Abstract
Objective:
The objective of this study is to propose a model of matching errors for identifying mismatches in alignments of large anatomical ontologies.
Methods:
Three approaches to identifying mismatches are utilized: 1) lexical, based on the presence of modifiers in the names of the concepts aligned; 2) structural, identifying conflicting relations resulting from the alignment; and 3) semantic, based on disjoint top-level categories across ontologies.
Results:
83% of the potential mismatches identified by the HMatch system are identified by at least one of the approaches.
Conclusions:
Although not a substitute for a careful validation of the matches, these approaches significantly reduce the need for manual validation by effectively characterizing most mismatches.
INTRODUCTION
Ontology alignment (or ontology matching) is an active field of research and many approaches to aligning ontologies have been developed in the past decade [1]. Like in other research communities, researchers from the ontology alignment community have set up a competitive evaluation: the Ontology Alignment Evaluation Initiative (OAEI 1), with the goal of comparing systems and algorithms and gaining insights from the best matching strategies [2]. Over the past years, ontologies of different sizes and from several different domains have been the object of the OAEI competition. Of particular interest to biomedicine is the alignment of two anatomical ontologies: the Foundational Model of Anatomy (FMA) and GALEN.
Most ontologies investigated in the OAEI competition are relatively small (e.g., the web directories from Google, Yahoo and Looksmart), averaging a few thousand concepts from non-specialized domains. A gold standard mapping between such ontologies can therefore be established relatively easily by experts for evaluation purposes. In contrast, evaluating large, specialized ontologies remains a challenge [3]. Typically, the organizers simply analyze the overlap between the results produced by the ontology alignment systems. In other words, each match is characterized by the list of systems that identified it. The assumption is that matches identified by many systems have a greater chance of corresponding to actual matches, while matches identified by fewer systems may be less reliable.
In previous work, we challenged this assumption [4]. Based on the manual review of matches specific to one alignment system, we showed that, the specific matches identified by alignment systems taking advantage of domain knowledge tend to be reliable, while those identified by general purpose alignment systems are usually not. During this manual review, we identified patterns of mismatches. For example, the match {Thigh, Right thigh} is incorrect because of laterality distinctions, although the two strings exhibit a relatively high lexical similarity. Recent work by Johnson et al. on mapping errors in biological ontologies differs from ours in that they focus on lexical mapping techniques and the on identification of what specifically causes the error [5].
The objective of this study is to propose a model of matching errors for large anatomical ontologies and to apply this model to the evaluation of the matches identified by the HMatch ontology alignment system between the FMA and GALEN during the 2006 OAEI competition.
BACKGROUND
Anatomical ontologies
The two anatomical ontologies under investigation in the 2006 OAEI campaign are the Foundational Model of Anatomy (FMA) [6] and the Generalized Architecture for Languages, Encyclopedias and Nomenclatures in medicine (GALEN) [7]. The FMA and GALEN were created using different knowledge representation formalisms: frames for the FMA and description logics for GALEN. In order to facilitate the alignment, the organizers converted the FMA and the anatomy subset of GALEN into OWL Full, the most expressive version of the Web Ontology Language. The resulting representation includes the class hierarchy and relations between classes for both ontologies. Additionally, concept names (including synonyms) and textual definitions for classes are represented for the FMA. The datasets provided by the organizers contain 72,560 concepts for the FMA (with 44,597 synonyms), and 9,566 concepts for GALEN (anatomy subset), of which 1,035 are anonymous.
Alignment systems
The alignment system investigated in this study is HMatch, a general purpose ontology alignment system, which we did not have the opportunity to evaluate in our previous analysis [4]. As part of the evaluation, we also use our own system, the Anatomical Ontology Alignment System (AOAS), specifically designed for anatomical ontologies. The goal of both systems is to identify equivalent concepts across ontologies. A brief description of the two systems follows.
HMatch is an algorithm for dynamically matching distributed ontologies. The overall similarity between two concepts combines linguistic and contextual affinity. Lexical affinity is generally based on Word-Net. However, due to the limited coverage of Word-Net for specialized domains such as anatomy and scalability issues, an n-gram algorithm was used instead. Contextual affinity compares the properties of two concepts, as well as the set of concepts to which they are related. Various properties and relationships are assigned different weights, depending on their importance. Due to differences in the representation of anatomical entities in OWL between the FMA and GALEN, the contextual affinity could not be used in this experiment. Finally, a global threshold of .6 is applied to the global similarity value in order to select the matches [8, 9].
AOAS is a domain-specific ontology matching system for anatomical entities. Its lexical component compares concept names using a model of lexical resemblance developed for biomedical terms and exploits additional synonyms from an external resource: the Unified Medical Language System® (UMLS®). The presence of shared hierarchical paths among concepts across ontologies is then used as positive evidence for the mappings identified lexically. AOAS also identifies incompatible concepts, which receive negative structural evidence [3, 10].
MATERIALS
The result files for the OAEI 2006 campaign for anatomy were downloaded from the participants’ web sites. The reporting format required from the organizers imposes four fields: entity1, entity2, measure (of confidence) and relation. All mappings identified by the 2 systems are between equivalent concepts (relation: =). Incompatible mappings despite lexical similarity (negative evidence) are also reported by AOAS (relation: !=). A measure of confidence (0-1, continuous) is attached to each mapping and thresholds determined heuristically are used to select valid mappings. The 7,259 mappings reported by HMatch have a confidence measure between .6 and 1. AOAS identified 3,132 mappings, of which 3,029 were supported by structural evidence. There are 2,343 mappings common to both systems, representing 32% of HMatch mappings and 75% of AOAS mappings.
METHODS
The methods we propose for identifying mismatches are guided by our experience in validating mappings. The first method operates at the lexical level and identifies differences in pairs of concept names indicative of a mismatch. The second method operates at the structural level and examines known relations between the two concepts matched in the two ontologies. Finally, we apply semantic constraints to identify matches between semantically incompatible concepts.
Identifying mismatches lexically
We observed that in a large number of cases of mismatches, concept names differ only by one word, most often an adjective. Names for anatomical entities are generally noun phrases. Adjectival modifiers often represent distinctive features such as direction (ascending, descending) and orientation, absolute (left, right; anterior, posterior; inferior, superior; upper, lower) or relative (proximal, distal; medial, lateral). Analogously, ordinal adjectives are used to distinguish features of the twelve thoracic vertebrae (first, …, twelfth).
In a previous study [11], we used the property that adjectival modifiers usually introduce a hyponymic relationship to suggest a possible hyponymic relation between modified and nonmodified terms (e.g., Right thigh and Thigh) and co-hyponym relation between terms modified by different modifiers (Ascending Palatine Artery and Descending palatine artery). As a corollary here, we suggest that the entities named by two terms differing by adjectival modification are unlikely to be equivalent. More generally, the underlying linguistic principles form the basis for acquiring semantic relations from text [12].
In practice, for each pair of concepts given as matches by HMatch, we analyze all pairs of terms composed of one synonym from the FMA concept and the (unique) name in GALEN. We analyze each pair of concept names for the following lexical properties suggestive of a relation other than equivalence between the two terms:
Proper substring. One concept name is a proper substring of the other (e.g., Mandible / Base of mandible)
Modifier/Ø. The two concept names differ only by one word, present in one and not in the other (e.g., Medulla / Ovarian medulla)
Modifier1/Modifier2. The two concept names differ only by one word, but each name has a different modifier2 (e.g., Internal Carotid Artery / Left internal carotid artery; Vestibulocochlear Nerve / Vestibulo-cochlear vein)
Identifying mismatches structurally
We observed that a large number of mismatches involved concepts between which there exist relations, often hierarchical, in one ontology or the other. For example, HMatch appropriately identifies a match between Kidney in GALEN and Kidney in the FMA (with a confidence of 1.0). However, it also identifies a match between Kidney in GALEN and Right kidney in the FMA (with a confidence of .82). The FMA asserts the relation Right kidney ISA Kidney. Therefore, it is not possible that Right kidney in GALEN is equivalent to both Kidney and Right kidney in the FMA, because Right kidney is distinct from and more specific than Kidney in the FMA. In addition to the hierarchical relations asserted in both ontologies, we use as a reference (i.e., tentative gold standard) the equivalence relations resulting from the alignment created by another alignment system (here, AOAS).
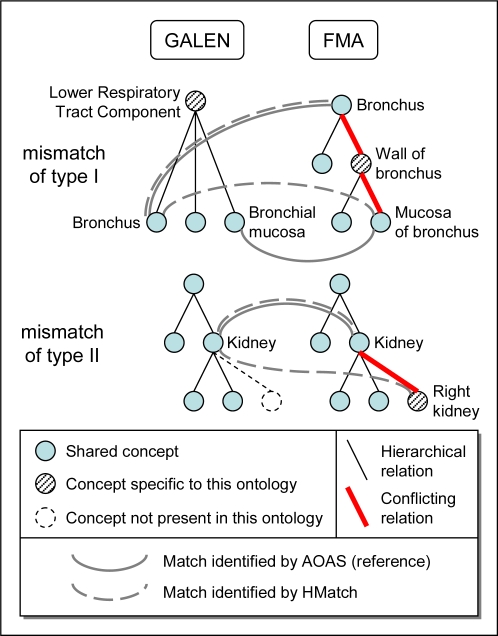
In practice, for each match {A,B′} identified by HMatch, we first check the existence of reference matches established by AOAS for these two concepts (e.g., {A,A′} and/or {B,B′}). Then, we use the transitive closures of ISA and PART OF relations in each ontology to find a path between the two concepts A and B in one ontology and A′ and B′ in one other. The existence of such a path, as well as the existence of a sibling relation between A and B or A′ and B′ (i.e., a common subsumer for A and B or A′ and B′) indicates an inconsistency between the two alignments. For example, the matches {A,A′} from AOAS and {A,B′} cannot be both valid if A′ and B′ are siblings or hierarchically related. Two types of mismatches, illustrated in Figure 1, can be distinguished, based on the existence of equivalence relations identified for these two concepts by AOAS.
Figure 1.
Types of mismatches identified structurally
Type I
The match {A,B′} is identified by HMatch, but AOAS identifies the two matches {A,A′} and {B,B′} instead. If there exists a sibling or subsumption relation between A and B (resp. A′ and B′), the equivalence between A and B′ is inconsistent. For example, HMatch identified a match between Bronchus in GALEN and Mucosa of bronchus in the FMA (Figure 1). However, AOAS has identified Bronchus and Mucosa of bronchus as distinct concepts, each having an equivalent in the other ontology. (Of note, HMatch also identified Bronchus in GALEN and Bronchus in the FMA as matches.) Examining the relations between Bronchus and Mucosa of bronchus, we find the following. In GALEN, Bronchus and Bronchial Mucosa are siblings, both subsumed by Lower Respiratory Tract Component. In the FMA, Mucosa of bronchus is part of Wall of bronchus which is part of Bronchus. These relations conflict with the equivalence relation suggested by HMatch between Bronchus and Mucosa of bronchus. When no conflicting relations are found, this approach is inconclusive. For example, the mismatch between the two distinct anatomical entities Lip and Limb cannot be established by this method.
Type II
The match {A,B′} is identified by HMatch, but AOAS identifies a match for only one of the two concepts ({A,A′} or {B,B′}) instead. Here again, if there exists a sibling or subsumption relation between A and B (resp. A′ and B′), the equivalence between A and B′ is inconsistent. The example mentioned earlier of a mismatch between Kidney in GALEN and Right kidney in the FMA falls under this category. In this case, the fact that Kidney subsumes Right kidney in the FMA is inconsistent with the equivalence relation suggested by HMatch between these two concepts. When no conflicting relation is found, this approach is inconclusive. For example, the mismatch between the two distinct muscles Palatoglossus and Ceratoglossus cannot be established by this method.
Identifying mismatches semantically
Semantic mismatch is based on the disjointness of top-level categories across ontologies [3]. For example, the match {Inguinal Pain, Inguinal ring} identified by HMatch is semantically invalid. In GALEN, Inguinal Pain is subsumed by Abdominal Pain Symptom, with Process as its top-level subsumer. The top-level subsumer for Inguinal ring in the FMA is Anatomical structure. Process and Anatomical structure are disjoint top-level categories across systems.
RESULTS
Mismatches identified lexically
Proper substring
2,908 (40%) of the 7,259 matches in HMatch are such that one concept name is a proper substring of the other. This phenomenon is not always indicative of a mismatch. 233 (10%) of the 2,343 matches identified in common by HMatch and AOAS exhibit this property. In fact, anatomical names often have short synonyms. Example of ellipses (valid matches) include Horizontal Fissure / Horizontal fissure of right lung, Splenius Cervicis Muscle / Splenius cervicis, Nucleus Caudatus / Caudatus and Prostate Gland / Prostate.
Modifier/Ø
2,460 (34%) of the 7,259 matches in HMatch are such that the two concept names differ only by one word, present in one and not in the other. In 94% of the cases, one term is also a proper substring of the other. The modifiers left and right alone account for almost one half of the differences. Modifiers with a frequency of at least 10 are listed in Table 1. Examples of mismatches include Arytenoid Cartilage / Arytenoid cartilage zone, Epitympanic Recess / Epitympanic recess proper, Arteriole/ Arteriole wall, Taste Bud / Taste bud cell and External Acoustic Meatus / External acoustic meatus nerve.
Table 1.
List of modifiers frequently occurring in mismatches
| Modifier | # | Modifier | # | Modifier | # |
|---|---|---|---|---|---|
| left | 637 | nerve | 14 | second | 11 |
| right | 546 | body | 14 | lumen | 11 |
| zone | 47 | muscle | 13 | head | 11 |
| proper | 46 | hair | 13 | fifth | 11 |
| wall | 26 | venous | 12 | fascia | 11 |
| cell | 18 | skin | 12 | root | 10 |
| third | 14 | first | 12 | fourth | 10 |
Modifier1/Modifier2
1,235 (17%) of the 7,259 matches in HMatch are such that the two concept names differ only by one word, but each name has a different modifier. Of the 1,072 unique pairs of modifiers, only 15 occur with a frequency higher than 2, the most frequent being left / muscle, nerve / vein and tree / trunk. Examples of mismatches include Sub-scapular Nerve / Subscapular vein, Pubococcygeus Muscle / Left pubococcygeus, Ophthalmic Artery / Ophthalmic nerve and External Ear / Internal ear.
Mismatches identified structurally
Of the 7,259 matches identified by HMatch, 4,001 can be analyzed structurally with respect to the reference matches from AOAS, because there is a match in the reference alignment for at least one of the 2 concepts matched by HMatch (distinct from the match in HMatch). A total of 2,271 mismatches were identified (57%), 107 of type I and 2,164 of type II. Most conflicts are identified on the basis of ISA relations (76%), most often from the FMA. In 56 cases, a conflict is identified in both ontologies. The examples presented in Figure 1 are quite typical of the conflicts identified by this method.
Mismatches identified semantically
The semantic constraints defined across ontologies contributed to identify 202 mismatches among the 7,259 matches identified by HMatch. This small number of conflicts reflects the semantic homogeneity of ontologies restricted to a narrow subdomain such as anatomy.
DISCUSSION
Characterizing HMatch misatches
Because AOAS was previously evaluated, we are confident that the matches it identified are valid. Therefore, the 2,343 matches identified by both systems are not considered in what follows. The 4,916 matches identified by HMatch, but not by AOAS are either mismatches from HMatch or missed matches from AOAS. Applied to this set of matches, the lexical and structural methods were able to characterize 4,094 of them (83%) as potential mismatches, thus significantly reducing the need for manual review. As shown in Table 2, 1,872 mismatches (38%) were identified by both lexical and structural methods. The semantic constraints (not reflected in Table 2) contributed only modestly to the identification of mismatches (202 mismatches identified by this method).
Table 2.
Lexical and structural characterization of mismatches
| Structurally | |||||
|---|---|---|---|---|---|
| Conflict | Inconcl. | n/a | Total | ||
| Lexically | Modifier/Ø | 1,712 | 554 | 194 | 2,460 |
| Mod1/Mod2 | 160 | 636 | 439 | 1,235 | |
| Inconclusive | 399 | 540 | 282 | 1,221 | |
| Total | 2,271 | 1,730 | 915 | 4,916 | |
Evaluating matches
The objective of the methods presented here is not the precise evaluation of the matches produced by a system. We rather attempted to identify the gross mismatches (e.g., {Kidney, Right kidney}) commonly produced by alignment systems not specifically designed for anatomical ontologies. The contribution of this paper is thus to reduce, not eliminate, the need for manual review. The method can be adapted for precision or recall. For precision, mismatches will require both lexical and structural identification, which still significantly reduces the need for manual review in this experiment. Alternatively, the list of modifiers used for the lexical analysis can be restricted to those modifiers indicative of distinctions known to be incompatible with equivalence relations (e.g., laterality, orientation, etc.).
Limitations
The structural validation typically requires another alignment used as a reference for matches. This requirement is appropriate in the context of the OAEI campaign where several alignments are generally available. Alignments with limited recall but high precision such as AOAS’ are particularly useful for structural validation. Limited structural validation can still be performed in the absence of a reference alignment. In fact, the conflicts shown in Figure 1 could still be identified without the reference provided by AOAS. In this case, the lexical analysis could be used to identify the mismatch between the two matches {Kidney, Kidney} and {Kidney, Right kidney} involved in a conflict. The lexical analysis proposed here, while effective, is limited to terms differing by one word (deletion or exchange) and does not distinguish modifiers from the head of the term. A lexico-syntactic analysis would increase recall and precision.
Missed matches
A partial review of the 822 matches on which the lexical and structural methods are either inconclusive or not applicable revealed some valid matches missed by AOAS. The lexical similarity techniques used by AOAS are much more conservative than the n-gram technique employed by HMatch. For example, because no synonym is provided, AOAS fails to identify the equivalence between Tibial Inter Condylar Eminence and Intercondylar eminence of tibia, correctly identified by HMatch. A simple review of the pairs of modifiers identified during the lexical analysis would help identify such missed matches.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and by the Natural Science Foundation of China (No.60496324), the National Key Research and Development Program of China (Grant No. 2002CB312004), the Knowledge Innovation Program of the Chinese Academy of Sciences, MADIS of the Chinese Academy of Sciences, and Key Laboratory of Multimedia and Intelligent Software at Beijing University of Technology.
Footnotes
The modifier is not necessarily a modifier in the grammatical sense. The word by which the two names differ can be the head of the noun phrase as in Vestibulocochlear Nerve / Vestibulocochlear vein.
References
- 1.Noy NF. Tools for mapping and merging ontologies In: Staab S, Studer R, editors Handbook on Ontologies: Springer-Verlag; 2004. pp. 365–384. [Google Scholar]
- 2.Euzenat J, Mochol M, Shvaiko P, Stuckenschmidt H, Šváb O, Svátek V, et al. First results of the Ontology Alignment Evaluation Initiative 2006. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI 2006) 2006:73–90. [Google Scholar]
- 3.Zhang S, Bodenreider O. Experience in aligning anatomical ontologies. International Journal on Semantic Web and Information Systems. 2007;3(2):1–26. doi: 10.4018/jswis.2007040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S, Bodenreider O.Lessons learned from cross-validating alignments between large anatomical ontologies Medinfo 2007. in press [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson HL, Cohen KB, Hunter L. A fault model for ontology mapping, alignment, and linking systems. Pac Symp Biocomput. 2007;12:233–268. [PMC free article] [PubMed] [Google Scholar]
- 6.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9(2):139–71. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]
- 8.Castano S, Ferrara A, Messa G. ISLab HMatch results for OAEI 2006. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI 2006) 2006:126–135. [Google Scholar]
- 9.Castano S, Ferrara A, Montanelli S. Matching ontologies in open networked systems: Techniques and applications. Journal on Data Semantics. 2006;V:25–63. [Google Scholar]
- 10.Zhang S, Bodenreider O. NLM anatomical ontology alignment system: Results of the 2006 ontology alignment contest. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI 2006) 2006:145–156. [Google Scholar]
- 11.Bodenreider O, Burgun A, Rindflesch TC. Assessing the consistency of a biomedical terminology through lexical knowledge. International Journal of Medical Informatics. 2002;67(1–3):85–95. doi: 10.1016/s1386-5056(02)00051-5. [DOI] [PubMed] [Google Scholar]
- 12.Maedche A, Staab S. Mining ontologies from text. Knowledge Engineering and Knowledge Management, Proceedings. 2000:189–202. [Google Scholar]