Abstract
Pharmacogenomic studies are studies designed to elucidate the relationships between drugs and genes on the genomic scale. Given the rapidly increasing amount of microarray data in international repositories, and the implicit drug information contained in PubMed, MeSH and UMLS, we propose automatic methods for identifying drug-related microarray experiments from NCBI GEO by the semantic connections between these data resources. In our study, we find that 51.5% of microarray experiments are associated with at least one PubMed identifier, 22.1% of these contain a MeSH term that relates to the UMLS Pharmacologic Substances semantic sub-tree. Our work shows an abundance of publicly available gene expression data available to enable the discovery of novel drug indications, drug classifications and other pharmacogenomic studies.
INTRODUCTION
The study of how drugs affect cellular and physiological processes has been aided by major advances in genomic technologies. One such technology is the development of RNA gene expression microarrays1. Ten years after their invention, these arrays are commonly used in biomedical research, primarily because they allow for the quantitative measurements of tens of thousands of genes simultaneously. The high throughput capability provided by gene expression arrays make them particularly attractive for pharmacogenomic studies.
Two recent studies highlight the utility of gene expression arrays for chemical and therapeutic discoveries. In one study, Stegmaier et al.2 used gene expression signatures to develop a high-throughput screening assays for 1,660 chemical compounds involved in inducing terminal differentiation in cellular models of acute myeloid leukemia (AML). Recently, Lamb et al.3 compiled the gene expression signatures of 164 small molecule compounds on 564 arrays into a reference database. Query signatures from other drugs and diseases were then pattern-matched to the reference signatures for “connections” among drugs, genes, and diseases. In this fashion, Lamb et al. successfully identified new mechanisms of action and indications for existing drugs.
While innovative and high-impact, the vast amount of resources required to undertake these two large-scale studies precludes the participation of most laboratories. Given the rapidly increasing amount of gene expression data in international repositories, we propose automatic methods for identifying drug-related microarray experiments from gene expression databases by the semantic connections between these data resources. The data extracted using these methods could be further used for meta-analysis as well as enable the discovery of novel drug indications and classifications.
There is currently an abundance of public gene expression repositories. The Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information4, the ArrayExpress at the European Bioinformatics Institute5, and the Stanford Microarray Database (SMD) at Stanford University6 are a few examples of such publicly available databases. Unfortunately, annotations in all of these repositories are stored in free-text form, thus, making the identification of desired experiments difficult. For our study, we chose to use NCBI GEO. As of this writing, GEO holds 108,371 samples from 5,037 experiment sets over 3,070 types of microarrays, and triples in size on an annual basis.
One major drawback to GEO is the lack of a controlled vocabulary used to describe the context of gene expression experiments: annotations are stored in free-text. While these contextual annotations can be parsed to identify drug and other experimental details, parsing is fraught with inaccuracy7. We previously showed that some GEO experiments are linked to a corresponding publication by a PubMed identifier, and each of those publications is manually assigned Medical Subject Headings (MeSH) terms (from a controlled vocabulary) 8. We hypothesize that enough annotations exist in GEO, MeSH, and UMLS to enable a comprehensive extraction of pharmacogenomic experiments in GEO. Our overall goal is to identify shared drug signatures among the various drugs.
METHODS
Gene Expression Omnibus (GEO)
The GEO data used for this study was downloaded in November 2006. Downloaded data was then parsed and stored into a relational database. There are three levels of annotation hierarchies in GEO. Each biological experiment is stored as a GEO series (GSE). A single GSE is composed of one or more GEO sample(s) (GSMs), or single microarray measurements. The GSMs have corresponding annotation related to the experiment such as title, PubMed identifier and the design of the experiment. Both the GSEs and GSMs are annotated by each individual submitter in free-text format.
To allow for comparison across experiments, additional annotations on experimental subsets are added by NCBI curators describing the experimental design and axes in GEO datasets (GDSs). While a GSE essentially contains only a list of the component microarrays, a GDS additionally contains the experimental axes tested in the experiment. Furthermore, the experimental axes, such as agent, tissue, protocol, and cell line used in the experiment are from a controlled vocabulary.
Unified Medical Language System (UMLS)
We downloaded the Unified Medical Language System (UMLS) 2006AD version and stored it into a relational database. The UMLS consists of three major components: the Metathesaurus, the Semantic Network, and the SPECIALIST Lexicon. For our study, we used information stored in the Metathesaurus and the Semantic Network. The Metathesaurus contains terms from 100+ medically-related vocabularies including MeSH. There are more than 1.3 million concepts in the 2006AD Metathesaurus. The Semantic Network is a separate knowledge source that captures the relationships between the concept terms within and among the different vocabularies in the Metathesaurus. Our method integrates information from both the Metathesaurus and the Semantic Network.
GEO to MeSH to UMLS Mapping
We have recently discussed the utility of using PubMed, MeSH terms and UMLS semantic types to find disease-related genomic experiments in GEO8. In this study, we started with this approach of relating the GEO experiments to MeSH terms by their associated PubMed annotations.
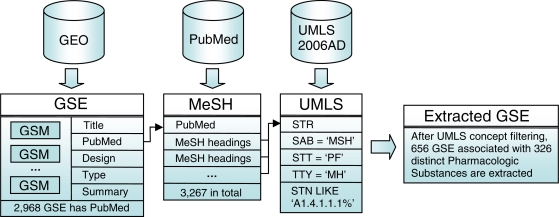
Our approach (Figure 1) first finds the PubMed identifiers in the annotations of GEO series (GSE), which are the collections of related microarray samples (GSM) within a single experiment. The PubMed identifiers represent the publication in which the GSEs were reported. PubMed identifiers are related to MEDLINE records which are manually annotated with Medical Subject Headings (MeSH) by curators at NCBI.
Figure 1.
Flowchart for extracting experiments that have associated MeSH terms that are under the UMLS semantic sub-tree of Pharmacologic Substances.
Once the MeSH terms are extracted, we map these terms to UMLS concepts using the UMLS2006AD Concept Names and Sources (MRCONSO) table with exact term matching. In order to identify the drug-related concepts, the semantic type (from the Semantic Types (MRSTY) table) of the MeSH terms are evaluated. Concepts with semantic types falling under the semantic subtree Pharmacologic Substances (Semantic Type Number [STN] A1.4.1.1.1), which we can easily identify by the prefix in STN field are kept.
Some of our extracted drug concepts are general categories of drugs, instead of specific drug names (e.g. Antineoplastic Agents). To identify only those specific drugs that are extracted with our method, we filtered out these general concepts, leaving only specific names. For each of the concepts (CUI) extracted, we used the Related Concepts table (MRREL) table to count the number of child concepts in MeSH, and removed them from consideration. A filter is applied to the extracted MeSH concepts that have more than twenty-five MeSH child (RELA[relationship]=’isa’) concepts. The intuition for this method is that general drug categories usually have many associated child concepts, so by setting a threshold to this arbitrary count, we can filter out these general concepts.
Control Terms Used In Drug-related GEO Experiments
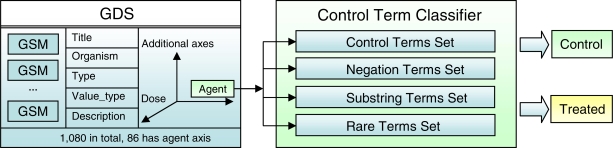
An important issue when identifying drug-related microarray experiments is to additionally identify treated and control states for the gene expression experiments. This can be done by investigating the different field annotations in GSMs. However, these GSMs annotations (from submitters) are in free-text form which is difficult to parse and automate. A more accurate solution is to use additional structured annotations available curated by NCBI curators describing the experimental design and axes in GDSs.
From the annotations in GDSs, we train our classifier to identify the control subset of microarrays based on the descriptions in experimental axis agent. These subset annotations are generally a short free-text description of the agent (e.g. chemical or drug) applied in the experiment, or of the control state (Figure 2). We used simple text mining methods of lower casing and stemming all the terms to build up a set of common control terms. Substring detection was then performed on all terms to see if they contain any of the control terms. Furthermore, our approach also identifies the negation prefix in the descriptions, common terms such as no, none, not, empty, mock, in-, un-, non-, to determine whether it is a control state. Finally, a list of the rare terms used to describe the control state is also compiled. These terms are seldom used, but needs to be constantly monitored when new instances of GDSs are added to make sure the method covers all new instances of control terms.
Figure 2.
Flowchart for identifying the control subset of microarrays based on descriptions in experimental axis.
RESULTS
We extracted 3,267 MeSH terms from the 2,968 GSE (51.5% of total) that have PubMed identifiers. Further mapping of these MeSH terms to UMLS concepts identified 656 GSE (11.4%) that have associated MeSH term(s) within the UMLS Pharmacologic Substances semantic sub-tree. This initially gave us 326 drug concepts, some of which are general drug categories.
After eliminating general drug categories, a total of 213 specific drugs were extracted relating to 326 GEO series. The terms remaining with the highest frequency of associated GEO samples are shown in Table 1. As described in the Method section, additional dataset annotation for experimental axes in GDS can be used to automatically identify the control state. Since not every GSE has an associated GDS, there is a reduction in the number of total GEO samples that we can retrieve with the association from GDS. In all, we identified a total of 212 GDSs that are associated with Pharmacologic Substances. Eighty-six of these have an experimental axis of agent.
Table 1.
The top 10 drug-related concepts identified by UMLS Concept Unique Identifier (CUI) [first column] and Drug string term [second column] ordered by total number of associated microarray samples (GSMs) [third column]. The fourth column indicates the number of MeSH child concepts that is associated with the term concept.
| CUI | Drug Term | GSM Count | Child Count |
|---|---|---|---|
| C0011015 | Daunorubicin | 1041 | 3 |
| C0042679 | Vincristine | 1028 | 1 |
| C0020281 | Hydrogen Peroxide | 741 | 1 |
| C0021745 | Interferon Type II | 695 | 1 |
| C0021747 | Interferons | 657 | 4 |
| C0031436 | Phenothiazines | 437 | 22 |
| C0014964 | Ethambutol | 437 | 1 |
| C0949665 | Fluoroquinolones | 437 | 0 |
| C0025677 | Methotrexate | 375 | 1 |
| C0014912 | Estradiol | 325 | 12 |
The experimental axis agent field consists of terms describing either the drug agent applied, or a control state. The distribution of the terms used is a long tail, where the most common terms (used more than twice) appear in more than 85% of the cases. Examples of the control terms and the rule that classifies them correctly are shown in Table 2.
Table 2.
A list of the terms that submitters use to describe the control state of drug-related experiments in GEO. The first column lists the terms used. The second column is the number of times it appears in the description. The third column shows the rules that are used to classify the term as a control state.
| Control Term | Count | Rule |
|---|---|---|
| Control | 84 | This is the control terms set, a set of common terms manually determined as control state. Stemming is applied to the terms to cover different forms or tenses submitters may use. 56% of the overall control terms are in this set. |
| Vehicle | 6 | |
| Saline | 5 | |
| Air | 4 | |
| Baseline | 2 | |
| Placebo | 2 | |
| Untreated | 45 | These are the termsthat fit within thecategory negation terms set. Terms like untreated, none and unstimulated are obvious. The other descriptions have a negation prefix: no, non-, inactive, mock or empty. This set covers 36.4% of the control terms. |
| None | 7 | |
| Unstimulated | 2 | |
| No sorbitol | 1 | |
| Empty adenovirus | 1 | |
| Non-preconditioned | 1 | |
| Mock stimulation | 1 | |
| Sesame oil vehicle | 1 | This is the substring terms set. These are the terms that have a substring of a term that is in the control terms set. This set covers 6.5% of the control terms. |
| Negative control | 1 | |
| Control-unsychronized | 1 | |
| GFP | 1 | This is the rare terms set. These are rare terms submitters have used to describe the control state. This list will need to be routinely updated for new terms. This set covers 1% of the control terms |
| PBS | 1 | |
| DMSO | 1 |
In our study, we successfully identified 213 distinct drugs and their associated microarray experiments from GEO. The most common categories for these identified drugs are antineoplastic agents covering 2,184 microarray samples, and enzyme inhibitors covering 1,187 samples. The breadth of the number of drugs identified by our approach will allow for meta-analysis for shared gene expression signatures within a specific drug as well as between drugs. These analyses will enable the discovery of novel drug indications and drug classifications, based on common gene expression signatures.
DISCUSSION
International repositories provide a rich resource of drug studies from various experiments. However, the experiment conditions are mostly represented in free-text form, which poses a challenge to process them automatically. In our study, we described a method of using PubMed identifiers of experiments, MeSH terms of the publications, and UMLS concepts for the exact MeSH terms to extract the pharmacogenomic-related experiments.
Out of a total of 2,968 GSE experiments in GEO we mapped 11.4% of them to 326 different drug concepts. Further filtering identified 213 different drug compounds. Additionally, we introduced an automatic method of identifying the control states within an experiment. We discovered that 11.7% of the pharmacogenomic experiments extracted have additional dataset annotations that can be processed automatically with this method. Identifying the control state of the microarray experiments is essential toward generating gene expression signatures for the drugs.
Compared with the 164 drug compounds used by Lamb et al, there is an overlap of 28 drugs from our set. For example, dexamethasone was identified in our set as well as in Lamb’s set, but tunicamycin was only present in our set. Moreover, there is only an overlap of 98 drugs with Stegmaier’s set of 1,660 compounds. For example, methotrexate was identified in our set as well, but daunorubicin was only present in our set. The small overlap among the drugs lists indicates that our extraction yielded roughly 100 drugs that were studied by neither Lamb nor Stegmaier group. The extracted datasets can be used in meta-analyses for the detection of common gene expression signatures, or meta-signatures, shared within the same drug or between different drugs. Our future work will include validating drug-signatures across the overlaps. Beyond this, we anticipate that these meta-signatures will augment our existing knowledge on drugs to aid the discovery of new drug indications and classifications as shared signatures may predict new uses for the drugs or new classifications of drugs.
With the exponential growth of microarray samples in international repositories, our method shows great potential in identifying drug-related experiments as they are deposited in GEO. The identification of the pharmacogenomic experiments is done in an automated way, hence making our method fully scalable to the growth of information. The extracted data can be further used to facilitate future pharmacogenomic studies.
AUTHOR CONTRIBUTION STATEMENT
YL and AB conceived the idea for the study. YL, AC, RL, and PY contributed to the design and planning of the method to extract drug experiments from MeSH. YL contributed to the design for identifying controlled state and analyzed the data. AC curated the extracted drug terms. YL wrote the manuscript. AC and AB revised the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
The work was supported by grants from the Lucile Packard Foundation for Children’s Health, National Library of Medicine (T15 LM007033), National Institute of General Medical Sciences (R01 GM079719), Howard Hughes Medical Institute, and the Pharmaceutical Research and Manufacturers of America Foundation.
REFERENCES
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995 Oct 20;270(5235):467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Stegmaier K, Ross KN, Colavito SA, O'Malley, Stockwell BR, Golub TR. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat Genet. 2004 Mar;36(3):257–63. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 3.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313(5795):1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 4.Edgar R, Barrett T. NCBI GEO standards and services for microarray data. Nat Biotechnol. 2006 Dec;24(12):1471–2. doi: 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, et al. ArrayExpress--a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007 Jan;35:D747–50. doi: 10.1093/nar/gkl995. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demeter J, Beauheim C, Gollub J, Hernandez-Boussard T, Jin H, Maier D, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007 Jan;35:D766–70. doi: 10.1093/nar/gkl1019. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butte AJ, Kohane IS. Creation and implications of a phenomegenome network. Nature Biotechnol. 2006 Jan;24(1):55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butte AJ, Chen R. Finding disease-related genomic experiments within an international repository: first steps in translational bioinformatics. Proc AMIA Symp. 2006:106–10. [PMC free article] [PubMed] [Google Scholar]