Abstract
Access to clinical evidence is a critical component of the practice of evidence-based medicine. Advanced retrieval systems can supplement precompiled secondary sources to assist physicians in making sound clinical decisions. This study explores one particular issue related to the design of such retrieval systems: the effective organization of search results to facilitate rapid understanding and synthesis of potentially relevant information. We hypothesize that grouping retrieved MEDLINE® citations into semantically-coherent clusters, based on automatically-extracted interventions from the abstract text, represents an effective strategy for presenting results, compared to a traditional ranked list. Experiments with our implemented system appear to support this claim.
Introduction
Physicians engaged in the practice of evidence-based medicine (EBM) combine their medical expertise with the best available external evidence to support their clinical decisions.1 There are at least two hurdles in seeking evidence at the point of care: locating relevant articles and synthesizing their findings through critical appraisal of article content. Since the proper practice of EBM is time-consuming, clinicians are often advised to use existing resources that summarize best practices in a highly-distilled form.2 Well-respected, manually-created secondary sources cover a wide spectrum of clinical questions and are increasingly used at the point of care.3
It is hard to imagine, however, that all possible information needs can be anticipated and addressed in advance. Furthermore, secondary sources are perpetually falling out of date due to rapid advances in medical research. One possible solution lies in advanced retrieval techniques such as document summarization and question answering,4 which can potentially reduce the time and effort needed to find answers to questions not covered in secondary sources. Such systems can supplement pre-digested resources as valuable tools for decision making at the point of service, and can also assist specialists in compiling such sources to begin with. This work focuses on the challenge of organizing search results into a more easily digestible form for consumption by clinicians and other information seekers.
The need to organize search results in a manner that facilitates more rapid decision-making is becoming more acute, as an increasing amount of the medical literature is accessible on-line. Most current retrieval systems (e.g., PubMed) return results in a linear list, usually sorted in decreasing order of relevance or chronologically in reverse. While such a presentation is clearly useful and easy to understand, it does not provide clinicians with a broad overview of the information space. For example, when trying to find the answer to a question about drug therapy, it is difficult to determine how many different treatment options are available, since multiple citations may discuss the same drug. A simple linear list may obscure important relationships between different results, preventing physicians from effectively synthesizing findings from multiple sources. We hypothesize that an interface that organizes search results into semantically-coherent clusters represents a more effective vehicle for delivering clinical evidence in support of decision-making.
Expanding on our previous exploration of this hypothesis,5 we developed a semantic clustering algorithm that groups MEDLINE citations based on automatically-identified interventions in abstract text. Within the framework of evidence-based medicine, the notion of intervention (e.g., drugs, therapeutic procedures, etc.) encompasses answers to therapy- and diagnosis-related questions, which is the focus of this work. We compare results retrieved by PubMed, presented as a list, with the same results organized into semantically-meaningful clusters. Experiments show that a cluster-based interface potentially brings more relevant citations to the immediate attention of the physician.
Background
Organization of search results into dynamically-created categories or clusters based on users’ queries has been shown to help patients and their families gain quick and easy access to information about breast cancer using DynaCat.6 The system defines several query types, e.g., treatment – side effects; criteria for generating categories for each query type; and valid category labels for a given category. It then uses UMLS semantic types7 to describe valid category labels and assigns documents to categories based on mapping of keywords to the UMLS semantic types. Keywords describing the documents are selected by their authors and human indexers, which increases the amount of manual knowledge required to create meaningful classifications. Although targeted towards laymen, DynaCat nevertheless confirms the value of categorized search results.
An interesting approach taken by DynaCat is the recognition of generic query types, or patterns of questions that share similar characteristics and can take advantage of similar automated processing techniques. Recent analysis of 197 clinical questions posed in the context of three clinical scenarios validates this generic query approach.8 In our work, we assume four broad question types: therapy, diagnosis, etiology, and prognosis, as in the abovementioned study, but focus specifically on therapy and diagnosis questions since they constitute up to 80% of questions asked by general practitioners.9 We believe that the same semantic-clustering algorithm can be successfully applied to both types of questions.
Since the majority of therapy and diagnosis questions focuses on different aspects of interventions, for example, the efficacy of therapeutic procedures,8 we developed an algorithm for clustering MEDLINE citations based on automatically-identified interventions. We utilize a previously-developed knowledge extractor that relies on MetaMap10 to automatically identify focal interventions present in abstract text.11 Concepts in UMLS are used as cluster labels to indicate the content of each cluster, thus providing the user with an overview of common themes.
The purpose of this work is to study the effectiveness of a semantic-clustering algorithm that automatically organizes MEDLINE citations into topically-coherent clusters. Sample output from our implemented system is shown in Figure 1. In this example about oppositional and defiant behaviors, our system discovered broad categories of interventions that may be of interest to the physician. Each category is associated with a cluster of abstracts from MEDLINE about that particular treatment option. Drilling down into a cluster, the user is presented with extractive summaries of abstracts that highlight the interventions under study and summarize the relevant clinical findings. This is accomplished by displaying the top-ranking outcome sentence, automatically identified using classification techniques we have previously developed.11 Drilling down into details further, the physician can pull up the complete abstract text, and finally the electronic version of the entire article (if available). In the example shown above, the physician can see several treatment approaches to Oppositional Defiant Disorder. Focusing on the first cluster, the physician sees summarized evidence for psychosocial interventions. The second and third clusters provide information about psychopharmacological agents.
Figure 1.
Sample system response.
Methods
Test collectio
Independent of this study, we gathered 59 clinical questions from two on-line sources: the Family Practice Inquiries Networka and Parkhurst Exchange.b The distribution of questions in our collection roughly follows the distribution of real-world clinical information needs. Each question is accompanied by the recommendations of specialists, which serve as “ground truth” for assessing system output. Twenty two questions pertaining to therapy and twelve questions pertaining to diagnosis were employed in our experiments. Twenty therapy and ten diagnosis questions were set aside for testing, and the remaining four questions were used for system development.
For each of the questions, the second author, an experienced searcher, manually constructed PubMed queries, extensively leveraging Clinical Queriesc and other advanced features, e.g. MeSH headings, as appropriate. This insured a high-quality baseline, representing possibly the best results available to clinicians searching MEDLINE. The top fifty hits (or all hits if the query retrieved less than 50 results) were retained for further processing.
Citations retrieved for each question were then evaluated for relevance by the second author, who consulted the recommendations associated with each question (provided by the specialists). This process required background medical knowledge, but did not require specialization in the wide range of specialties covered in our set of questions. Each retrieved citation was judged on a four-point scale, from 0 to 3. Abstracts containing at least one aspect of the answer were scored as 3; topically relevant abstracts potentially leading to answers were scored as 2; marginally relevant abstracts were given a score of 1, and irrelevant abstracts were given a score of 0.
Intervention Extraction and Semantic Clustering
Extraction of the main interventions in each abstract begins with the identification of all entities belonging to Chemicals & Drugs, Devices, and Procedures semantic groups,12 which form the set of possible interventions for therapy and diagnosis questions. Each candidate is scored based on its position in the abstract, the number of times it occurs, and presence of certain cue phrases.11 The intervention with the highest score is selected as the main intervention (allowing for multiple selections in the case of ties).
Once the interventions are identified, retrieved MEDLINE citations are organized into clusters according to a hierarchical agglomerative clustering algorithm13 with semantic distance as the link function. The system starts by assigning each intervention (and the associated abstract) to its own cluster, and then iteratively merges clusters whose interventions share a common UMLS hypernym, ascending the UMLS hierarchy in the process. For example, rofe-coxib would be grouped with ibuprofen because they are both Anti-Inflammatory Agents according to UMLS. The process stops when no new clusters can be formed. To avoid forming clusters under labels that are too general to be of interest, we truncated the tops of the UMLS hierarchies. For example, the MeSH category CHEMICALS & DRUGS was removed from consideration. An abstract may appear in multiple clusters if it contains more than one main intervention. For example, if the abstract compared the efficacy of two interventions that belong to different semantic groups. The abstracts within each cluster are sorted in the original PubMed presentation order (chronologically in reverse). Clusters themselves are sorted by size (in number of abstracts).
As the clinician drills down into a cluster, she sees citations that discuss a common theme. To increase the number of results that can be viewed simultaneously, our system displays a short summary of each abstract, consisting of the identified intervention (in brackets) and the main outcome sentence, as shown in Figure 1. Clinical outcomes summarize the major findings of a study, and can be automatically identified using machine learning techniques.11 Naturally, the outcome sentence is insufficient for basing a clinical decision, but it can serve as an entry point into the medical literature, which the physician can explore further in depth.
Assessing System Output
To test our hypothesis that semantic clustering represents an effective technique for organizing search results, we compared clustered output with the original PubMed results. Our evaluation methodology was as follows: under different experimental conditions, how “good” are the first three abstracts that a physician is likely to examine? Our choice of three abstracts was motivated by the finding that doctors are willing to spend perhaps two minutes looking for relevant information.14
Faced with the original PubMed results, physicians are most likely to examine the first three abstracts, given an expectation that “better” results will be placed higher in the list. Thus, for the baseline condition, we assess the quality of the top three PubMed hits (details below).
For citations that have been semantically clustered, which abstracts is the clinician likely to examine? One possibility is that she will examine the first abstract in the first, second, and third clusters (which are ordered by size). Thus, we can assess the quality of these three citations (the “Semantic1” condition).
However, the advantage of semantic clustering is that it provides a better overview of the information space, freeing physicians from a linear browsing order. Furthermore, since clinicians are highly-trained individuals, they are unlikely to browse a cluster that is “obviously irrelevant”. To simulate this behavior, we implemented an oracle condition, in which an oracle told the clinician what the three most relevant clusters were. She then examines the first abstract in these three clusters. In our experiments, the second author played the part of the oracle. We believe that this oracle condition approximates reality because given a small number of choices, clinicians are likely able to identify relevant intervention classes even if they did not previously know the answer. We term this the “Semantic2” condition.
Instead of sampling three abstracts from different clusters, a physician could examine three abstracts from the best cluster. This simulates the scenario where she recognizes a promising class of intervention and wants to learn about it in depth. We term this the “Semantic3” condition.
To further support our hypothesis regarding semantic clustering, we developed another experimental condition based on lexical clustering. An existing tool15 was employed to cluster MEDLINE citations purely based on keyword content, i.e., by representing each document as a high dimensional vector in which each unique term serves as a feature. The term with the highest tf.idf weight is selected as the cluster label; clusters created in this manner are sorted by size. For evaluation purposes, we assumed that the clinician examines the first abstract in the top three clusters. We term this the “Lexical” condition.
To simplify evaluation, we collapsed the original four-point judgments made on the abstracts into binary relevance judgments. Citations with an original score of 0 or 1 were considered “non-relevant”, while those with a score of 2 or 3 were considered “relevant”. Based on judgments that were made independent of system development (thus guarding against possible assessor bias), we are able to assess the relevance of the first, second, and third abstract that a clinician is likely to have examined under the different experimental conditions. The Wilcoxon signed rank test was employed to evaluate statistical significance of all results.
Results
Table 1 presents the evaluation results for all questions (therapy and diagnosis) in our test set (those not used in system development). For each condition, we report the fraction of the first, second, and third examined abstract that was relevant. The next column shows the overall accuracy of the system across the top three examined abstracts. The final column shows the total number of relevant abstracts retrieved. We can see that even a simple strategy of examining clustered results (Semantic1) yields access to more relevant abstracts. Under the Semantic2 and Semantic3 conditions, which more closely mirror physicians’ real-world behavior, nearly double the number of relevant abstracts would be encountered. This means that our semantic clustering algorithm is able to better bring relevant citations to the attention of physicians, compared to a linear result list.
Table 1.
Fraction of relevant abstracts retrieved for all questions. The last column shows the total number of relevant citations retrieved for each condition (out of 90).
| Condition | Rank | Total | Number | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| Baseline | 0.33 | 0.36 | 0.30 | 0.33 | 30 |
| Lexical | 0.30 | 0.36 | 0.36 | 0.33 | 30 |
| Semantic1 | 0.40 | 0.40 | 0.36 | 0.38 | 35 |
| Semantic2 | 0.63 | 0.70 | 0.53 | 0.62 | 56 |
| Semantic3 | 0.63 | 0.53 | 0.56 | 0.58 | 52 |
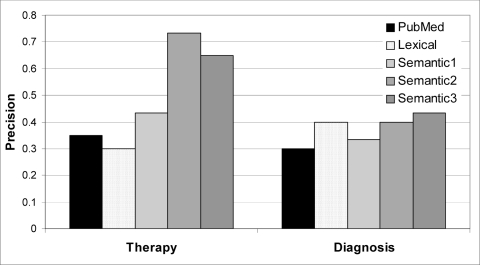
Figure 2 breaks down the precision figures over all three examined abstracts into therapy and diagnosis questions; see below for a discussion.
Figure 2.
Precision of various experimental conditions broken down by question type.
Discussion
Despite our efforts in carefully crafting PubMed queries, only a third of the top three retrieved abstracts were relevant, as demonstrated by our baseline condition in Table 1. Of those abstracts, only 16% contained an answer (i.e., scored a 3). These results confirm the barriers to practicing evidence-based medicine that many previous studies have found.
Lexical clustering, or grouping abstracts based solely on their keyword content, did not improve upon the PubMed baseline. Furthermore, the method did not yield meaningful cluster labels for most of the questions, and hence it is difficult for a clinician to get an accurate overview of the potentially relevant information. For example, “straw”, “nic”, and “history” were the top cluster labels generated for the question “What is the most effective nicotine replacement therapy?” For the question “Do TCAs or SSRIs have any effect on decreasing tinnitus?” the retrieved abstracts were so similar that only one cluster labeled “depression” was created. In other words, lexical clustering provides little in the way of organizing principles to help the user navigate a complex information space. Thus, an oracle condition for lexical clustering is not meaningful.
The Semantic1 condition, in which the first abstract in the first three clusters were examined, performed better than both the baseline and lexical clustering conditions, but the results are not statistically significant. Nevertheless, the advantages provided by our semantic clustering algorithm are evident from inspection of actual system output. For the question about tinnitus described above, the top three clusters were: “Drug groups primarily affecting the central nervous system”, “musculoskeletal medications”, and “Tranquilizing Agents”.
For the Semantic2 and Semantic3 conditions, we see that the examined abstracts are much higher in quality, given the larger fraction of relevant citations. The differences in performance between these conditions and the baseline are statistically significant, which appears to confirm the effectiveness of our approach. Since the cluster labels are descriptive, a physician can easily hone in on the most promising answers. It is fairly clear that clustering based on semantic concepts (interventions, in our case) outperforms clustering based purely on keywords. This result reaffirms the value of semantic text processing and ontological resources such as UMLS, without which our work would not be possible.
Overall performance on therapy questions was better than on diagnosis questions. This might be explained by the mixture of questions about diagnostic tests and differential diagnosis in our collection. We discovered that organizing results by intervention is less suitable for differential diagnosis questions. In retrospect, it appears that clustering by disorder would have yielded a better organization.
Conclusions and Future work
This work focuses on one particular aspect of medical information retrieval: the organization of search results to support physicians practicing evidence-based medicine. Specifically, we compared an approach based on semantic clustering to the baseline results produced by PubMed. Experiments suggest that our technique brings more relevant abstracts into prominent positions, based on an expectation of how physicians are likely to interact with clustered results. Nevertheless, whether this translates into more sound clinical decisions remains to be seen, and this critical question will be the focus of future work.
Acknowledgments
This work was supported in part by the U.S. National Library of Medicine. The first author thanks Esther and Kiri for their loving support.
Footnotes
References
- 1.Sackett DL, Rosenberg WM, Gray JA, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn’t. BMJ. 1996 Jan 13;312(7023):71–2. doi: 10.1136/bmj.312.7023.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White B. Making evidence-based medicine doable in everyday practice. Fam Pract Manag. 2004 Feb;11(2):51–8. [PubMed] [Google Scholar]
- 3.Honeybourne C, Sutton S, Ward L. Knowledge in the Palm of your hands: PDAs in the clinical setting. Health Info Libr J. 2006 Mar;23(1):51–9. doi: 10.1111/j.1471-1842.2006.00621.x. [DOI] [PubMed] [Google Scholar]
- 4.Voorhees EM. Overview of the TREC 2003 Question Answering Track. Proceedings of the Twelfth Text REtrieval Conference (TREC 2003) 2003. pp. 54–68.
- 5.Demner-Fushman D, Lin J. Answer extraction, semantic clustering, and extractive Summarization for clinical Question Answering. Proceedings of ACL 2006. 2006:841–848. [Google Scholar]
- 6.Pratt W, Fagan L. The usefulness of dynamically categorizing search results. J Am Med Inform Assoc. 2000 Nov-Dec;7(6):605–17. doi: 10.1136/jamia.2000.0070605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seol YH, Kaufman DR, Mendonca EA, Cimino JJ, Johnson SB.Scenario-based assessment of physicians’ information needs Medinfo 200411(Pt 1)306–10. [PubMed] [Google Scholar]
- 9.Timpka T, Ekstrom M, Bjurulf P. Information needs and information seeking behaviour in primary health care. Scand J Prim Health Care. 1989 Jun;7(2):105–9. doi: 10.3109/02813438909088656. [DOI] [PubMed] [Google Scholar]
- 10.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 11.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- 12.McCray AT, Burgun A, Bodenreider O.Aggregating UMLS semantic types for reducing conceptual complexity Medinfo 200110(Pt 1)216–20. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Karypis G. Evaluation of hierarchical clustering algorithms for document datasets. Proceedings of CIKM. 2002;2002:515–524. [Google Scholar]
- 14.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians’ clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005 Mar-Apr;12(2):217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leuski A. Evaluating document clustering for interactive information retrieval. Proceedings of CIKM. 2001;2001:41–48. [Google Scholar]