Abstract
Oncologists managing cancer patients use radiology imaging studies to evaluate changes in measurable cancer lesions. Currently, the textual radiology report summarizes the findings, but is disconnected from the primary image data. This makes it difficult for the physician to obtain a visual overview of the location and behavior of the disease. LesionViewer is a prototype software system designed to assist clinicians in comprehending and reviewing radiology imaging studies. The interface provides an Anatomical Summary View of the location of lesions identified in a series of studies, and direct navigation to the relevant primary image data. LesionViewer’s Disease Summary View provides a temporal abstraction of the disease behavior between studies utilizing methods of the RECIST guideline1. In a usability study, nine physicians used the system to accurately perform clinical tasks appropriate to the analysis of radiology reports and image data. All users reported they would use the system if available.
Introduction
Oncologists caring for cancer patients are often faced with large amounts of complex clinical data. When making treatment decisions, the Oncologist looks for changes in the measurable disease. Measurable disease includes cancer lesions that are large enough to be identified via imaging studies, observed through physical examination, or measured in blood tests. In monitoring a patient’s response to treatment, the Oncologist looks for changes in the size, number and metabolic activity of measurable cancer lesions. One method is to compare cancer lesions identified in a new imaging study with those in prior studies.
Imaging studies can contain hundreds of images, yet only a few have significant findings. In the current clinical workflow, the Radiologist reviews the imaging study, identifies the lesions and reports the findings in a textual report. The Radiologist can use several methods for describing the location of the lesion: anatomical descriptions, image number and/or image annotation (the proverbial “X marks the spot”). Radiology workstations allow Radiologists to create and save image annotations in the form of arrows or lines to mark the regions of interest (ROI). In addition to marking the ROI, these tools provide calipers for measuring the size of the lesions, which the Radiologist may make note of in their report. Comparison with prior studies allows the Radiologist to make a qualitative impression of the overall increase or decrease in the size and number of lesions observed.
The Oncologist uses the report, image annotations, and raw imaging data to aid diagnosis and treatment decisions. However, there is no current technology to enable the Oncologist to directly access those regions of images corresponding to each abnormality described in the radiology report. The Oncology must use the textual descriptions and image number references in the report as a guide to re-identify the lesions of interest. Some PACS systems allow all annotated images to be grouped together, decreasing the search space, but unfortunately, Radiologists do not annotate all the regions of interest. Like the Radiologist, the Oncologist uses a qualitative assessment of the data within the context of the larger clinical picture to make an impression of the behavior of the disease.
Currently, no method exists for visually summarizing all of the lesions of interest into a single view that shows the location of the disease in the body. Such a summary view could aid physician understanding of the status of the patient’s disease by providing an overview of the location and number of measurable cancer lesions. The summary view could be used to navigate directly to the respective axial image that was identified by the Radiologist to contain the lesion of interest. When presented with multiple radiology studies of the same patient, the summary view of each study could aid understanding of disease progression over time. Additionally, rules for evaluating lesion data trends can also provide further abstraction in summarizing disease behavior over time. In summary, navigation using a summary view provides opportunities to not only improve efficiency of image search, but to improve overall comprehension of disease trends.
We hypothesized that the use of visualization techniques to summarize and analyze large amounts of radiographic imaging data will assist clinicians in accurately and efficiently assessing a patient’s disease state. We thus set out to create LesionViewer with the following goals:
Provide a summary view of all lesions identified in a radiographic study with navigation from this view directly to the image and metadata corresponding to a given lesion
Provide a temporal view of lesions across imaging studies with navigation directly to images of a specific lesion in all studies, to allow for direct comparison
Provide quantitative assessment of changes in disease behavior via analysis of lesion number and size using the RECIST guideline1
Background
In the context of clinical trials, researchers often attempt to quantify the change in the measurable disease in order to accurately report the response to treatment. The National Cancer Institute has published the Response Evaluation Criteria in Solid Tumors (RECIST) guideline1 as a method for quantifying changes in measurable cancer lesions. The guideline uses the change in the number and size of measurable target lesions to determine a patient’s response to treatment. The guidelines’ methodology is as follows:
Define a set of target lesions: a maximum of 10 representative lesions with greatest diameter at least 10 mm in size
Count remaining non-target lesions: all other measurable lesions of any size
Create composite length score: the sum of the longest diameters (SLD) of target lesions
Calculate percent change in SLD from study to study
Calculate change in number of target and non-target lesions from study to study
Outcome definitions as per RECIST guideline:
Complete Response (remission): disappearance of all lesions
Partial Response (regression): no new lesions AND at least 30% decrease in the SLD
Progressive Disease (progression): increase in number of lesions OR at least 20% increase in SLD
Stable Disease (stable): no new lesions AND between 30% decrease and 20% increase in SLD
Application of the RECIST method in clinical trials provides a quantitative assessment of changes in disease activity. Opportunity, however, exists to more generally apply these methods to everyday clinical practice. Nevertheless, applying RECIST is very time consuming and physicians are unlikely to use it in the context of the current clinical workflow.
Visualization techniques can be used to reduce the data space of hundreds of images and text reports into concise images or data structures, providing users with patterns that aid them in decision-making2. Clinical data visualization techniques have been described for multiple types of patient data including laboratory and medication administration data3,4, but have not yet been described in the radiology imaging domain. Similarly, temporal abstraction methods have also been used to automatically summarize and visualize large amounts of clinical data5. These systems utilize structured data and temporal reasoning algorithms to make clinical conclusions. Since radiology data is usually reported in textual reports, such temporal abstraction algorithms, which require highly structured data, have not been applied to this domain.
DICOM (Digital Imaging and Communications in Medicine) is a comprehensive set of standards for storing and processing medical imaging data. The file format definition includes data structures for storing ROI’s, their coordinates and associated textual metadata. While literature has described radiology imaging systems displaying metadata associated with regions of interest6,7, there has not yet been a system that utilizes such data to create an interactive summary visualization of the patient’s tumor burden. Furthermore, the DICOM standard does not currently specify a method for uniquely identify individual lesions on the images across studies. The lack of such an identification method limits the ability to navigate directly to all relevant images across studies and to automatically compare lesion size over time.
Methods
After receiving a waiver from Stanford’s Institutional Review Board (IRB), we obtained de-identified data from 5 cancer patients treated at Stanford Hospital and Clinics. Patients were selected that had at least two CT scans of the Chest, Abdomen and Pelvis taken at least one month apart, and type of cancer was unknown to system developers. We obtained the DICOM images and radiology report for each study. All DICOM images were converted to the JPEG format for purposes of implementation. From the reports and images, we identified and recorded the following data for each cancer lesion:
Longest Lesion Diameter
Axial image number corresponding to the axial slice with longest lesion diameter
Coordinates of the center of the lesion on axial and coronal images
Text description of anatomical location and type of lesion
In order to track lesions over time, each lesion was assigned a unique LesionID number. When a lesion was observed across studies, it was given the same LesionID number. These lesion attributes were recorded in XML files maintained for each imaging study.
The temporal abstraction algorithm implemented the RECIST methods. The algorithm used the lesion number and changes in lesion size from one study to the next to determine if there has been a 1) complete response to treatment, 2) partial response to treatment, 3) stable disease, or 4) disease progression.
Our prototype was implemented using an XML data structure and the Python scripting language. To create the graphical interface we utilized Python’s Tkinter library, Imaging Library and MegaWidgets.
Results
Implementation
We implemented LesionViewer employing the methods described above. The Anatomical Summary View depicted in Figure 1 provides a visualization of a series of studies. Each lesion in the study is represented by a red dot that marks its location. Mousing over a lesion triggers the appearance of a popup box describing the basic characteristics of the lesion, including its ID number, anatomical location, lesion type, and dimensions. Clicking on a lesion triggers the appearance of all corresponding axial images in which the lesion is present.
Figure 1.
Anatomical Summary View with three CT scans of the Chest, Abdomen and Pelvis. A lesion located in the second study is moused over, triggering the appearance of a pop-up box describing the lesion’s basic characteristics. The same lesion is later clicked. Since that lesion is present in all three studies, its axial slices from all the studies appear at the bottom of the screen.
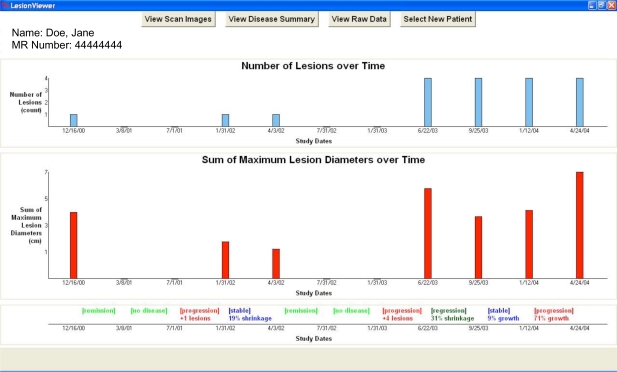
The Disease Summary View shown in Figure 2 employs a temporal abstraction method based on RECIST to visualize disease trends. Two graphs are displayed, one measuring the total number of lesions identified in each study, and the other displaying the SLD for each study over time. At the bottom of the screen is the disease state timeline that charts the state of the disease during the intervals between each study date. Again, using RECIST, the abstraction method determines whether the disease is in a state of remission, regression, stability, progression, or if no disease is present at all. Apart from the disease state labels, the specific quantitative change in the disease condition responsible for triggering the disease label is also displayed. Due to space limitations, not pictured is the Raw Data Table feature that summarizes all the imaging data associated with the lesions for a given patient.
Figure 2.
Disease Summary View with eleven studies. The top graph charts the number of lesions over time. The bottom graph charts the composite length score over time. (See text for explanation of composite length score.) The timeline at the bottom displays the disease state (automatically generated using the RECIST guidelines) during each interval between studies.
Evaluation
While Oncologists make up the primary user base for this tool, we wanted to see if other types of clinicians who care for cancer patients, such as Internists, would find the tool intuitive and useful. We thus asked nine physician volunteers from Stanford Hospital to evaluate LesionViewer (see Table 1).
Table 1.
Results of pre-task User Survey about current clinical practice
| Level of Training | Number of users | Perceived Skill using Radiology Workstation (average rating) | Time to review radiology report and images (average) (minutes) | Familiarity with RECIST Guidelines |
|---|---|---|---|---|
| Internal Medicine Attending | 3 | 3.3 | 7.6–10 (8.8) | None |
| Medical Oncology Fellow | 3 | 3.3 | 8.3–15 (11.6) | All |
| Medical Oncology Attending | 3 | 3.3 | 8.6–18.3 (13.5) | All |
| TOTAL | 9 | 3.3 | 8.2–14.4 (11.3) |
Prior to beginning the system evaluation the users completed a questionnaire regarding their skills, workflow habits and knowledge. Each user was asked to rate their perceived skill using the radiology workstation at their clinic. (1=expert skill of radiologist, 2=know many advanced features, 3=know some advanced features, 4=know basic features for viewing images, 5=don’t know how to use radiology system). Overall, the physicians perceived they had moderate skill giving an average rating of 3.3. The physicians also estimated that it took on average between 8.2 and 14.4 minutes to review the radiology report and images for a given patient in their clinic. Finally, only the Oncologists had prior knowledge of the RECIST guideline methods.
The system usability study consisted of direct observation of participants using LesionViewer to complete a multiple choice and fill-in-the blank questionnaire of 17 clinical tasks across 4 of the clinical cases used to build the prototype system. Comprehension tasks included determining the change in the number of lesions, SLD and disease state from one time point to another. Data review tasks were designed to test the user’s ability to navigate to the primary imaging data using the interface features. Of the 9 users, 4 had perfect scores and the remaining 5 users performed 16 out of 17 tasks correctly. Four users incorrectly calculated the difference between two dates and one user incorrectly assessed disease behavior due to lack of feature awareness. There was no difference in performance between the Internists and Oncologists. Users took approximately 10 minutes to complete the 4 cases.
After completing the clinical tasks associated with the clinical cases, users were given a structured questionnaire detailing their satisfaction with the system. Users rated the ease of performing tasks using LesionViewer on a 5-point scale (1=Very Easy, 2=Easy, 3=Somewhat Easy, 4=Somewhat Difficult, 5=Difficult). The nine users gave an average usability score of 2. All nine users stated that they would use the system in clinical practice if it were integrated into their current radiology workstation and felt it would save them time.
Discussion
We built LesionViewer with the goal of assisting clinicians in the task of comprehending and reviewing the results of imaging studies. Our tool was evaluated by nine users who found the system intuitive, and enthusiastically reported that they would adopt such a system in their everyday practice. Tasks were completed accurately and efficiently.
The Usability Study was designed to determine if our visualization strategy was intuitive and could be used to perform clinical tasks associated with patient care. It was not designed to compare our system with current techniques. As our users are busy clinicians who volunteered their time, we wanted to limit our study to less than 30 minutes. A formal comparative evaluation measuring differences in accuracy and time between methods is intended as future work.
The methods applied in LesionViewer could be used to extend the data elements of the DICOM standard to support computational reasoning about regions of interest across studies. We have demonstrated one such computational algorithm with the RECIST guideline to evaluate an individual patient, but others including cohort identification would also be supported if applied to a database of patient imaging studies. Furthermore, the methods in LesionViewer could be extended to include other imaging modalities such as Mammography, MRI, and PET.
Conclusion
We have shown that LesionViewer provides an effective summarized view of the location and course of disease in cancer patients. LesionViewer does this by creating an electronic link between the Radiologist’s findings and the raw imaging data, summarized through multiple novel visualization techniques. LesionViewer also provides a method for automatically applying the RECIST guidelines to assess disease behavior. This tool could enable a new and more accurate paradigm for assessing and communicating cancer imaging results.
References
- 1.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000 Feb 2;92(3):205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 2.Readings in Information Visualization: Using Vision to Think, Chapter 1. Stuart K Card, Jock D Mackinlay, Ben Shneiderman, Morgan Kaufmann Publishers; pp. 1–34. [Google Scholar]
- 3.Powsner SM, Tufte ET. Graphical Summary of patient status. Lancet. 1994;344(8919):386–389. doi: 10.1016/s0140-6736(94)91406-0. [DOI] [PubMed] [Google Scholar]
- 4.Plaisant C, Mushlin R, Snyder A, Li J, Heller D, Shneiderman B. LifeLines: using visualization to enhance navigation and analysis of patient records. Proc AMIA Symp. 1998:76–80. [PMC free article] [PubMed] [Google Scholar]
- 5.Shahar Y, Cheng C. Model-Based Visualization of Temporal Abstractions. Computational Intelligence. 2000;16(2):279–306. [Google Scholar]
- 6.Lee S, Lo C, Wang C, Chung P, Chang C, Yang C, Hsu P. A computer-aided design mammography screening system for detection and classification of microcalcifications. Int J Med Inform. 2000 Oct;60(1):29–57. doi: 10.1016/s1386-5056(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 7.Pietka E, Pospiech-Kurkowska S, Gertych A, Cao F. Integration of computer assisted bone age assessment with clinical PACS. Comput Med Imaging Graph. 2003;27(2–3):217–28. doi: 10.1016/s0895-6111(02)00076-9. [DOI] [PubMed] [Google Scholar]