Abstract
We introduce the notion that research administrative databases (RADs), such as those increasingly used to manage information flow in the Institutional Review Board (IRB), offer a novel, useful, and mine-able data source overlooked by informaticists. As a proof of concept, using an IRB database we extracted all titles and abstracts from system startup through January 2007 (n=1,876); formatted these in a pseudo-MEDLINE format; and processed them through the SemRep semantic knowledge extraction system. Even though SemRep is tuned to find semantic relations in MEDLINE citations, we found that it performed comparably well on the IRB texts. When adjusted to eliminate non-healthcare IRB submissions (e.g., economic and education studies), SemRep extracted an average of 7.3 semantic relations per IRB abstract (compared to an average of 11.1 for MEDLINE citations) with a precision of 70% (compared to 78% for MEDLINE). We conclude that RADs, as represented by IRB data, are mine-able with existing tools, but that performance will improve as these tools are tuned for RAD structures.
Introduction
For many of us engaged in biomedical research, especially those working in non-industrial settings like academic medical centers, research administration is a chore, a necessary evil that steals time from “real work.” Administrative burdens often are most acute when dealing with regulatory administration, like the Institutional Review Board (IRB) or the conflict of interest compliance office.1–3 Yet, for a host of sound ethical, legal, and managerial reasons, research administration and its attendant processes of data collection is, and will remain, a fact of life for the biomedical researcher.
We argue here that there is an interesting and useful way to turn this sow’s ear into a silk purse: we introduce the novel perspective of viewing research administration databases (RADs) as an important target for biomedical informatics research, especially in an era where translational research methods are increasingly gaining sway. We offer a specific, practical pilot feasibility example of how a RAD (an electronic IRB repository) can be mined for semantic content using natural language processing (NLP) techniques for data unavailable from any other source, and suggest two lines of future work in this underdeveloped but potentially fruitful area of research.
Background
Today no one would dispute the value to researchers of what is collectively referred to as administrative data (e.g., admission and discharge ICD9/10 codes, CPT procedural codes, etc.). These data are used routinely in healthcare quality improvement research and across a wide variety of epidemiological, health services, and public health informatics research. They also form the core of important new initiatives such as biosurveillance and patient safety (e.g., they drive the AHRQ’s patient safety indicators4). Administrative data were originally collected to improve a management process (in this case, billing collection and revenue reporting). Only later did their significant research potential become apparent. We argue that the same is true of research administrative data.
RAD data sources in general
The universe of data at an academic medical center is expansive; there is considerable overlap between the content (and the intended use) of data sources as diverse as electronic medical record systems, enterprise-wide data warehouses, business and accounting systems, research data stores, research administrative systems, library information retrieval systems, and the like. The lines that distinguish these systems are blurring as academic healthcare embraces the translational research model promulgated by the National Institutes of Health Roadmap Initiative.5 As data and knowledge are shared under the translational model, the traditional silos represented by these various databases are being integrated or interconnected.6
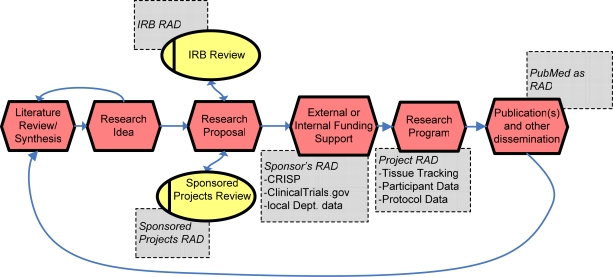
We illustrate in Figure 1 the fundamental research process flow, starting from a literature-inspired research idea through to publication, and indicate the obvious points where research administrative data are captured. The shaded rectangles in Figure 1 show several common RAD data sources. The PubMed search engine,7 which has opened up the tremendous resources of the National Library of Medicine’s MEDLINE literature archive, may seem out of place here, an unlikely RAD. However, in a very direct way, it closes the research process flow loop.
Figure 1.
Simplified research cycle showing the major research administrative data sources (in rectangles).
IRB management systems, an important specific RAD data source
IRBs serve the critical role of reviewing research that uses human subjects, ensuring compliance with a complicated set of regulatory and ethical mandates. This is an information intensive process, and increasingly IRBs are turning to automation to assist in managing that information. The stress on e-solutions for IRBs specifically, and research administration generally, receives widespread institutional and federal support.8 While no single group tracks the extent of IRB automation, an informal poll at a recent meeting of the largest IRB professional organization, PRIM&R,9 indicated that about 20% of the IRBs represented have substantial automation.10 Many of these, such as the e-IRB at the University of California San Diego, are home-grown software projects. Others, like ours at the University of Utah, use a commercial base system (two commercial examples are ClickCommerce11 and API’s BRANN12).
Methods
Data source
Supported by a grant from the National Center for Research Resources (2S07RR0181839), the University of Utah developed and implemented an electronic IRB management system, called ERICA. It is built on a Web-based, object oriented document management architecture,11 and contains discrete data fields and a tightly controlled document revisions database. In this paper, we focus on two discrete text fields, the IRB protocol abstract and title. In ERICA, a protocol abstract is a short, unstructured, free-text summary of proposed research being submitted to the IRB. A protocol title is also an unstructured free text field of arbitrary length, but the data entry “box” is only one line long, encouraging brevity.
Data extraction and semantic processing
We extracted all IRB abstracts/titles submitted from system startup through January 31, 2007 (n=1,876), including all studies using human subjects from all departments on the campus. Since the goal of this pilot work is to assess the feasibility of semantic processing on these raw research administrative texts, we used the National Library of Medicine’s SemRep system to extract medically relevant semantic relations (also known as predications). SemRep is a symbolic natural language processor tuned to extract semantic relations for medical treatments, drug interactions, and pharmaco-genomics. The SemRep system is well described elsewhere.13, 14 In brief, it works by passing text through a shallow parser (a stochastic tagger fed by tokens from a lexical analyzer built on the UMLS SPECIALIST Lexicon), whose output is processed by a sophisticated set of biomedical indicator rules. These rules assert semantic relations in the form:
Argument1 RELATION Argument2
We use the term LHS (left-hand side) to refer to Argument1 and RHS (right-hand side) to refer to Argument2. Relations specify the semantic connection between the LHS and RHS arguments. For example, in the sentence “Doppler echocardiography can be used to diagnose left anterior descending artery stenosis in patients with type 2 diabetes” SemRep returns the relation (one of four):
Echocardiography, Doppler DIAGNOSES Acquired stenosis
To suit SemRep, which is designed to process literature citations, the IRB sample was re-formatted into a minimal MEDLINE format including: a pseudo-identifier (PMID, the MEDLINE id), title (TI), and the abstract (AB). These were submitted as a batch to the ‘Batch SemRep’ interface on the Semantic Knowledge Representation Web site.15
To assess the precision of SemRep on the IRB texts, we extracted the 1,625 TREATS relations and manually reviewed 20% of these chosen at random (n = 325 sentences; after removing the non-health studies as they were detected in the manual review, final n = 296). An IRB expert reviewer and physician (JFH, a former IRB chairman) reviewed these sentences to determine true- and false-positives in the subset. Recall was not assessed in this pilot work owing to manpower and time constraints, but a representative example is cited in the Results section.
Results
SemRep’s overall performance in extracting relations from the IRB corpus is shown in Table 1. That table also shows typical SemRep performance on a convenience sample of 14,921 MEDLINE citations that had already been analyzed for another project. These were comprised of a mix of PubMed queries looking at 15 diseases/syndromes, 9 drugs, and 1 diagnostic technique.
Table 1.
Crude Frequency of Extracted Relations (IRB Texts vs. SemMed sample set)
| IRB Texts | Sample MEDLINE | |
|---|---|---|
| Total # of texts | 1,876 | 14,921 |
| Total # of relations found (abstract + title/abstract only) | 11,876/10,516 | 166,178 |
| Avg. # relations per text (abstract + title/abstract only) | 6.3/5.3 | 11.1 |
| % texts with no relations found | 19.2% | 1.3% |
An example of SemRep’s output is shown below for the first text in the corpus. The sentences (1) and (3) are two of seven (including the title) that occurred in the text; the relations (2), (4), (5), (6), and (7) are all the relations SemRep found in those two sentences (it found relations only for these two sentences):
(1) “These ECG effects are of interest because a few years ago the FDA issued a warning that high doses of droperidol can cause changes in the ECG that can lead to potentially dangerous, abnormal heart rhythms.”
(2) Droperidol CAUSES Potentially abnormal finding
(3) “There is widespread agreement among experts that these drug induced ECG changes are very unlikely to occur in a severe form at the low dose used in the prevention and treatment of PONV.”
(4) Pharmaceutical Preparations CAUSES Electrocardiogram change
(5) Low dose CONCEPTUAL_PART_OF Prevention
(6) Low dose CONCEPTUAL_PART_OF Therapeutic procedure
(7) Pharmaceutical Preparations TREATS Postoperative Nausea and Vomiting
In the sub-sample of the 296 TREATS relations used to assess precision, SemRep returned 206 true positives/90 false positives for a precision of 70%. Table 2 illustrates the sort of semantic relations and LHS/RHS arguments SemRep found in the IRB text corpus. The table lists the top 10 most frequent values for these three groups, and also lists a few rare values to illustrate the range of both relations and arguments SemRep was able to detect. An example of a false negative, text containing a relation that SemRep might have found that is pertinent in the IRB context but did not, is
Table 2.
Distribution of Relation Semantic Types and Their Arguments (n = 11,876 relations from 1,876 IRB texts; nb: these LHS, Relations, and RHS columns are not from the same asserted predications)
| Top 10 LHS Arguments | Count (% of relations) | Top 10 Relations | Count (%) | Top 10 RHS Arguments | Count (%) |
|---|---|---|---|---|---|
| Therapeutic Procedure | 444 (3.7) | PROCESS_OF | 3,073 (25.9) | Patients | 1,898 (16) |
| Pharmaceutical Preparations | 128 (1.1) | TREATS | 1,625 (13.7) | Child | 504 (4.2) |
| Universities | 116 (1.0) | PART_OF | 1,276 (10.7) | Population Group | 225 (1.9) |
| Scientific Study | 101 (0.9) | LOCATION_OF | 981 (8.3) | Woman | 180 (1.5) |
| Disease | 92 (0.8) | USES | 556 (4.7) | Individual | 175 (1.5) |
| Placebos | 85 (0.7) | ISA | 452 (3.8) | Clinical Research | 159 (1.3) |
| Primary Operation | 78 (0.7) | ISSUE_IN | 401 (3.4) | Therapeutic Proced. | 150 (1.3) |
| Operative Surgical Procedures | 75 (0.6) | CONCEPTUAL_PART_OF | 388 (3.3) | Disease | 121 (1.0) |
| Malignant Neoplasms | 72 (0.6) | AFFECTS | 368 (3.1) | Infant | 112 (0.9) |
| Biologic Development
|
69 (0.6)
|
METHOD_OF
|
355 (3.0)
|
Adolescent
|
99 (0.8)
|
| Sample Rare LHS Arguments | Count | Sample Rare Relations | Count | Sample Rare RHS Arguments | Count |
| herpes simplex virus type 2 gd subunit vaccine induced antibody | 1 | NEG_LOCATION_OF | 1 | preformed anti hla antibody | 1 |
| Homeopathic Remedies | 2 | NEG_METHOD_OF | 2 | Personality inventories | 2 |
| Chronic Childhood Arthritis | 4 | COMPLICATES | 4 | juvenile delinquent | 4 |
(8) “This study will help describe the effect that intravenous injection of dolasetron and droperidol has on the electrocardiogram (ECG).”
An informed human reader would infer that either dolasetron or droperidol AFFECTS the ECG.
Discussion
SemRep, which was designed to extract semantic relations from literature citations, does surprisingly well at finding meaningful relations in the IRB corpus. If the IRB texts that produced no relations are removed, the crude frequency jumps from 6.3 to 7.8 relations per text, which compares favorably to the 11.1 relations per text found in the MEDLINE citations, which averaged about twice the size. Why would so many (19.2% of the IRB texts) produce no relations at all? In large part, this is due to a dilution effect: the IRB corpus contains texts for all human studies projects on campus, including those conducted in non-biomedical departments such as psychology, sociology, education, family and consumer studies, economics, etc. The exact proportion of these “main campus” studies is surprisingly hard to extract from ERICA, but according to the University of Utah IRB director, about 25–30% of all IRB studies come from outside the health campus.10
SemRep also demonstrated a credible precision of 70% in extracting semantic relations from the IRB texts. For MEDLINE citations, SemRep exhibits a precision of ~78%.13 The categories of arguments and relations extracted are illuminating. The low percentage of any single LHS (max ~ 4%) suggests that early research (i.e., early in Figure 1) is focused on a broad array of research themes. The concentration of RELATIONS on process, treatment, and genomics (note: the majority of the PART_OF relations are genomic) suggests what kind of research is underway at Utah. Finally, that a quarter of the relations’ RHSs are specific references to human studies makes sense in the IRB human-subjects context. Note from Table 1 that titles are a very rich source of relations, adding about one relation each to the crude frequency.
Limitations
SemRep is extensible, but we chose to use it “out-of-the-box,” clearly a limitation on determining how much semantic knowledge can be extracted from these IRB texts. The lack of a characterization of the recall in this study clearly is also problematic. Finally, without a better way to summarize and visualize the relations extracted, the utility of the IRB relations will be difficult to realize. The NLM is researching actively both summarization and visualization of SemRep output.13, 16
Future work
In addition to remediating the limitations noted above, we plan to extend SemRep to tune it for RAD-oriented processing. For IRB texts it does surprisingly well as-is, an indication of how similar IRB texts are to citations, but its extraction performance can be improved.
The semantic content of IRB data could be used in several ways. IRB semantic mining could be used to characterize presearch, the portion early in the research life cycle where a nascent research idea is just emerging. Exactly in the same way PubMed lays out what has been done (i.e., the culmination of a research idea brought to fruition over a course of many years, from conception to funding to execution of work to publication), presearch data provide a roadmap of where biomedicine is heading. PubMed highlights the state of scientific innovation as it was three to five years ago. Presearch highlights the state of scientific innovation as it is now. That information would be useful to translational research efforts, say to allocate/forecast resources at an institutional or cross-institutional level in an informed way. Another example is helping identify and connect related research projects conducted by two or more otherwise isolated research groups. This is an important translation research goal, one that is surprising hard to realize at the presearch stage because the IRB is often the sole common point of intersection in the formative stages.
Conclusion
We asserted that research administrative databases, largely overlooked as a data source by informaticists, could constitute an important new field of inquiry in biomedical informatics. The first step to prove that assertion is to prove that these sources can be mined at all. We presented a case study of mining IRB abstracts and titles for their semantic content, and showed that, even out-of-the-box, a mature semantic knowledge extraction tool (SemRep) does a very good job on the RAD source of an IRB.
Acknowledgments
This work was supported by an NLM senior biomedical informaticists training grant, (1F38LM8478-1) and by the Intramural Research Program of the NIH, NLM. The authors are indebted to the staffs at both the Cognitive Science Branch of the Lister Hill Center at the NLM and at the University of Utah IRB for their assistance in data collection.
References
- 1.Green LA, Lowery JC, Kowalski CP, Wyszewianski L. Impact of institutional review board practice variation on observational health services research. Health Serv Res. 2006 Feb;41(1):214–30. doi: 10.1111/j.1475-6773.2005.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McWilliams R, Hoover-Fong J, Hamosh A, Beck S, Beaty T, Cutting G. Problematic variation in local institutional review of a multicenter genetic epidemiology study. JAMA. 2003 Jul 16;290(3):360–6. doi: 10.1001/jama.290.3.360. [DOI] [PubMed] [Google Scholar]
- 3.Vick CC, Finan KR, Kiefe C, Neumayer L, Hawn MT. Variation in Institutional Review processes for a multisite observational study. Am J Surg. 2005 Nov;190(5):805–9. doi: 10.1016/j.amjsurg.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Zafar A. The AHRQ National Resource Center for Health Information Technology (Health IT) Public Web Resource. AMIA Annu Symp Proc. 2006;1154 [PMC free article] [PubMed] [Google Scholar]
- 5.NIH 2007. cited February 20, 2007.; NIH Roadmap Initiatives.. Available from: http://nihroadmap.nih.gov/initiatives.asp
- 6.Zerhouni EA. Clinical research at a crossroads: the NIH roadmap. J Investig Med. 2006 May;54(4):171–3. doi: 10.2310/6650.2006.X0016. [DOI] [PubMed] [Google Scholar]
- 7.PubMed. cited 2007 February 27, 2007.; Available from: www.pubmed.gov
- 8.Federal-Demonstration-Partnership cited 2007 July 15, 2007.; Available from: http://www.thefdp.org/IRB_Task_Force.html
- 9.PRIM&R cited 2007 February 27, 2007.; Available from: http://www.primr.org/
- 10.Stillman J.University of Utah IRB Director. In: communication p, ed. email ed. Salt Lake City, UT: 2007 [Google Scholar]
- 11.ClickCommerce. cited 2007 February 27, 2007.; Available from: http://www.clickcommerce.com
- 12.API cited 2007 February 27, 2007.; Available from: http://www.apibraan.com/
- 13.Rindflesch T, Fiszman M, Lubbus B. Semantic interpretation for the biomedical research literature. In: Cheh H, editor. Medical informatics: knowledge management and data mining in biomedicine. New York, NY: Springer Science + Business Media; 2004. pp. 399–422. [Google Scholar]
- 14.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003 Dec;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 15.SemRep. cited 2007 February 08, 2007.; Available from: http://skr.nlm.nih.gov/batch-mode/index.shtml
- 16.Fiszman M, Rindflesch T, Kilicoglu H. Summarizing drug information in medline citations. AMIA Annu Symp Proc. 2006:254–8. [PMC free article] [PubMed] [Google Scholar]