Abstract
Objective:
Identify the set of features that best explained the variation in the performance measure of TREC 2006 Genomics information extraction task, Mean Average Passage Precision (MAPP).
Methods:
A multivariate regression model was built using a backward-elimination approach as a function of certain generalized features that were common to all the algorithms used by TREC 2006 Genomics track participants.
Results:
Our regression analysis found that the following four factors, were collectively associated with variation in MAPP: (1) Normalization of keywords in the query into their respective root forms (2) Use of Entrez gene thesaurus for synonymous terms look-up (3) Unit of text retrieved using respective IR algorithms and (4) The way a passage was defined by the respective run.
Conclusion:
These reasonably likely hypotheses, generated by an exploratory data analysis, are informative in understanding results of the TREC 2006 Genomics passage extraction task. This approach has general value for analyzing the results of similar common challenge tasks.
Introduction
Challenge evaluations in fields such as information retrieval (IR) create a common task for different research groups using a common test collection so they may compare different algorithms to determine which perform best on a common standard of documents and information needs (also called topics or queries). In the IR field, the best-known known challenge evaluation is the Text REtrieval Conference (TREC), sponsored by the National Institute of Standards and Technology (NIST). TREC operates on an annual cycle and provides an infrastructure for evaluation and cross comparison of various IR methodologies [1]. TREC is organized into several tracks including a Genomics track that traditionally focused on retrieval of relevant documents reflecting the real information needs of biomedical researchers.
In 2006, the TREC Genomics Track focused on retrieval of short passages that answered a biological question, while providing users the textual context of the retrieved answers [2]. The main idea is thus to save the user’s time usually spent in manually locating the answer of interest in the full text and to let the user decide whether or not a passage is relevant by providing him/her with supporting evidence to the “answer”. For example, a biologist who seeks to know the antibodies that have been used to detect the protein TLR4, a paragraph like the following that succinctly addresses the need while providing the context for supporting evidence could be invaluable: “An aliquot of cytoplasmic protein (20–100 μg) was utilized for Western blotting with specific primary antibodies (Santa Cruz Biotechnology) to TLR4 (sc-10741)”. [TREC 2007 genomics protocol]. The document collection for this task comprised of 162,259 full-text articles from 49 journals published electronically by Highwire Press [www.highwire.org]. Gold standard relevance judgments were prepared by a panel of expert biologists who manually reviewed passages, which represented the “answers” to the queries. The precision at each passage that was judged relevant was calculated as the fraction of characters that overlapping with those in the gold standard divided by the total number of relevant characters nominated in all passaged. These individual passage precisions were then averaged over all the topics for this run to measure the Mean Average Passage Precision (MAPP) score [2].
One of the problems with a common task such as the TREC Genomics Track is in comparing the results of different groups’ experimental runs. The many participants submitted runs using many different approaches, and they did not necessarily conduct systematic component- (or feature-) level “leave-one-out” experiments to help determine which features provided real benefit. Individually, the systems had to be taken as a whole, and therefore did not provide detailed information about their individual components. However, by examining a large group of systems together with descriptions of their components, patterns could be discerned that provide insights into the task under study and estimates of the positive and negative effects of system components on performance measures.
In this paper, we focus on identifying the set of system features that were highly correlated with a system run’s high or low MAPP performance. Viewed at a high level of abstraction, all the submissions took some variation of the following steps for extracting passages for a given query:
Document preprocessing and representation. In this initial phase, documents are processed for cleaning out text perceived as uninformative, converted into a format that provides easier cross-linkage and then represented as an inverted file of indices, just the way Medline articles are represented using MeSH terms for faster and automated retrieval.
Query expansion. In this phase, the user’s query is augmented by first identifying the most informative keywords and then expanding it using their corresponding synonyms. The intuition behind doing a query augmentation is the expectation that synonymous terms in the expanded version of the query might increase the chances of retrieval of more related documents.
Document retrieval. A range of standard IR algorithms, their variations and an ensemble of approaches [3] were employed for retrieval of text that best matched the query. The unit of text that was retrieved using these algorithms varied across the runs, however, ranging from a sentence [4] to the whole document [5][6] while most runs chose a paragraph (defined by the track administrators) [3][7][8] as their unit of retrieval.
Passage extraction: Passages were extracted as a subset of sentences from the top few text units that were retrieved in the previous phases using a variety of techniques with the general assumption that most relevant passages are found in most relevant documents. The majority of the runs did this task of passage extraction by first identifying the sentence that had the highest density, also known as coverage factor [4], of query terms and then expanding in both upstream and downstream directions until a sentence with no query term was encountered [5][6][7][8]. Few runs have defined their passages using HMMs [9], by representing sentences as hidden states for passage relevance, and minimal interval semantics [10][11]. Once a passage was extracted, it was rescored by certain runs [5][6][7] while some chose to rank them according to the rank of their parent text units that were retrieved and scored in the previous phase.
Methods
To gain insights into the effect of different algorithms employed for MAPP, an exploratory data analysis (EDA) approach was taken. A multivariate regression model in SPSS [12] is built for this purpose using a backward elimination approach to identify the factors that were most relevant to explaining the variability in MAPP. Data were collected by manually reviewing 26 TREC 2006 Genomics Track publications, distilling the overall approaches and grouping the variants into eighteen buckets of informational attributes that we suspect to be correlated with retrieval performance (see Table 1). In all, 45 data points were collected after eliminating from the dataset redundant approaches, which we define as approaches that are otherwise similar except for differences in some parameter value of the retrieval algorithm.
Table 1.
The table shows the general steps taken by participants at each phase of the retrieval and the features we have extracted for use in multivariate regression modeling.
| Step | General approaches | Features extracted |
|---|---|---|
| Document preprocessing and indexing | -html to plain text by eliminating the tags | -Stemming
-Stop-words filtering |
| -html to xml | ||
| -filtering out certain sections, such as references and acknowledgements | ||
| -conversion of html to records of a relational database structure[IIT] | ||
| -Stemming and stopwords filtering | ||
| Query expansion | -Identification of keywords using automated, manual and interactive methods |
|
| -Synonyms lookup using online biomedical dictionaries such UMLS, Entrez Gene, MeSH, HUGO, MetaMAP etc. | ||
| -Assigning weights to keywords in the query | ||
| -Normalizing keywords into their root forms | ||
| Document retrieval | -Use IR algorithms such as tf-idf, BM25, I(n)B2,dtu.dtn,Jelinek- Mercer smoothing, KL- divergence, SVM classifiers and an ensemble of standard algorithms | - Retrieval Algorithm
- Unit of Text retrieval |
| -Retrieve different units of text, such as document, paragraph, subset of paragraphs and a sentence, using these algorithms | ||
| Passage retrieval | -Use one of the following for passage extraction:
|
-Passage Definition
-Passage rescoring |
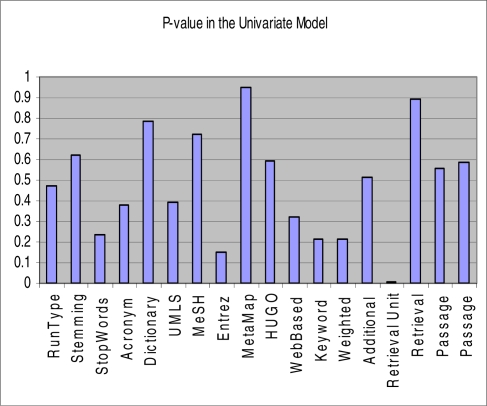
For regression analysis, we first checked for the normality of MAPP and found it to be highly skewed. Normality was improved by using a square root transformation and hence, we used square root MAPP (SqrtMAPP) as the dependent variable of interest throughout our regression analysis. A univariate regression analysis was then performed to weed out the attributes that were not relevant to the dependent variable of interest, SqrtMAPP (see Figure 1). Type-I error rate, α was set to 0.3 as the critical value with the intuition that attributes that might not appear highly significant in explaining variation in MAPP by themselves might turn out to be significant in presence of a few other significant factors in the multivariate model, and hence need to be retained. Attributes that did not seem highly significant in their corresponding univariate models but were strongly assumed to be relevant from a priori knowledge were also retained for multivariate regression analysis.
Figure 1.
The figure shows the p-values of the attributes in their corresponding univariate models.
Results
From the univariate regression analysis, we found: filtering of stop words, use of Entrez gene thesaurus for synonyms look-up during the query expansion, normalization of keywords identified in the query into their root forms, unequal weights assignment to keywords reflecting the importance relative to each other and unit of text retrieved using the IR algorithm were linearly associated with SqrtMAPP as their corresponding p-values were less than our threshold αvalue 0.3, and hence were assumed to be important in explaining the variability in MAPP. In addition, we have considered: algorithm for retrieval, definition of passage and rescoring of passage, although not significantly associated with SqrtMAPP on their own, for multivariate analysis as we suspect that they might be helpful, with a synergistic effect from the presence of other significant variables in the multivariate model, in predicting the dependent variable.
We then started building a multivariate regression model with the eight parameters (five which were significant in the univariate models and three which we suspected to be important) from the univariate models, and proceeded backwards by eliminating one parameter at a time that was insignificant until the model had all significant parameters. When encountered with a choice between two insignificant parameters for elimination from the model, we chose the one which we suspected to be unrelated to the dependent variable. With this intuition, we have proceeded with the backwards elimination regression approach and arrived at the following regression model (see Table 2):
Table 2.
The table shows the p-values corresponding to the parameters in the final multivariate regression model obtained from backwards elimination approach using SPSS. The Beta column represents the coefficients estimate of each of these parameters in the model.
| S.No | Attribute Name | p-value | Beta |
|---|---|---|---|
| 1 | Keyword Normalization | 0.056 | No = −0.044 |
| Yes = 0 [ref] | |||
| 2 | Entrez Gene | 0.071 | No = 0.041 |
| Yes = 0 [ref] | |||
| 3 | Retrieval Unit | 0.002 | Document = −0.167 |
| Legal-span = −0.056 | |||
| Sentence = −0.208 | |||
| Trimmed-paragraphs = 0[ref] | |||
| 4 | Passage Definition | 0.032 | HMMs based estimate = 0.009 |
| Legal-span = 0.068 | |||
| Minimal-interval-semantics = 0.072 | |||
| Sentence = 0.115 | |||
| Trimmed- paragraphs = 0 [ref] |
SqrtMAPP = 0.282 – 0.044 * KeywordNormalization [=no] + 0.41 * EntrezGene [=no] –0.167*retrievalUnit [=document] – 0.056 * retrievalUnit [=legal-span] – 0.208 * retrievalUnit [=sentence] + 0.009 * PassageDefinition [=HMMs-based-estimate] – 0.068 * PassageDefinition [=legal-span] + 0.072 * PassageDefinition [=minimal-interval-semantics] + 0.115 * PassageDefinition [=sentence]
Note that the parameter values not shown in the above model are used as reference values.
Discussion
From our final regression model, we infer that lack of normalization of keywords, usage of document or a legal span or a sentence, and defining legal span as a passage were negatively correlated with MAPP, Not using the Entrez gene thesaurus [14], and using HMMs or minimal interval semantics for passage extraction were positively correlated with and explain 48.2% (adjusted R2 measure obtained from SPSS output tables) of variability in SqrtMAPP. This is logical as blind and unfiltered use of synonymous terms lookup can often penalize the precision of the retrieval [10], and retrieving a whole document or just a sentence, as opposed to paragraph or a subset of paragraph, probably decreases the chances of accurately locating the most important stretch of sentences that could be labeled as a passage. Also, use of minimal interval semantics or HMMs for passage extraction might improve results as a passage may contain a sequence of relevant sentences bounded by irrelevant ones within the retrieval unit. Also note from the model that passages that have been defined as a single sentence were most correlated with MAPP reflecting the fact that short stretch of text was most rewarded with good precision rating while longer text that had a higher proportion of non-relevant characters was penalized.
The regression model constructed here is an explanatory aid in understanding the affect of different system features on MAPP. However, it is likely not the optimal model and may not have much predictive value it and of itself for several reasons. There are inherent problems with multivariate regression model building using backwards elimination [13]. We lacked a large data collection and categorized system features after the systems were completed, which together results in the data available on individual features being rather noisy. Using a different set of attribute values that could be a priori assigned by system developers and including a higher number of data points, could lead to a model with higher significance and predictive value. Despite these shortcomings, our approach can be a very informative way to determine what system features should be studied more closely in building an optimized biomedical question answering system,. Given that the current best performance with this task is about MAPP = 0.14 [2], there is sufficient room for improvement to warrant further study.
Conclusion
Using multivariate regression analysis, we found that four factors, among a set of seventeen factors, were collectively associated with changes in mean average passage precision: (1) Normalization of keywords in the query into their respective root forms (2) Use of Entrez gene thesaurus for synonymous terms look-up (3) Unit of text retrieved using respective IR algorithms and (4) The way a passage was defined by the respective run. Exploratory data analysis, the task of constructing reasonably likely hypotheses by looking at the patterns in data can be quite informative to understanding results of common challenge tasks such as those in the TREC Genomics track.
Acknowledgments
The authors gratefully acknowledge Nichole Carlson PhD., Dan Rubado and Grant 0325160 of the US National Science Foundation.
References
- 1).National Institute of Standards and Technology [homepage on the Internet]. Gaithersburg, MD: National Institute of Standards and Tecnology. [Updated 2007 March 13; cited 2007 March 14]. Available from: http://trec.nist.gov/overview.html
- 2).Hersh W, Cohen AM, Roberts P, Rekapalli HK. TREC 2006 Genomics Track Overview. Proceedings of the Text Retrieval Conference 2006; Gaithersburg, MD. 2006. [Google Scholar]
- 3).Demner-Fushman D, Humphrey SM, Ide NC, Loane RF, Smith LH, Tanabe LK, Wilbur WJ, Aronson AR, Ruch P, Ruiz ME. Finding Relevant Passages in Scientific Articles: Fusion of Automatic Approaches vs. an Interactive Team Effort. Proceedings of the Text Retrieval Conference 2006; Gaithersburg, MD. 2006. [Google Scholar]
- 4).Bergler S, Schuman J, Dubuc J, Lebedev A. BioKI, A General Literature Navigation System at TREC Genomics 2006. Proceedings of the Text Retrieval Conference 2006; Gaithersburg, MD. 2006. [Google Scholar]
- 5).Trieschnigg D, Kraaij W, Schuemie M. Concept Based Document Retrieval for Genomics Literature. Proceedings of the Text Retrieval Conference 2006; Gaithersburg, MD. 2006. [Google Scholar]
- 6).Yu N, Lingpeng Y, Jie Z, Jian S, Donghong J.I2R at TREC 2006 Genomics Track. Proceedings of the Text Retrieval Conference 2006: Gaithersburg, MD; 2006.
- 7).Wan R, Takigawa I, Mamitsuka H, Anh NV.Combining Vector-Space and Word-Based Aspect Models for Passage Retrieval. Proceedings of the Text Retrieval Conference 2006: Gaithersburg, MD; 2006.
- 8).Goldberg AB, Andrzejewski A, Van Gael J, Settles B, Zhu X, Craven M.Ranking Biomedical Passages for Relevance and Diversity: University of Wisconsin, Madison at TREC Genomics 2006. Proceedings of the Text Retrieval Conference 2006: Gaithersburg, MD; 2006.
- 9).Jiang J, He X, Zhai CX.Robust Pseudo Feedback Estimation and HMM Passage Extraction: UIUC at TREC 2006 Genomics Track. Proceedings of the Text Retrieval Conference 2006: Gaithersburg, MD; 2006.
- 10).Dorff KC, Wood MJ, Campagne F.Twease at TREC 2006: Breaking and Fixing BM25 Scoring With Query Expansion, A Biologically Inspired Double Mutant Recovery Experiment. Proceedings of the Text Retrieval Conference 2006: Gaithersburg, MD; 2006.
- 11).Boldi P, Vigna S.Efficient lazy algorithms for minimal-interval semantics Proceedings of the 13th Symposium on String Processing and Information Retrieval, number 4209 in Lecture Notes in Computer Science, pages 134–149.Springer-Verlag; 2006 [Google Scholar]
- 12).SPSS for Windows. 2006. Version 15. Chicago: SPSS Inc. [computer program in CD-ROM].Available in SPSS Inc. SPSS web page available in:http://www.spss.com/
- 13).Kleinbaum D, Kupper LL, Muller KE, Nizam A. Testing hypothesis in Multiple Regression. In: Mazow Kugushev A., editor. Applied Regression Analysis and Other Multivariable Methods. 3rd ed. Crawfordsville: Duxbury Press; 1998. pp. 136–159. [Google Scholar]
- 14).Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Research. 2005 Jan;33:D54–58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]