Abstract
The @neurIST ontology is currently under development within the scope of the European project @neurIST intended to serve as a module in a complex architecture aiming at providing a better understanding and management of intracranial aneurysms and subarachnoid hemorrhages. Due to the integrative structure of the project the ontology needs to represent entities from various disciplines on a large spatial and temporal scale. Initial term acquisition was performed by exploiting a database scaffold, literature analysis and communications with domain experts. The ontology design is based on the DOLCE upper ontology and other existing domain ontologies were linked or partly included whenever appropriate (e.g., the FMA for anatomical entities and the UMLS for definitions and lexical information). About 2300 predominantly medical entities were represented but also a multitude of biomolecular, epidemiological, and hemodynamic entities. The usage of the ontology in the project comprises terminological control, text mining, annotation, and data mediation.
Keywords: Medical Informatics Applications, Ontology design, Intracranial aneurysm, Subarachnoid hemorrhage, Terminology
Introduction
The @neurIST project is part of the 6th European framework program and aims for the development of an integrated IT infrastructure to manage and process intracranial aneurysms and subarachnoid hemorrhage. Its main objective is to integrate data from disparate sources and various disciplines that are characterized by a high fragmentation and heterogeneity both in terms of format as well as scale. The envisaged benefits for clinicians, scientists and patients include the diagnosis support and treatment planning – particularly in regard to individualized rupture risk assessment – and an easier and quicker access to knowledge in the domain, provided by an integrated software platform. In this project 29 academic and business partners from 12 countries are collaborating1.
One important activity in the @neurIST project is the development of a description logic-based ontology in the Web Ontology Language OWL (http://www.w3.-org/2004/OWL/) to represent all relevant concepts associated with cerebral aneurysms and subarachnoid bleedings respecting the sometimes differing views of the involved disciplines and scientific areas. Therefore, the ontology is not only concerned with the purely medical entities of interest like anatomical, pathological or medical procedural entities but also with biomolecular, epidemiological and hemodynamic entities. Furthermore, the ontology is committed to integrate the various levels of disease descriptions (e.g., views of clinicians, geneticists, and epidemiologists) with various sources of information (e.g., literature, clinical databases, imaging databases, and terminologies). The @neurIST projects’ objective to integrate data from a wide range of both medical and scientific disciplines on a large spatial and temporal scale has to be mirrored in the ontology as well and is one of its most important and challenging features.
Another important aspect in the @neurIST ontology developmental process is the identification of the requirements from the project partners towards the ontology and the ability to provide access to it terms of usability: The adoption and extension of the ontology can be severely impaired if domain experts are not provided with views on the ontology that are adequate in their context reducing complexity and focusing on their particular interest. To deliver a straightforward access to the ontological resources in the scope of the project, both specific tools as well as a web-based ontology browser are under active development.
Term acquisition and the conceptual space of the @neurIST ontology
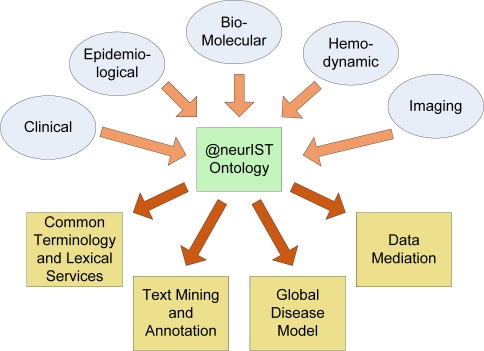
Because of the idea to integrate different scientific disciplines and views in regard to cerebral aneurysms and subarachnoid hemorrhage the @neurIST ontology thus needs to represent entities from those disciplines on a wide spatial and temporal scale (Figure 1). Various strategies served in the initial identification of relevant entity types and relations:
Through a patient database scaffold designed by medical experts, a standard set of documentation fields was defined for the description of patients and their management, the evaluation of treatment outcomes and the assessment of associations between patient characteristics, imaging, genetic results and outcomes. This initial data dictionary constituted the basis of the @neurIST Clinical Reference Information Model (CRIM) and is also consistent with two large studies which investigated intracranial aneurysms and subarachnoid hemorrhage2,3. The CRIM is essential as a source for intracranial aneurysm associated clinical and experimental data acquisition and hence a adequate starting point for ontology development.
Domain specific terms of high frequency, extracted from literature, served to identify relevant items in the domain and subsequently add them to our ontology. The ranking was done on a corpus of MEDLINE abstracts (n ≈ 25,000) retrieved on the basis of a Pub-Med query for “Intracranial Aneurysm OR Subarachnoid Hemorrhage”. Titles and abstracts were tagged with parts-of-speech using the TeMis tagger (http://www.temis-group.com/). Noun phrases “(NP)” were extracted using the basic pattern “(adjective or noun or proper word)” plus “(noun or proper word)”. The resulting noun phrases were counted and those counts again submitted to a normalization process against a corpus retrieved via the PubMed query “author==Smith” on the basis of the Kullback-Leibler relative entropy. For the noun phrases that were present in the test corpus but missing in the normalization corpus, pseudocounts (i.e., terms which do not occur in the second corpus but only in the first one are counted with n=1) were introduced.
Domain experts such as biologists, neurosurgeons, pathologists, and engineers all provided technical terms with the respective state-of-the-art-knowledge on the meaning of terms and relations among the entity types. Up to now the process of term acquisition is not standardized and mainly based on personal communication. Ways to improve and simplify this important step are currently under investigation.
Figure 1.
Schematic representation of the various disciplines providing input to the conceptual space of the @neurIST ontology and its main application areas.
So far about 2300 entities have been identified and incorporated into the @neurIST ontology. The extension of the ontology in certain areas has shown to be quite intricate because of the inherent complexity of the ontology combined with a limited understanding of the domain experts with regard to the actual application purpose of ontologies in technical systems.
As technical framework for the ontology development the Protégé ontology editor (http://protege.-stanford.edu/) was utilized employing its plugins to simplify OWL ontology development. The @neurIST ontology is fully conformant with OWL-DL. Therefore the reasoner Pellet (http://pellet.-owldl.com/) is continuously used to check the consistency of the ontology and to classify it.
Ontology components and architecture
The @neurIST ontology is based on and adapted to several existing ontologies that are commonly accepted as standard in the field (Figure 2). For example, the choice of an appropriate upper ontology is of crucial decision in the development of any more complex ontology. In contrast to ‘lightweight’ ontologies which focus on a minimal terminological structure (i.e., often just a pure taxonomy) fitting the needs of a specific and mostly smaller community, the main purpose of foundational ontologies is to negotiate meaning on a larger scale, either to enable effective cooperation among multiple artificial agents or to establish consensus in a mixed society where such artificial agents need to cooperate with human beings4.
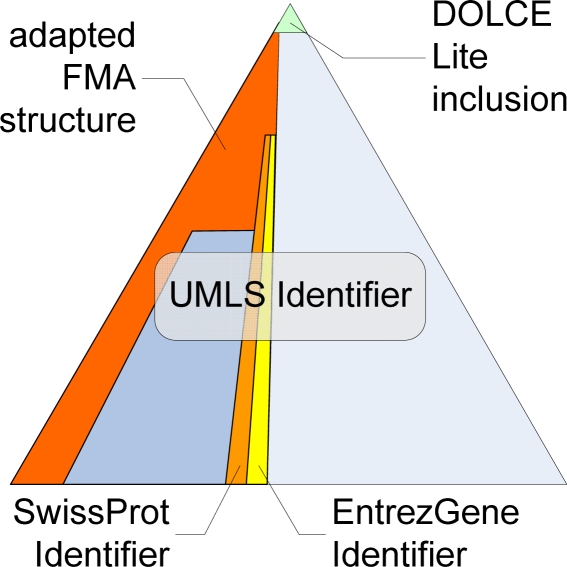
Figure 2.
Links and inclusions of biomedical reference terminologies and ontologies into the @neurIST ontology
We have chosen the Descriptive Ontology for Linguistic and Cognitive Engineering (DOLCE) as the basic ontological framework for our ontology and included its DOLCE-Lite-Plus version via the OWL import mechanism. DOLCE stands out in comparison to other upper ontology candidates which had been evaluated as a possible top-level ontology for the @neurIST project: DOLCE has a “clear cognitive bias” and its authors “do not commit to a strictly referentialist metaphysics related to the intrinsic nature of the world: …”. The categories of DOLCE are seen as “as cognitive artifacts ultimately dependent on human perception, cultural imprints and social conventions”5.
In our view, the philosophical background of DOLCE is especially important in the domain of clinical medicine where many entities are dependent on either social practice or individual human perception. The choice to use DOLCE also depends on the estimation that an ontology with a “cognitive bias” is more appropriate for representing a conceptual space that covers several scientific domains with different views on the term disease, than the realism-based approach of the BFO (Basic Formal Ontology)6. While DOLCE aims at capturing the ontological categories underlying natural language and human commonsense (i.e., concepts), BFO rather claims that “each ontology … represents some partition of reality” and does deny the validity of (man-made) concepts in any ontology intended to represent reality7.
Following this, all derived entity types of the @neurist ontology where categorized according to the DOLCE upper ontology and are therefore subclasses of one of the DOLCE basic entities: endurant (independent essential wholes, e.g., object-and substance like entities), perdurant (events, processes, activities, and states), quality (entities which can be perceived or measured, e.g. color, length) and region (abstract entities, i.e., spatial, temporal and abstract regions).
For the representation of anatomical entities of interest, the respective parts of the Foundational Model of Anatomy were included in our ontology as well and (re)classified along the lines of the DOLCE top-level concepts. The FMA is a domain ontology representing the complete, canonical human body through explicit declarative knowledge about anatomy and makes its anatomical information available in a frame-based format to ontology engineers and developers of applications for education, clinical medicine, electronic health record, and biomedical research8. The intent is to assure – through the FMA – consistency and standards in the representation of anatomical entities. Although the FMA comprises a very detailed anatomical model, the representation of anatomical entities common in the neuron-surgical domain needs to be further extended for the @neurIST ontology.
Information of several other biomedical terminological resources and databases are either linked into the @neurIST ontology (i.e., via OWL imports) or parts of them were directly and manually incorporated. The UMLS metathesaurus provides mainly taxonomic information about concepts, as well as synonyms and definitions for a large number of entities9. The entities of the ontology were mapped to UMLS Concept Identifiers (CUIs) whenever this was feasible and using MetaMapTransfer MMTX (http://mmtx.nlm.nih.gov). The mapping results were manually checked and corrected in case of any wrong automatic mapping which occurred in a high percentage especially in the field of anatomical entity types. The mapping to UMLS CUIs provides a preliminary classification of entity types according to the top level categories of the UMLS Semantic Network as well as a mapping to existing biomedical vocabularies containing adequate entities. Biomolecular entities are directly linked to SwissProt IDs, Entrez-Gene IDs and Gene Ontology IDs.
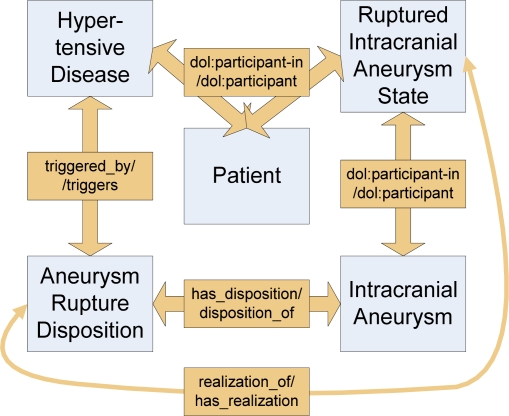
The semantics of entity types are given in terms of relations to other entity types which are defined as description logic/OWL restrictions. The basic types of relations are also provided by the DOLCE ontology defining e.g., mereological relations and participation relations5,10; domain specific relations were additionally created where necessary following as much as possible the recommendations and definitions of the Relation Ontology as part of the Open Biology Ontologies (OBO) Consortium4. As an example, Figure 3 shows the relations defined between the entity types in the risk model of our ontology.
Figure 3.
Part of the risk model of the @neurIST ontology exemplifying the usage of DOLCE and domain-specific relations. Displayed is the state of a patient with hypertensive disease and an existing intracranial aneurysm – the patient is in an intracranial aneurysm state. Entity type “Patient” participates in the process of “Hypertensive Disease (“dol:participant-in” is the corresponding DOLCE relation). “Hypertensive Disease” is a known risk factor for the rupture of intracranial aneurysms which is by “Hypertensive Disease” triggers “Aneurysm Rupture Disposition” with the domain specific relation “triggers”. The “Aneurysm Rupture Disposition” will manifest itself with a certain probability (“has_realization”) as “Ruptured Intracranial Aneurysm State”. The manifestation state of intracranial aneurysm has again as a “dol:participant” the type “Intracranial Aneurysm”.
Requirements, Use Cases and Usability
The development of any domain ontology should be driven by a comprehensive collection of both use cases and requirements towards the ontology.
Primarily, the ontology is intended to serve as a terminology in all the parts of the project. It identifies the allowed terms and their respective meanings in the clinical and experimental documentation as well as in the development of databases and graphical user interfaces. The @neurIST ontology gives both textual and formal (i.e., description logic) definitions for all required entities and will be used as a standardizing instrument throughout the project where ambiguities are frequent (e.g., clinical terms). To provide an adequate terminological coverage, the ontology is furthermore linked to a separate lexical resource which is not part of the ontology proper. This resource provides both preferred terms as well as synonyms and will be at least partly multilingual.
An important activity in the @neurIST project is knowledge discovery. Therefore, our ontology will provide several term lists which can be employed in text mining and annotation. Semantic analyses are supported via relations between the entities.
One of the most ambitious objectives of the @neurIST project is the inclusion of distributed computing on a large scale. The roles of the ontology in this scenario are data mediation and service binding. On the one hand the ontology is targeted at mediating between heterogeneous databases on a semantic level. On the other hand it is planned to serve as a knowledge repository for the binding of distributed services.
A main difficulty that arose during the ontology development is its inherent complexity which in turn may lead to issues in extensibility and usability. With current ontology editing tools it is difficult to present the ontology in such a simple way that does not impair the understanding of domain experts. This is particularly problematic since the domain ontology development is an interdisciplinary approach during which the communication with a domain expert about the ontology contents and its relation to the project needs are heavily dependent on his or her understanding and hence are crucial. Therefore specific tools and a web-based ontology browser are currently being developed to alleviate this problem and to help with the extension, quality control and usage of the @neurIST ontology.
The just mentioned web-based ontology browser provides the following features:
Easy web-based navigation: the entity type hierarchy can be browsed.
Enhanced search: based on the definition texts and the linked lexical resources, a textual search provides a ranked and easy access for domain experts into the complex hierarchy.
The logical expression of formal definitory restrictions are „translated“ into natural language suitable for domain experts.
A graphical representation of complex semantic relations provides an overview of the dependencies between entity types.
Conclusion
In parallel with the objective of the @neurIST project to integrate the heterogeneous data on a large spatial and temporal scale and originating from different sources the described ontology must incorporate conceptual spaces of such various domains as clinical medicine, molecular biology, imaging, physiological simulation and epidemiology. On the sound foundations of the DOLCE upper ontology we succeeded in representing about 2300 relevant entity types so far. The @neurIST ontology will be used in several other parts of the project: as a controlled vocabulary for documentation and user interfaces, as basis for text-mining and annotation and also in disseminated software architecture for data mediation. Further development and usage of the ontology in the @neurIST project will be heavily influenced by the development of tools and user interfaces to access the ontology in a straightforward fashion.
Acknowledgments
This work was generated in the framework of the @neurIST Integrated Project, which is co-financed by the European Commission through the contract no. IST-027703. http://www.aneurist.org
References
- 1.Alejandro Frangi.@neurIST. Integrated Biomedical Informatics for the Management of Cerebral Aneurysms http://www.aneurist.org accessed 2007-3-14.
- 2.International Subarachnoid Aneurysm Trial (ISAT) Collaborative Group. International Subarachnoid Aneurysm Trial (ISAT) of neurosurgical clipping versus endovascular coiling in 2143 patients with ruptured intracranial aneurysms: a randomized trial. The Lancet. 2002;360:1264–74. doi: 10.1016/s0140-6736(02)11314-6. [DOI] [PubMed] [Google Scholar]
- 3.The International Study of Unruptured Intracranial Aneurysms Investigators. Unruptured Intracranial Aneurysms-Risk of Rupture and Risks of Surgical Intervention. New England Journal of Medicine. 1998;339:1725–33. doi: 10.1056/NEJM199812103392401. [DOI] [PubMed] [Google Scholar]
- 4.Smith B, Ceusters W, Klagges B, et al. Relations in Biomedical Ontologies. Genome Biology. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masolo, Claudio, Borgo, Stefano, Gangemi, Aldo, Guarino, Nicola, Oltramari, AlessandroWonderWeb Deliverable D18Ontology Library (final). 2003
- 6.Grenon P, Smith B, Goldberg L. Biodynamic Ontology: Applying BFO in the Biomedical Domain. In: Pisanelli DM, editor. Ontologies in Medicine. Amsterdam: IOS Press; 2004. pp. 20–38. [PubMed] [Google Scholar]
- 7.Grenon Pierre.BFO in a Nutshell: A Bicategorial Axiomatization of BFO and Comparison to DOLCEGrenon, Pierre. IFOMIS Reports. 2003. Leipzig, IFOMIS, Universität Leipzig.
- 8.Rosse C, Mejino JLV. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons P. Parts A Study in Ontology. Oxford; Oxford University Press; 1987. [Google Scholar]