Abstract
The Clinical E-Science Framework (CLEF) project is building a framework for the capture, integration and presentation of clinical information: for clinical research, evidence-based health care and genotype-meets-phenotype informatics. A significant portion of the information required by such a framework originates as text, even in EHR-savvy organizations. CLEF uses Information Extraction (IE) to make this unstructured information available. An important part of IE is the identification of semantic entities and relationships. Typical approaches require human annotated documents to provide both evaluation standards and material for system development. CLEF has a corpus of clinical narratives, histopathology reports and imaging reports from 20 thousand patients. We describe the selection of a subset of this corpus for manual annotation of clinical entities and relationships. We describe an annotation methodology and report encouraging initial results of inter-annotator agreement. Comparisons are made between different text subgenres, and between annotators with different skills.
Introduction
Although large parts of the medical record exist as structured data, a significant proportion exists as unstructured free texts. This is not just the case for legacy records. Much of pathology and imaging reporting is recorded as free text, and a major component of any UK medical record consists of letters written from the secondary to the primary care physician (GP). These documents contain information of value for day-to-day patient care and of potential use in research. For example, narratives record why drugs were given, why they were stopped, the results of physical examination, and problems that were considered important when discussing patient care, but not important when coding the record for audit.
CLEF1 uses information extraction (IE) technology2 to make information available for integration with the structured record, and thus to make it available for clinical care and research3. IE aims to extract automatically from documents the main events and entities, and the relationships between them, and to represent this information in structured form. IE has immense potential in the medical domain. One of the earliest IE applications was the analysis of discharge summaries in the Linguistic String Project4, and it has since seen application in various clinical settings.
Although much IE research has focused on fully automated methods of developing systems (pioneering work is reported in5), most practical IE still needs data that has been manually annotated with events, entities and relationships. This data serves three purposes. First, an analysis of human annotated data focuses and clarifies requirements. Second, it provides a gold standard against which to assess results. Third, it provides data for system development: extraction rules may be created either automatically or by hand, and statistical models of the text may be built by machine learning algorithms.
Biomedical corpora are increasingly common. For example, the GENIA corpus of abstracts has been semantically annotated with multiple entities. It does not, however, include relationships between them6. Other authors have reported semantic annotation exercises specific to clinical documents, but these are generally restricted to a single type of entity7. This paper reports on the construction of a gold standard corpus for the CLEF project, in which clinical documents are annotated with both multiple entities and their relationships. To the best of our knowledge, no one has explored the problem of producing a corpus annotated for clinical IE to the depth and to the extent reported here. Our annotation exercise uses a large corpus, covers multiple text genres, and involves over 20 annotators. We examine two issues of pertinence to the annotation of clinical documents: the use of domain knowledge; and the applicability of annotation to different sub-genres of text. Results are encouraging, and suggest that a rich corpus to support IE in the medical domain can be created.
The CLEF Corpus
Our development corpus comes from CLEF’S main clinical partner, the Royal Marsden Hospital (RMH). RMH is Europe’s largest specialist oncology centre. The entire corpus consists of both the structured records and free text documents from 20234 patients. The free text documents consist of three types: clinical narratives (with sub-types as shown in Table 1); histopathology reports; and imaging reports. Patient confidentiality is ensured through a variety of technical and organisational measures, including automatic pseudonymisation and manual inspection.
Table 1.
% of narratives in random sample
| Narrative subtype | % of standard | Neoplasm | % of standard |
|---|---|---|---|
| To GP | 49 | Digestive | 26 |
| Discharge | 17 | Breast | 23 |
| Case note | 15 | Haematopoetic | 18 |
| Other letter | 7 | Respiratory etc | 12 |
| To consultant | 6 | Female genital | 12 |
| To rererrer | 4 | Male genital | 8 |
| To patient | 3 |
Gold standard document sampling
Given the expense of human annotation, the gold standard portion of the corpus has to be a relatively small subset of the whole corpus of 565000 documents. In order to avoid events that are either rare or outside of the main project requirements, it is restricted by diagnosis, and only considers documents from those patients with a primary diagnosis code in one of the top level sub-categories of ICD-10 Chapter II (neoplasms). In addition, it only contains those sub-categories that cover more than 5% of narratives and reports. The gold standard corpus consists of two portions, selected for slightly different purposes.
Whole patient records
Two applications in CLEF involve aggregating data across a single patient record. The CLEF chronicle builds a chronological model for a patient, integrating events from both the structured and unstructured record8. CLEF report generation creates aggregated and natural language reports from the chronicle9. These two applications require whole patient records for development and testing. Two whole patient records were selected for this portion of the corpus, from two of the major diagnostic categories, to give median numbers of documents, and a mix of document types and lengths.
Stratified random sample
The major portion of the gold standard serves as development and evaluation material for IE. In order to ensure even training and fair evaluation across the entire corpus, the sampling of this portion is randomised and stratified, so that it reflects the population distribution along various axes. Table 1 shows the proportions of clinical narratives along two of these axes. Initial annotation of the random sample is focused on 50 each of clinical narratives, histopathology reports, and imaging reports. The final numbers of documents annotated is expected to be greater than this.
Annotation Schema
The CLEF gold standard is a semantically annotated corpus. We are interested in extracting the main semantic entities from text. By entity, we mean some real-world thing referred to in the text: the drugs that are mentioned, the tests that were carried out etc. We are also interested in extracting the relationships between entities: the condition indicated by a drug, the result of an investigation etc.
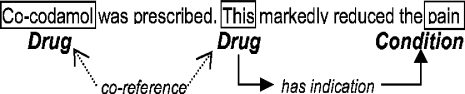
Annotation is anchored in the text. Annotators mark spans of text with a type: drug, locus and so on. Annotators may also mark words that modify spans (such as negation), and mark relationships as links between spans. Two or more spans may refer to the same thing in the real world, in which case they co-refer. This is also marked by the annotators. Some aspects of annotation are shown in Diagram 1.
Diagram 1.
Annotations, co-reference, relationships.
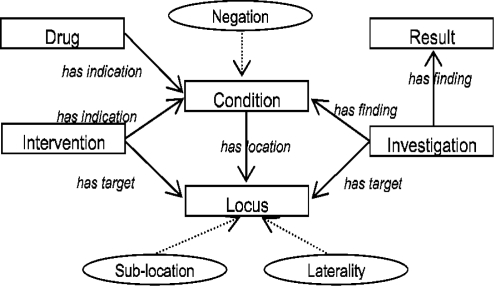
The types of annotation are described in a schema, shown in Diagram 2. The schema has been based on a set of requirements developed between clinicians and computational linguists in CLEF (there are no standard schemas or theories in this area). The schema types are mapped to types in the UMLS semantic network. For the purposes of annotation, the schema is modeled as a Protégé-Frames ontology10. Annotation is carried out using an adapted version of the Knowtator plugin for Protégé11. This was chosen for its handling of relationships, after evaluating several such tools.
Diagram 2.
CLEF annotation schema. Rectangles: entities; ovals: modifiers; solid lines: relationships.
An Annotation Methodology
The annotation methodology follows established natural language processing standards12. Annotators work to agreed guidelines; documents are annotated by at least two annotators; documents are only used where agreement passes a threshold; differences are resolved by a third experienced annotator. These points are discussed further below.
Annotation Guidelines
Consistency is critical to the quality of a gold standard. It is important that all documents are annotated to the same standard. Questions regularly arise when annotating. For example, should multi-word expressions be split? Should “myocardial infarction” be annotated as a condition, or as a condition and a locus? To ensure consistency, a set of guidelines is provided to annotators. These describe in detail what should and should not be annotated; how to decide if two entities are related; how to deal with co-reference; and a number of special cases. The guidelines also provide a sequence of steps, a recipe, which annotators should follow when working on a document. This recipe is designed to minimise errors of omission. The guidelines themselves were developed through a rigorous, iterative process, which is described below.
Double Annotation
A singly annotated document can reflect many problems: the idiosyncrasies of an individual annotator; one-off errors made by a single annotator; annotators who consistently under-perform. There are many alternative annotation schemes designed to overcome this, all of which involve more annotator time. Double annotation is a widely used alternative, in which each document is independently annotated by two annotators, and the sets of annotations compared for agreement.
Agreement Metrics
We measure agreement between double annotated documents using inter annotator agreement (IAA, shown below). Pairs of double annotations are rejected if agreement does not pass some threshold (currently 80% for entities).
IAA = matches / (matches + non-matches)
Results reported in this paper give a “relaxed” score. Partial matches (inclusive overlaps) are counted as a half match. In development, both strict (non-overlap) and relaxed (overlap) scores were calculated.
Together, these show how much disagreement is down to annotators finding similar entities, but differing in the exact spans of text marked.
The metrics used are equivalent to others more typically used in IE evaluations, as shown in Table 2. IAA also approximates the widely used kappa score, which is not appropriate in this case13.
Table 2.
Equivalence of agreement and IE metrics.
| Agreement metrics | IE evaluation metrics |
|---|---|
| Match | 2 x correct |
| non-match | spurious + missing |
| IAA | F measure |
Relationships were also scored using IAA. In development, two variations were used. First, all relationships found were scored. This has the drawback that an annotator who failed to find a relationship because they had not found one or both the entities would be penalized. To overcome this, a second IAA was calculated, including only those relationships where both annotators had found all entities involved. This allows us to isolate, to some extent, relationship scoring from entity scoring. Results reported in this paper use this second score.
Difference Resolution
Double annotation can be used to improve the quality of annotation, and therefore the quality of statistical models trained on those annotations. This is achieved by combining double annotations to give a set closer to the “truth” (although it is generally accepted as impossible to define an “absolute truth” gold standard in an annotation task with the complexity of CLEF’S). The resolution process is carried out by a third experienced annotator. All agreements from the original annotators are accepted into a consensus set, and the third annotator adjudicates on differences, according to a set of strict guidelines. In this way, annotations remain at least double annotated.
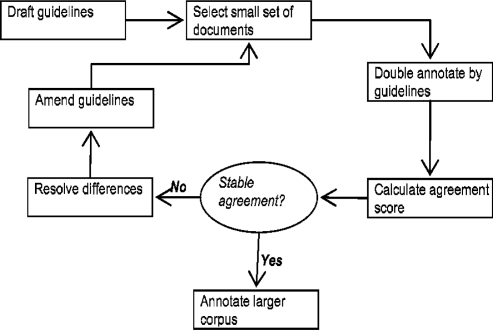
Developing the Guidelines
The guidelines were developed, or debugged, using an iterative process, designed to ensure their consistency. This is shown in Diagram 3. Two qualified clinicians annotated different sets of documents in 5 iterations (covering 31 documents in total). The IAA for these iterations are shown in Table 3. As can be seen, entity IAA remains consistently high after the 5 iterations, after which very few amendments were required on the guidelines. Relation IAA does not appear so stable on iteration 5. Difference analysis showed this to be due to a single, simple type of disagreement across a limited number of sentences in one document. Scoring without this document gave a 73% IAA.
Diagram 3.
Iterative development of guidelines
Table 3.
IAA (%) for each development iteration.
| Debug iteration | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Entities | Matches | 244 | 244 | 308 | 462 | 276 |
| Partial match | 2 | 6 | 22 | 6 | 1 | |
| Non-matches | 45 | 32 | 93 | 51 | 22 | |
| IAA | 84 | 87 | 74 | 89 | 92 | |
| Relationships | Matches | 170 | 78 | 116 | 412 | 170 |
| Partial match | 3 | 5 | 14 | 6 | 1 | |
| Non-matches | 31 | 60 | 89 | 131 | 103 | |
| IAA | 84 | 56 | 56 | 75 | 62 | |
Annotator Expertise
In order to examine how easily the guidelines could be applied by other annotators with varying levels of expertise, we also gave a batch of documents to our development annotators, another clinician, a biologist with some linguistics background, and a computational linguist. Each was given very limited training. The resultant annotations were compared with each other, and with a consensus set created from the two development annotators. The IAA matrix for this group is shown in Table 4. This small experiment shows that even with very limited training, agreement scores that approach acceptability are achievable. A difference analysis suggested that the computational linguist was finding more pronominal co-references and verbally signaled relations than the clinicians, but that unsurprisingly, the clinicians found more relations requiring domain knowledge to resolve. A combination of both linguistic and medical knowledge appears to be best.
Table 4.
IAA (%) for entities. D1 and D2: development annotators; C: clinician; B: biologist with linguistics background; L: computational linguist
| D2 | 77 | ||||
| C | 67 | 68 | |||
| B | 76 | 80 | 69 | ||
| L | 67 | 73 | 60 | 69 | |
| Consensus | 85 | 89 | 68 | 78 | 73 |
| D1 | D2 | C | B | L |
This difference reflects a major issue in the development of the guidelines: the extent to which annotators should apply domain specific knowledge to their analysis. Much of clinical text can be understood, even if laboriously and simplistically, by a non-clinician armed with a medical dictionary. The basic meaning is exposed by the linguistic constructs of the text. Some relationships between entities in the text, however, require deeper understanding. For example, the condition for which a particular dmg was given may be unclear to the non-clinician. In writing the guidelines, we decided that such relationships should be annotated, although this requirement is not easy to formulate as specific rules.
Different text sub-genres
The guidelines were mainly developed against clinical narratives. We were interested to see if the same guidelines could be applied to imaging and histopathology reports. We found that the guidelines could be quickly adapted with minimal change, to give excellent IAA after only two iterations, as is shown in Table 5. The fact that report IAA is better than clinical narrative IAA may reflect the greater regularity of the reports.
Table 5.
IAA (%) scores on different document types
| IAA | |||
|---|---|---|---|
| Iterations | Entities | Relationships | |
| Narratives | 5 | 92 | 62 |
| Imaging | 2 | 90 | 84 |
| Histopathology | 2 | 88 | 70 |
Annotation: Training and Consistency
We are currently training annotators ahead of the main annotation exercise. In total, 27 annotators are involved in debugging, annotation and review roles. They are drawn from practicing clinicians, medical informaticians, and final year medical students. They were given an initial 2.5 hours training session, focused on the annotation recipe and the guidelines.
After the initial training session, annotators were given two training batches to annotate, which comprised documents originally used in the debugging exercise, and for which consensus annotations had been created. IAA was computed between annotators, and against the consensus set. These results are shown for one group of annotators, in Table 6 for entities, and Table 7 for relationships.
Table 7.
IAA (%) for relationships, between 7 trainee annotators, two expert development annotators (D1 and D2), and a consensus C created from D1 and D2
| D2 | 63 | ||||||||
| 1 | 54 | 44 | |||||||
| 2 | 55 | 44 | 41 | ||||||
| 3 | 65 | 60 | 60 | 49 | |||||
| 4 | 74 | 64 | 54 | 59 | 62 | ||||
| 5 | 66 | 48 | 43 | 47 | 54 | 54 | |||
| 6 | 56 | 51 | 50 | 54 | 66 | 56 | 46 | ||
| 7 | 69 | 54 | 52 | 52 | 59 | 61 | 64 | 57 | |
| C | 87 | 74 | 52 | 52 | 61 | 68 | 57 | 61 | 71 |
| D1 | D2 | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
The matrices allow us to look at two factors. First, the IAA between annotators and the consensus set gives us a measure of consistency between annotators and our notion of “truth”. For entities, the trainee annotators clearly agree with the consensus as closely as the expert annotators do. For relations, they do not agree so closely. Second, the matrices allow us to examine the internal consistency between trainee annotators. Are they applying the guidelines consistently, even if not in agreement with the consensus? The wide range of relation IAA scores suggests that relationship annotation is inconsistent. Again, this may reflect the difficulty in applying highly domain-specific knowledge to relationships between entities. This is currently being addressed by feedback to the trainee annotators.
Conclusion
We have elucidated a methodology for the annotation of a gold standard for clinical IE, and shown that it is workable. Initial results show that promising levels of inter-annotator agreement can be achieved. We have examined the applicability of annotation to several clinical text sub-genres, and our results suggest that guidelines developed for one sub-genre may be fruitfully applied to others. Our work has raised several challenges. In terms of results, the greatest difficulty has been in achieving consistent relationship annotation. Organisationally, the co- ordination of many annotators has proved difficult. Access to the corpus is currently restricted: our final challenge is to develop a governance framework in which it can be made more widely available.
Table 6.
IAA (%) for entities, between 7 trainee annotators, two expert development annotators (D1 and D2), and a consensus C created from D1 and D2
| D2 | 77 | ||||||||
| 1 | 76 | 79 | |||||||
| 2 | 76 | 81 | 78 | ||||||
| 3 | 76 | 83 | 89 | 82 | |||||
| 4 | 75 | 84 | 83 | 81 | 85 | ||||
| 5 | 76 | 81 | 79 | 87 | 80 | 78 | |||
| 6 | 78 | 84 | 89 | 84 | 95 | 87 | 82 | ||
| 7 | 79 | 81 | 81 | 83 | 86 | 87 | 82 | 88 | |
| C | 85 | 89 | 84 | 84 | 88 | 85 | 83 | 91 | 87 |
| D1 | D2 | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
Acknowledgments
CLEF is funded by the UK Medical Research Council. We would like to thank the Royal Marsden Hospital for providing the corpus; our annotators; and members of CLEF who have helped with logistics.
References
- 1.Rector A, Rogers J, Taweel A, et al. CLEF: joining up healthcare with clinical and post-genomic research. Proc 2nd UK e-Science All Hands Meeting; 2003. pp. 264–267. [Google Scholar]
- 2.Cowie J, Lehnert W. Information Extraction. Commun ACM. 1996;39(1):80–91. [Google Scholar]
- 3.Harkema H, Roberts I, Gaizauskas R, Hepple M. Information extraction from clinical records. Proc 4th UK e-Science All Hands Meeting; 2005. [Google Scholar]
- 4.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1(2):142–160. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riloff E. Automatically generating extraction patterns from untagged text. Proc 13th Nat Conf on Artificial Intelligence; 1996. pp. 1044–1049. [Google Scholar]
- 6.Kim J-D, Ohta T, Tateisi Y, Tsujii J. GENIA corpus - a semantically annotated corpus for biotextmining. Bioinformatics. 2003;19( 1):180–182. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 7.Ogren PV, Savova GK, Buntrock JD, Chute CG. Building and evaluating annotated corpora for medical NLP systems. AMIA Annu Symp Proc. 2006;1049 [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers J, Puleston C, Rector A. The CLEF chronicle: patient histories derived from electronic health records. Proc 22nd Int Conf on Data Engineering Workshops; 2006. p. 109. [Google Scholar]
- 9.Hallet C, Power R, Scott D. Summarisation and visualization of e-health data repositories. Proc 5th UK e-Science All Hands Meeting; 2006. [Google Scholar]
- 10.Gennari JH, Musen MA, Fergerson RW, et al. The evolution of Protégé: an environment for knowledge-based systems development. Int J Hum Comput Stud. 2003;58:89–123. [Google Scholar]
- 11.Ogren PV. Knowtator: a Protégé plug-in for annotated corpus construction. Proc Human Language Technology. 2006:273–275. [Google Scholar]
- 12.Boisen S, Crystal MR, Schwartz R, Stone R, Weischedel R. Annotating resources for information extraction. Proc Language Resources and Evaluation. 2000;1211:1214. [Google Scholar]
- 13.Hripcsak G, Rothschild A. Agreement, F-measure and reliability in information retrieval. J Am Med Inform Assoc. 2005;12:296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]