannot be unequivocally inferred, the tumor will not be classified under any grade. For example, tumors that could be grade I or grade II tumors are classified nowhere. There is no explicit difference between the grades the tumor belongs to (here I and II) and those it cannot belong to (here III and IV).
Methods
The ontology we developed is based on the NCIT. A specific relevant part of the NCIT has been extracted using eleven terms corresponding to the names of the glioma tumors and nine terms that correspond to subclasses of atypia and mitotic activities. We first retrieved the NCIT classes corresponding to these terms and all their parents. For each of these classes, we followed all their relations and recursively retrieved the fillers and their parents.
Several operations have been necessary to address the issues mentioned in the previous sections and enhance the extracted portion of the NCIT. First we provided definitions for all WHO grades. Second, we added new classes (and new properties) for filling the granularity gap between the histologic features described in the WHO and the classes present in the NCIT. For handling the open-world-assumption, we also introduced the negations of each grade (namely nograde).
Two sets of classification tests have been created. The first set (15 tests) has been generated for representing plausible combinations of the histologic criteria. Each test corresponds to a prototypical tumor. The second set corresponds to eleven pathologic reports provided by the pathology department of the Rennes hospital. Each report was represented as a subclass of Disease_Grade_Modifie. This step was performed manually. Each report is read and the corresponding Tumor class has been built manually. For each test, the description of its histologic criteria was done by existential restrictions for indicating the presence of a criterion, and by cardinality restriction to zero for indicating the absence of a criterion. We have classified these reports using class-based reasoning. The reasoning process consisted in retrieving the inferred subclasses of a particular grade or in retrieving the inferred subclasses of a particular nograde.
Results
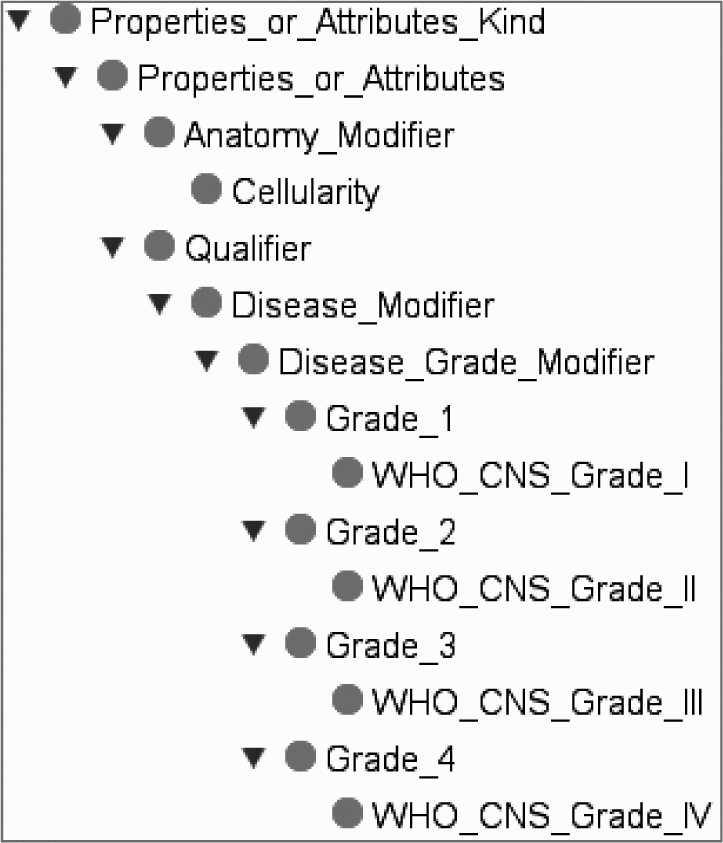
The generated ontology is composed of 243 classes, among which 33 are defined. Among the 243 classes, 234 classes correspond to NCIT classes, 5 classes have been added for the description of the histologic criteria and 4 classes have been added for the description of nogrades. We reused 24 class definitions from the NCIT and created the remaining 9. Figure 3 illustrates a part of the hierarchy of the grading system.
Figure 3.

A part of the Hierarchy of the grading system
New histologic classes
Five new classes are added to represent the histologic features.
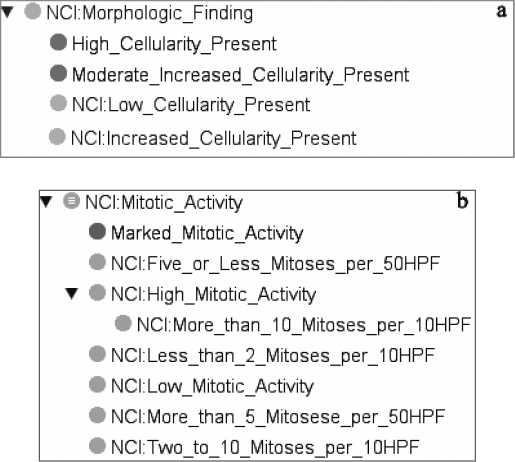
The cellular density criteria have different values in WHO that are moderately increased, increased and high. In the NCIT, two classes exist: Low_Cellularit_ Present and Increased_Cellularity_Present. We added two classes in the NCIT to represent high cellularity and moderate cellularity. All of these classes were made disjoint (Figure 4a).
Figure 4.
The Cellular Density (a) and the Mitotic Activity criteria (b)
The nuclear atypia criteria have different values in WHO that are occasional, distinct or marked. In the NCIT, one class exists: Marked_Nuclear_Atypia_Pre sent. We added two classes in the NCIT for the description of occasional nuclear atypia and distinct nuclear atypia. All of these classes were made disjoint.
The necrosis and vascular proliferation criteria are described by their presence or absence in the WHO grading system. We did not modify these two NCIT classes.
The mitotic Activity in the NCIT is described by seven classes (Figure 4b) whose values may be symbolic values such as Low_Mitotic_Activity or numeric ones such as More_than_5_Mitoses_per_50HPF. We have added one class for representing Marked_Mitotic_Activity (Figure 4). Figure 4b illustrates the new hierarchy of Mitotic Activity in our ontology.
New Properties
The relation used in the NCIT for describing histologic features is Disease_Has_Finding. This relation is used for all kinds of findings and this property can take more than one value. We have decided to create a property HasHistologiCriteria and five sub properties (HasVascularProliferation, HasN ecrosisActivity, HasMitoticActivity, HasAtypia and H asCellularDensity) for each histologic criterion. Each of them is a functional property so it can only have one value.
Classes for grading system
The OWL representation of each WHO grade has required an interpretation of the WHO definition. Although it is not mentioned explicitly, the WHO grades are exclusive. The corresponding NCIT WHO classes were made disjoint. For example, The WHO grade IV is represented by the intersection of six restrictions that correspond to the five histologic criteria and the Disease_Grade_Modifier class. Figure 5 illustrates the definition of WHO grades.
Figure 5.
Definition of the WHO Grades in our ontology
Classification tests
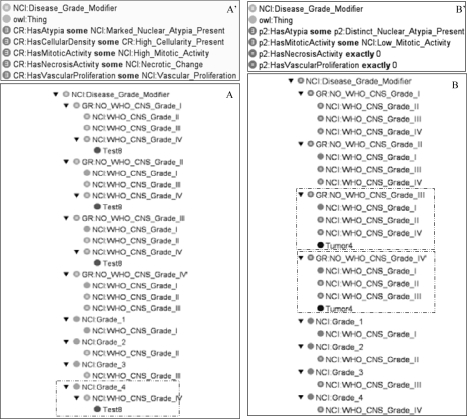
All of the generated tests (15) were classified correctly. Figure 6A illustrates the histologic feature of Test8 and the result of classification for this test. These constraints correspond to a WHO_CNS_Grade_IV. We show that Test8 is correctly classified under WHO_CNS_Grade_IV. Eleven clinical reports are used for the class-based reasoning. For ten of them, the WHO grades have been determinated. The tumor for which the grade could not be unequivocally determined (Tumor4, Figure 6B), was not classified under any of the four grades because of the open-world-assumption. However, it was classified under NO_WHO_CNS_Gr ade_III and NO_WHO_CNS_Grade_VI. We were then able to restrict the possible grades to 1 or 2. The files for the ontology and the test classification are available online (http://www.ea3888.univ-rennes1.fr/~marquet/ontology/GliomaClassification/). The classification is performed in 2 to 3 seconds on a regular desktop.
Figure 6.
Class-Based-Reasoning and description for the Test8 (A and A’) and Tumor4 (B and B’). Test8 is correctly classified under WHO_CNS_Grade_IV. The grade of tumor4 was not determined, however the grades to which it does not belong (grade III and Grade IV here) are provided.
Discussion
We have shown that automatic grading of glioma tumors according to the WHO grading system can be performed using a set of diseases and histologic features extracted from the NCIT and extending it. We provided definitions for the WHO grading system and added five classes and seven relationships to the set of classes extracting from NCIT.
Among the biomedical ontologies available, the NCIT has been created with the goal of providing a controlled vocabulary of the cancer domain which can be used by specialists in the various sub-domains of oncology. Several authors [5, 6] analyzed the NCIT as a terminology and as an ontology. These authors have shown that only a small portion of the NCIT can be used for automatic classification. In addition, these studies describe problems concerning relations. For example, the is-a relation reflects different types of partition of reality and they noticed an inconsistent use of the OWL-qualifiers allValuesFrom and someValuesFrom in the NCIT [6]. In the NCIT, the histologic criteria associated with glioma tumors are described with the OWL property Disease_Has_Finding. This property is used for describing all of the histologic features and does not enable to use the universal and cardinality restrictions such as those shown in Fig 5. In addition, the grade and range of this property do not permit to describe grades. For these reasons, we have created new properties for describing the histologic criteria. Despite these problems, we preferred to extend the NCIT in order to overcome these limitations rather than developing a new ontology from scratch. We made as few modifications as possible: namely new classes, new properties and new definitions. For the Mitotic Activity, we have chosen to add missing symbolic values. The partition of the Mitotic Activity provided by the NCIT does not correspond to the partition of the HFP (high-power fields) values used for the Mitotic Activity in the glioma. We believe our modifications to be temporary patches in places where the NCIT is still ontologically incomplete. Future versions will probably address these issues and be compatible with our solution.
Because all the tests from the first set were correctly classified, we believe our system to be correct. The second test has shown that our classification can be used for representing clinical reports. The nogrades were represented to classify tumors if the information found in the clinical reports is not complete.
The system always classifies tumors under a grade or under one or more NoGrades. A tumor can not be classified under the four noGrades at the same time indeed the histologic criteria are exclusive. We focus our study on the malignant grades. The Grade 1 has not been represented in our system because it is a benign tumor. Moreover, grade 1 can present occasionally an atypia, a vascular proliferation, a mitosis activity or a necrosis activity. The presence of these criteria presents a problem for automatic classification. Indeed, the WHO criteria are not clear for the WHO grade 1. Grade 1 is particular compared with the other grades. It has specific characteristics that are not taken into account by the 5 criteria of WHO such as the presence of rosenthal fibers and eosinophilic granular bodies. The Grade 1 representation requires adding other knowledge in ontology. Moreover, these last two criteria (rosenthal fibers and easoniphilic granular bodies) are not often described in the reports
We have chosen class-based reasoning for the test classification because the only means to represent negative restrictions at the instance level is to create artificial classes [7]. Instance-based reasoning seems to be better for representing pathologic reports.
However, these reports would have been difficult to represent and the result would have been the same. Related works focused on the representation of the TNM classification [7,8]. The TNM criteria involve the anatomical location of the primary tumor and of its possible metastasis as well as the involved lymph nodes. The TNM is based on a score based on three axes: the description of the tumor, the spreading into lymphatic nodes and the possible metastasis. The stage is determined according to the TNM score. For example, the stage 0 of the colon carcinoma corresponding to Tis, N0, M0. Some of the TNM criteria refer to anatomical entities as location landmarks, with different granularities between clinical descriptions and the definitions of the staging criteria. In the WHO classification of the glioma, as only the histological features are used to describe the grade of the tumor. We do not have to address such granularities issues.
Our study and other works [7, 8] have shown the feasibility of reusing (portions of) existing ontologies such as the NCIT, the Foundational Model of anatomy [7] and bioinformatics ontologies [9]. In addition this approach can be generalized to other types of tumors.
Our study shows that such reasoning tasks only require to add a few classes and definitions. This is possible because we leverage the knowledge represented in the ontology. Other approaches such as rule-based systems would require this knowledge to be embedded in rules, leading to systems more complicated to design and to maintain.
Acknowledgments
This work was supported by a grant from the Région Bretagne (20046805)
References
- 1.David FG, Mc Carthy BJ. Epidemiology of Brain Tumors. Curr Opin Neurol. 2000;13:635–40. doi: 10.1097/00019052-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Golbeck J, Fragoso G, Hartel F, Hendler J, Oberthaler J, Parsia B. The national cancer institute’s thésaurus and ontology. Journal of web semantics. 2003;1(1):75–80. [Google Scholar]
- 3.Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, Cavenee WK.The WHO classification of tumors of the nervous system J Neuropathol Exp Neurol 2002613215:25 [DOI] [PubMed] [Google Scholar]
- 4.Hartel FW, Corronado S, Dionne R, Fragoso G, Golbeck J. Modeling a description logic vocabulary for cancer research. Journal of Biomedical Informatics. 2005;38:114–129. doi: 10.1016/j.jbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Ceusters W, Smith B, Golberg L. A terminological and ontological analysis of the NCI thesaurus. Methods of Information in Medecine. 2005:498–507. [PubMed] [Google Scholar]
- 6.Kumar A, Smith Barry. Oncology ontology in the NCI thesaurus. Proceedings of the Artificial Intelligence in Medicine Europe Conference AIME. 2005:213–220. [Google Scholar]
- 7.Dameron O, Roques E, Rubin DL, Marquet G, Burgun A. Grading lung tumors using OWL-DL based reasoning. 9th International Protégé Conference; 2006. [Google Scholar]
- 8.Kumar A, Lina Yip Y, Smith B, Marwede D, Novotny D. An Ontology for Carcinoma Classification for Clinical Bioinformatics. Medical Informatics Europe (MIE 2005), Geneva. :635–640. [PubMed] [Google Scholar]
- 9.Kumar A, Yip YL, Smith B, Grenon P. Bridging the gap between medical and bioinformatics: an ontological case study in colon carcinoma. Comput Biol Med. 2006;36(7–8):694–711. doi: 10.1016/j.compbiomed.2005.07.001. [DOI] [PubMed] [Google Scholar]