Abstract
In this paper we present a novel approach to integrate non-structured and structured sources of biomedical information. We part from previous research on database integration conducted in the context of the EC funded INFOGENMED project. In this project we developed the ONTOFUSION system, which provides a robust framework to integrate large sets of structured biomedical sources. Methods and tools provided by ONTOFUSION cannot be used to integrate non-structured sources, since the latter usually lack a logical schema. In this article we introduce a novel method to extract logical schemas from text-based collections of biomedical information. Non-structured sources equipped with a logical schema can be regarded as regular structured sources, and thus can be bridged together using the methods and tools provided by ONTOFUSION. To test the validity of this approach, we carried out an experiment with a set of five cancer databases.
Introduction
Recent genomic-based approaches to medicine are generating exciting challenges for computer scientists, such as the development of new methods to collect, integrate, and access medical and genetic information1. In this scenario, we have recently completed the EC funded INFOGENMED2 project. This project, finished in September 2004, was focused in the creation of a virtual laboratory to integrate and access heterogeneous and distributed sources of biomedical information.
The main result of INFOGENMED was the ONTOFUSION3 system. ONTOFUSION provides methods and tools to integrate large sets of structured sources. The latter can be defined as data repositories equipped with a logical schema that summarizes their information contents. The integration is achieved by performing two basic operations: mapping and unification. The mapping process aim to manually translate the sources’ logical schema into a conceptual schema built upon a global domain model that contains standardized terminology. By contrast, the fully automated unification process merges the mapping schemas into a unified schema that encapsulates the whole information space of the underlying sources.
However, over the last years, biomedical researchers are showing a growing interest in non-structured sources— e.g. plain text or HTML-based document collections. Conversely to structured sources, non-structured sources do not provide a logical schema describing their contents. As stated above, these sources are collections of plain text documents or HTML pages, so the only existing structures—if any— are chapters, sections, or paragraphs.
Methods and tools provided by ONTOFUSION cannot be used to integrate non-structured and structured sources due to the lack of a logical schema in non-structured databases. To solve this problem, we propose a four-phase method to automatically acquire a logical schema given a concrete source. Once a logical model has been extracted for a non-structured source, it can be regarded as a structured source and therefore can be integrated using the tools provided by ONTOFUSION.
This paper is organized as follows. In the next section we describe the methods we have developed to integrate structured and non-structured sources. We emphasize in the four-phase method for logical schema acquisition from non-structured sources. Next, we present and discuss the results of an integration experiment we have conducted with several structured and non-structured cancer databases, and finally we draw the conclusions.
Methods
The ONTOFUSION System
ONTOFUSION follows a domain model-based approach to integrate structured sources. This includes relational or object-oriented databases, and in general, any structured source that can be accessed by means of the Open Database Connectivity (ODBC) interface.
In the context of ONTOFUSION, databases are represented by domain models (DMs). DMs are conceptual models that capture the portion of the domain of interest to which a given source belongs. DMs are obtained by mapping objects from the sources’ logical schema to objects belonging to a global domain model (GDM) that contains all relevant objects named with standardized terminology.
Once a DM has been generated with the mapping tool3 for each source to be integrated, an automated unification process is performed. This process outputs a unified DM that summarizes the information space of all underlying sources. As can be seen, using these operators— mapping and unification—we obtain a hierarchy of DMs that describes the whole information space provided by the sources at different levels of granularity.
Mapping and unification processes cannot be applied to non-structured sources since they lack a logical schema. Therefore, it is not possible to build a DM to model the source since there is no logical description of the source to be translated. To solve this problem we propose a four-phase process that extracts a logical model from the text of the documents stored in the non-structured source. In the next paragraphs we describe the proposed method.
Logical schema acquisition
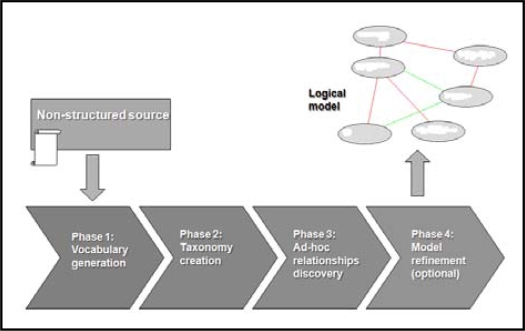
To extract the logical schema of a given source, we have developed a four-phase process whose activities are shown in figure 1.
Figure 1.
Overview of the logical schema acquisition method
As can be seen, the process required input is the raw document collection. The result is a logical model composed of a hierarchy of concepts and a set of ad-hoc relationships between concepts.
In the first phase the goal is the extraction of all the concepts contained in the documents that may be relevant to the domain of interest. To accomplish this task we have used classic natural processing language techniques such as tokenizers, probabilistic part-of-speech taggers, and transition networks. Concepts generated during this phase are noun phrases (NPs) that can be composed of one or more words. We used three distinct transition networks to capture three different types of NPs: i) simple NPs (a succession of adjectives followed by a common or proper name), ii) conjunctive NPs (a conjunction or disjunction of adjectives followed by a common or proper name), and iii) adverbial NPs (an adverbial form followed by a succession of adjectives followed by a common or proper name). As shown in figure 2, each document is decomposed into phrases by a phrase generator. Each phrase is tokenized, and each token (word) is tagged with the part-of-speech (POS) it belongs to. POS tags are then used by the transition networks to generate the NPs. To assess the relevance of a given NP, we search for it in a vocabulary server powered by ONTOFUSION which provides access to a unified repository that integrates the Unified Medical Language System, the Gene Ontology, and the Human Gene Nomenclature. If the NP can be found in the vocabulary server, it is marked as potentially relevant, and it is assigned its preferred string if available in the vocabulary server. Otherwise it is removed from the vocabulary. After the pruning, the remaining NPs are labeled with their corresponding TF-IDF4 weights to assist human experts to recognize non-relevant terms in the refinement (fourth) phase.
Figure 2.
Flowchart of the vocabulary generation algorithm
The second phase is aimed to discover hierarchical relationships between pairs of concepts discovered during the first phase. These hierarchical relationships can be classified into two types: i) hyponymy (relationships among a generic concept and its related more specific concepts), and ii) meronymy or partitive relationships (relations among a whole and its parts). To discover hierarchical relationships we have adopted a “pattern-matching”5 approach. We have manually created a knowledge base with more than 100 hyponymy and meronymy patterns. The pattern-matcher has been built upon a production rules-based inference engine.
The third phase is focused in the discovery of non-hierarchical relationships between pairs of concepts. To carry out this task we have adopted an approach based on collocations6. Word collocations can be defined as short-distance occurrences of two or more words in a text. Since our method is based in concepts rather than words, we have defined concept collocations as short-distance occurrences of two NPs in a document. To discover ad-hoc relationships between pairs of concepts we proceed as follows. For each pair of NPs in the vocabulary, we perform a hypothesis testing under the following null hypothesis: “Given two concepts, the occurrence of one of them is independent of the occurrence of the other in the same text”. The statistic used to perform this hypothesis testing is the T-Score, based on the Student’s T. This statistic compares the observed and expected co-occurrence frequencies between both NPs. T-Score values greater than 2 indicate that the null hypothesis must be rejected, and thus an ad-hoc relationship holds between the concepts. It should be made clear that observed frequencies are not computed along the whole text, but in small contexts called concept collocates. A concept collocate is a 2s+1 sized list of NPs centered in a given NP. The parameter s is an integer span factor that denotes the size of the context. Typical values for s are between 5 and 15. Observed co-occurrence frequencies between a given NP cj and the rest of NPs in the vocabulary are computed along its set of concept collocates. The latter includes all concept collocates for the NP cj that have been extracted from all the documents in the collection. Expected co-occurrence frequencies are computed analogously, but in this case the corpus is composed of large samples of documents obtained from the MEDLINE online database. The output of this activity is a matrix R(n x n), being n the number of distinct NPs discovered during the first phase. Each element ri,j ∈ R— i.e. the value of the T-Score—represents the strength of the relationship between NPs ci and cj. At the current state, our method does not extract relationships role names. The latter can be manually assigned by domain experts during the next phase.
The fourth phase is an optional activity targeted to the manual refinement of the extracted logical schema. The refinement task should preferably be conducted by experts in the domain of application assisted by a knowledge engineer. Refinement tasks include the removal of irrelevant or incorrect concepts and relationships (both hierarchical and ad-hoc), the addition of new concepts or relationships of interest not captured during the previous phases, or the enhancement of the model using standard vocabularies and/or ontologies. Data recorded during the previous phases, such as TF-IDF weights, or elements in the R matrix, are available to curators during this activity to facilitate the refinement task. Once the refinement process— if any— has been completed, we obtain a logical model of the sources’ information contents composed of a concept hierarchy, and a set of ad-hoc relationships. The original source, equipped with the extracted logical model, is now equivalent to a structured source, and thus it can be integrated using ONTOFUSION.
Integration of structured and non-structured sources
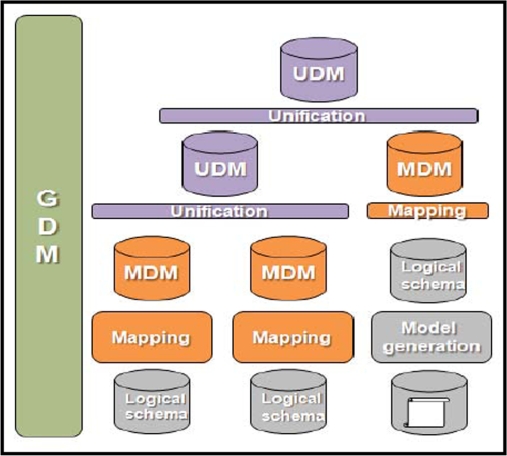
Once the non-structured sources have an available logical model, it is now possible to integrate them using the mapping and unification primitives. The integration process is depicted in figure 3.
Figure 3.
Overview of the integration process
As can be seen in figure 3, once the logical schema of the non-structured database is available, we can proceed with the mapping and unification processes. Once the mapping process has been performed, we obtain a mapping domain model (MDM) built using concepts from the global domain model (GDM). The MDM also establishes translation correspondences between entities in the MDM and objects belonging to the logical schema.
Once all the sources have been translated into MDMs, they can be integrated into one or more unified domain models (UDMs) using the automated unification algorithm3.
Users can navigate and query any available UDM or MDM using the schema browser provided by ONTOFUSION [3]. The result set presented to the user will be composed of two different types of results: i) an unsorted list of instances coming from the structured sources, and ii) a ranking of documents belonging to non-structured sources sorted in descending order of relevance.
Queries targeted to structured sources are handled by ONTOFUSION wrappers using the native query processing capabilities provided by their corresponding database management systems. Regarding the document retrieval process in non-structured sources, we have developed a concept-based variant of the classic vector space model7 for document annotation and retrieval. We evaluated the performance of our document retrieval model using four test collections widely used by the information retrieval community. For three of these collections our method outperformed the vector space model, while for the remaining collection both methods performed similarly.
Evaluation
We have conducted an integration experiment using real sources. We used five cancer repositories, being two of them relational (structured) databases. The first database “Tumors_1” contains both clinical and genetic cancer-related data, while the second only contains clinical data about cancer. The remaining sources are subsets of text-based documents borrowed from three public medical and genetic databases. Table 1 shows detailed information about the structure and size of the sources used in the experiment.
Table 1.
Summary of sources used in the integration experiment
| Source | Type | Owner | # Tables/Docs | # Records |
|---|---|---|---|---|
| Tumors_1 | Relational | Institute of Health Carlos III (Spain) | 15 | 200 |
| Tumors_2 | Relational | Institute of Health Carlos III (Spain) | 6 | 50 |
| Subset of PUBMED | Text-based collection | National Center for Biotechnology Information (U.S.A.) | 50 | N/A |
| Subset of OMIM | Text-based collection | National Center for Biotechnology Information (U.S.A.) | 50 | N/A |
| Subset of PDB | Text-based collection | Rutgers, the State University of New Jersey (U.S.A) | 50 | N/A |
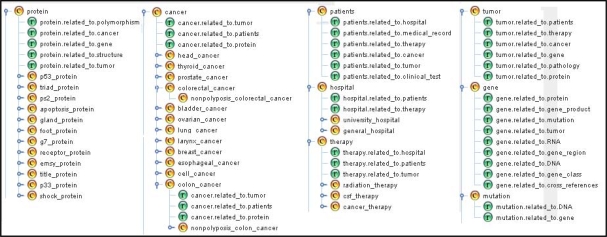
To create the GDM required by the mapping and unification processes, we applied our four phase method to a set of 1500 cancer-related documents selected from the PUBMED database. After the refinement task, we obtained a GMD composed of 2754 concepts, 865 hierarchical relationships, and 5650 ad-hoc relationships. Next we generated the logical schemas for each of the non-structured databases. Table 2 resumes the main features of such schemas. Two senior biomedical researchers and one computer scientist participated in the schema refinement task. Their feedback on the quality of the generated models was positive. However, they emphasized the need of a method to infer relationships role names to facilitate the refinement and integration tasks. After the refinement task we performed a mapping process to create a MDM for each source. Finally we merged the existing MDMs into a single UDM using the unification engine. This UDM was composed of 257 concepts, 106 hierarchical relationships, and 425 ad-hoc relationships. An extract of the schema of the generated UDM is depicted in Figure 4. As can be seen the unified conceptual schema is coherent and represents accurately a subset of the domain of interest.
Table 2.
Main features of extracted logical schemas for PUBMED, OMIM, and Protein Data Bank
| Concepts | Hierarchical Relationships | Ad-hoc Relationships | |
|---|---|---|---|
| PUBMED | 274 | 89 | 514 |
| OMIM | 548 | 156 | 927 |
| PDB | 824 | 134 | 1463 |
Figure 4.
Detail of the virtual schema associated to the UDM
Discussion
Many of the existing database integration systems have been created to integrate only one type of sources: either structured or non-structured. To the best of our knowledge, only systems such as TSIMMIS8 can integrate both types. The main difference with respect to our approach is that these systems convert non-structured sources into structured databases by extracting information (records) following the structure of a given conceptual schema. Conversely, our approach generates a logical schema that encapsulates the source, acting as a semantic mediator. Therefore, systems such as TSIMMIS always retrieve records of structured data, while our system outputs either records or full-text documents (or both) depending on the type of the underlying sources.
Conclusions
In this paper we propose a novel method to generate a logical model from a non-structured source of biomedical information. A non-structured source equipped with a logical model is equivalent to a structured source, and thus can be integrated using ONTOFUSION. The integration experiment we conducted using five real sources proved that our approach is adequate to integrate both kind of sources.
Acknowledgments
This research has been supported by the European NoE INFOBIOMED (IST-2002-507585) and the Advanced Clinic-Genomic Trials in Cancer (ACGT) project (IST-2005-026996) funded by the European Commission.
References
- 1.Sander C. Genomic medicine and the future of health care. Science. 2001;287(5460):1977–78. doi: 10.1126/science.287.5460.1977. [DOI] [PubMed] [Google Scholar]
- 2.INFOGENMED: A virtual laboratory for accessing and integrating genetic and medical information for health applications. EC funded project IST-2001-39013.
- 3.Pérez-Rey D, Maojo V, García-Remesal M, et al. ONTOFUSION: Ontology-based integration of genomic and clinical databases. Comput Biol Med. 2006;36(7–8):712–30. doi: 10.1016/j.compbiomed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. Addison-Wesley; Wokingham, UK: 1999. [Google Scholar]
- 5.Hearst M. Automatic Acquisition of Hyponyms from large text corpora. Proceedings of the 14th Conference on Computational Linguistics; Nantes (France). 1992. pp. 539–45. [Google Scholar]
- 6.Sinclair J. Corpus, Concordance, Collocation. Oxford University Press; Edinburgh, UK: 2000. [Google Scholar]
- 7.Salton G, Wong A, Yang CS. A vector space model for automatic indexing. Commun ACM. 1975;18(11):613–20. [Google Scholar]
- 8.García-Molina H, Papakonstantinou D, Quass D, et al. The TSIMMIS Project: Integration of Heterogeneous Information Sources. J Intell Inf Sys. 1997;8(2):117–32. [Google Scholar]