Abstract
Clinical knowledge, best evidence, and practice patterns evolve over time. The ability to track these changes and study practice trends may be valuable for performance measurement and quality improvement efforts. The goal of this study was to assess the feasibility and validity of methods to generate and compare trends in biomedical literature and clinical narrative. We focused on the challenge of detecting trends in medication usage over time for two diseases: HIV/AIDS and asthma. Information about disease-specific medications in published randomized control trials and discharge summaries at New York-Presbyterian Hospital over a ten-year period were extracted using Natural Language Processing. This paper reports on the ability of our semi-automated process to discover disease-drug practice pattern trends and interpretation of findings across the biomedical and clinical text sources.
INTRODUCTION
Narrative data are valuable sources of information regarding patient characteristics, including diseases and medications. When analyzed over time, clinical data (including narrative) may reflect changes in practice resulting from advancements in medical knowledge and care processes. For example, the reporting of the effectiveness of new drugs for a particular disease in randomized controlled trials (RCTs) may be followed by the adoption of those drugs in patient care. Moreover, as new evidence emerges in RCTs, the use of past therapies may decline. Trends reflecting emergence or disappearance of certain therapies may be markers for adherence to best evidence and improvements in quality of care. Automated or semi-automated tools to generate and visualize trends based on information captured in electronic patient records compared to the published literature could support a range of activities including performance measurement and quality improvement initiatives.
Our group has been developing automated techniques for acquiring disease-specific associations from both the biomedical literature and patient record1. The main goal of this previous work was to discover the commonalities and differences in disease-drug knowledge among these disparate text sources. The present study builds upon this work by extending these methods to explore changes in the medications associated with related diseases over time. This paper describes a semi-automated approach to detecting disease-drug trends based on information extracted from RCTs in Medline and discharge summaries spanning a ten-year period at New York-Presbyterian Hospital (NYP). As proof of concept, we report on initial results in trend detection using these disparate text sources for two diseases: acquired immunodeficiency syndrome and asthma.
BACKGROUND
Trend Detection in Text
There have been several reports of general approaches and systems for discovering or detecting trends in textual data2. In these studies, a trend has been defined as “a specific subsequence of the history of a phrase that satisfies the users’ query over the histories”3 or “a clinically significant pattern in a sequence of time-ordered data”4. In a survey on emerging trend detection, Kontostathis and colleagues reported on automated and semi-automated systems developed for identifying topic areas that were growing in interest and utility over time5. These included systems for discovering and visualizing trends among patent data (PatentMiner3) and topics covered in a specific corpus (TimeMines6). From an industrial viewpoint, these systems may be valuable for purposes such as comparing past and current activities of a company or becoming aware of new developments in science, technology, or business. Other studies have discussed studying temporal changes in literature to support scientific research (research fronts and intellectual bases)7–10 and evidence-based medicine11,12. These projects have involved the use of Medline and the Web of Science for investigating research activities for aspirin7, anthrax8, heart diseases and lung cancer9,12, and the global prevalence of diseases10.
Related Work
A number of studies have been concerned with extracting co-occurrence data from the biomedical literature (i.e., Medline) and clinical narrative. In several of the reports, MeSH or NLP systems have been used to extract disease-drug pairs and summarize drug information from these disparate text sources13–15.
MedLEE16–18 and BioMedLEE19,20 are NLP systems at our institution that have been used for a variety of clinical and biomedical applications. While MedLEE is used to extract and encode information in clinical narrative, BioMedLEE is focused on extracting and structuring biomedical entities and relations in biomedical literature, including phenotypic and genotypic information. Both systems produce a set of primary findings (e.g., problem, procedure, and medication) along with associated modifiers (e.g., certainty, status, change, and body location) for a given document (e.g., discharge summary or Medline article). For example, in the sentence “His past medical history is significant for asthma” from a discharge summary, MedLEE extracts asthma as a primary finding with type problem where modifiers include certainty and status with values high certainty and past history, respectively. Codes may be available for primary findings as well as certain modifiers and are represented as additional modifiers called code. For this study, MedLEE and BioMedLEE generated UMLS codes to clinical findings (e.g., C0004096 corresponds to Asthma and C0001927 corresponds to Albuterol).
Our group has been performing studies for extracting disease-finding and disease-drug associations from clinical and biomedical documents. Cao et al. applied MedLEE to a set of discharge summaries and developed a statistical methodology for the automated generation of medical problem lists21,22. We adapted and extended the methods used by Cao and colleagues in order to acquire disease-drug associations from both the patient record (i.e., discharge summaries from 2003 and 2004) and biomedical literature (i.e., Medline RCTs focused on drug therapy in the 2006 Medline baseline)1. This ‘disease-drug association’ study involved extracting disease and drug entities (based on MeSH and BioMedLEE for the literature and processing of clinical narrative by MedLEE), mapping trade name drugs to their generic names where possible using drug knowledge sources, and applying the various statistical techniques to produce lists of associated generic drugs for a set of eight diseases (acquired immunodeficiency syndrome, asthma, breast neoplasms, congestive heart failure, diabetes mellitus, Parkinson’s disease, pneumonia, and schizophrenia). We found that the overall approach was effective and for the most part, the associations were consistent between the disparate text sources. The present study builds upon this work by expanding the approach to explore disease-drug trends in the patient record and how they correspond with knowledge in the literature.
METHODS & RESULTS
Overview
This study was a retrospective, descriptive study involving the analysis of two primary data sources: (1) ten years of discharge summaries authored by physicians at NYP and (2) RCTs published in electronic form in the 2006 Medline baseline. Both text sources were analyzed using NLP to detect emerging or disappearing medications for our chosen diseases. We selected two diseases, which we felt exemplified different paces of change: one rapidly evolving clinical domain (HIV/AIDS) and one more established clinical domain (asthma). The principal study measures were: (1) proportion of discharge summaries mentioning the use of medications for patients with the diseases and (2) frequency of RCTs for drugs used for these diseases. To discover trends, we generated graphical representations of the results. Clinical experts (practicing physicians and informaticians) then reviewed the trends for face validity.
Step 1: Selection and Processing of Documents
Discharge summaries from 1994 through 2004 were included in this study. In particular, we processed reports at 2- to 3-year intervals resulting in document sets for the following years: 1994, 1996, 1999, 2002, and 2004. For each year, the MedLEE XML output was transformed to a tabular representation for loading into database tables to facilitate queries on primary findings in the reports (specifically, diseases and drugs) and their corresponding UMLS codes23. Similar to the disease-drug association study, only findings considered present or current were maintained (e.g., findings with certainty value of no or rule out and status value of past were filtered out). The database tables were then queried to identify reports specific to HIV/AIDS and asthma. Here, we used the UMLS codes ‘C0001175’ and ‘C0004096’ to represent each disease respectively (issues with respect to disease classes and codes are discussed further in the discussion section).
As with the discharge summaries, we extracted documents specific to the two diseases from the RCTs in the 2006 Medline baseline based on UMLS concepts assigned by BioMedLEE (performed in our previous disease-drug association study). Table 1 presents the total number of documents and disease-specific documents in the collection.
Table 1.
Total and Disease-Specific Documents
| Document | Year | Total | HIV/AIDS | Asthma |
|---|---|---|---|---|
| Discharge Summaries | 1994 | 29,178 | 543 | 1,582 |
| 1996 | 31,629 | 561 | 1,869 | |
| 1999 | 27,817 | 285 | 1,565 | |
| 2002 | 25,332 | 301 | 1,546 | |
| 2004 | 25,742 | 805 | 1,457 | |
| Total | 139,698 | 2,495 | 8,019 | |
| Medline RCTs | 2006 Baseline | 81,828 | 1,612 | 2,777 |
Step 2: Identification of Disease-Specific Drugs
In order to obtain a list of drugs to focus on for each disease, we referred to the associations identified through automated methods in our prior disease-drug association study. Based on the statistical techniques, stronger associations (i.e., those above the calculated cutoff) from the Medline RCTs and discharge summaries (from 2003 and 2004) were combined in order to create disease-specific drug lists. This resulted in a list of 45 drugs for HIV/AIDS (e.g., ritonavir, zidovudine, and azithromycin) and 43 for asthma (e.g., albuterol, fluticasone, and montelukast).
In an initial review of these lists (by domain experts and references to medical knowledge sources), we found that the drugs for HIV/AIDS included antifungal agents, anti-cytomegalovirus agents, nucleoside reverse transcriptase inhibitors, and protease inhibitors. These were broken into two groups: (1) antiretrovirals and (2) those for the treatment or prevention of opportunistic infections (OIs). For asthma, the list of drugs included beta agonists, costicosteroids, and leukotriene inhibitors. These were broken into two groups for: (1) acute management and (2) chronic management of asthma.
While the intended goal was to produce trends for generic name drugs, we found that the trade name drug Advair was included in the list for asthma (as it could not be mapped by the drug knowledge sources used), which is the combination of fluticasone and salmeterol. Discovery of this combination agent for asthma led to our inclusion of common combinations for HIV/AIDS (i.e., Combivir [lamivudine + zidovudine] and Trizivir [abacavir + lamivudine + zidovudine]) and asthma (i.e., Advair and Combivent [albuterol + ipratropium]).
Step 3: Detection of Disease-Drug Trends
With the relevant information extracted from the discharge summaries and Medline RCTs, the next step was to generate trends reflecting changes for both diseases using the drug lists from the previous step. For each year of discharge summaries, we calculated the proportion of disease-specific reports containing the drug (number of reports including both the disease and drug out of the total number of reports for a disease in that year) in order to generate trends showing the changes in patient care across the ten years with respect to drugs for the particular diseases. While proportions were calculated for the clinical narratives, frequency counts were used for plotting the number of Medline RCTs (from the 1960s through 2005) reporting on both the disease and drug.
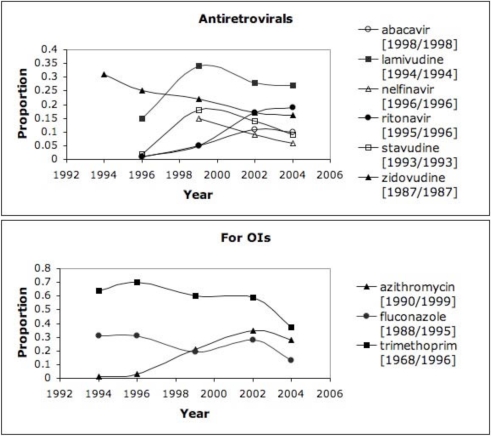
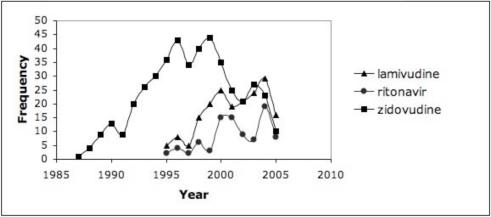
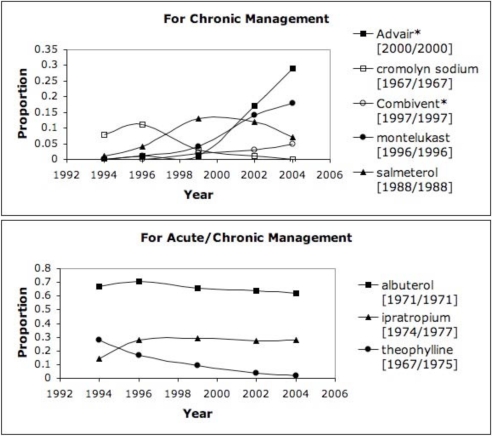
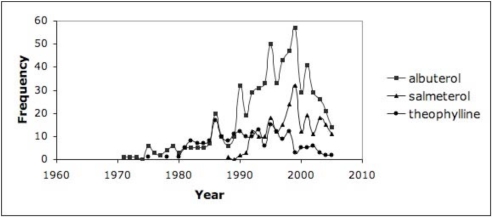
In order to highlight the more commonly used drugs in clinical practice, we set a threshold of 10% (proportion >= 0.1 in at least one year) as an initial assessment for the discharge summaries, which resulted in trends for 30–40% of the drugs for HIV/AIDS and asthma. Figure 1 highlights the trends from the discharge summaries for drugs related to HIV/AIDS (grouped as antiretrovirals or for OIs); Figure 2 shows the trends from RCTs for HIV/AIDS and antiretrovirals. Figure 3 depicts the drug trends for acute and chronic management of asthma from the discharge summaries while Figure 4 contains the trends for drugs for chronic management based on the RCTs. In order to further characterize and interpret the trends generated from the patient record with respect to the literature, we obtained the year the drug was first mentioned in all the RCTs (based on UMLS codes extracted by BioMedLEE) as well as those specific to the disease (included in the graphs for discharge summaries in Figures 1 and 3). Using these graphical representations and supplementary information about start years in the RCTs, the clinical experts manually reviewed the trends generated from the patient record and literature for face validity.
Figure 1.
DSUMs – HIV/AIDS. Proportion of HIV/AIDS-specific discharge summaries including antiretrovirals and drugs used for opportunistic infections (combination drugs indicated by ‘*’ and RCT start years are indicated by ‘[year/year for HIV/AIDS]’).
Figure 2.
RCTs – HIV/AIDS. Frequency of Medline RCTs reporting on HIV/AIDS and antiretrovirals from Figure 1.
Figure 3.
DSUMs - Asthma. Proportion of asthma-specific discharge summaries including drugs used for chronic or acute/chronic asthma management (combination drugs indicated by ‘*’ and RCT start years are indicated by ‘[year/year for asthma]’).
Figure 4.
Asthma - RCTs. Frequency of Medline RCTs reporting on drugs for chronic management of asthma from Figure 3.
Step 4: Interpretation of Disease-Drug Trends
The previous steps demonstrated the utility of information captured in discharge summaries for detecting trends in patterns of utilization of medications for two diseases. As an initial exploration, we observed the various increasing and decreasing trends and attempted to validate these changes as well as correlate them to studies in the biomedical literature (specifically, RCTs reporting on drug therapy and effectiveness for a disease).
Overall, we found that the trends were reflective of the changes in clinical knowledge and practice. Focusing on the antiretroviral drugs in Figure 1, the trends reveal an emergence of several newer drugs (abacavir and ritonavir appear in the Medline RCTs in 1998 and 1995, respectively) and decline of older drugs (zidovudine appears in 1987). Other decreasing trends are detected for lamivudine, stavudine, and nelfinavir starting around 1999. While causal relationships cannot be determined within the scope of this study, several reasons for decreasing trends of use are possible: (1) newer agents are more effective, (2) older agents have more side effects, and (3) development of resistance to older agents. With respect to the drugs depicted in Figure 1 for treating or suppressing OIs, a general decreasing trend is noted. This finding may be indicative of advancements in the therapies for HIV/AIDS leading to the decreasing prevalence of complications for which these drugs were intended (i.e., opportunistic infections). Finally, with respect to adoption of drugs, the estimated “lag time” or duration for translation into practice is on average one year (based on drugs with start years in both the RCTs and discharge summaries in the ten-year time window).
Analogous interpretations can be made for the drug trends associated with asthma. Newer drugs such as montelukast (appearing in 1996) became more frequent while older drugs begin to fall like theophylline (appearing in RCTs starting in 1967). While a slight decrease can be seen for albuterol and ipratropium, the trend is generally steady, which may be due to their continued effectiveness for managing asthma. On the other hand, the fall of theophylline, cromolyn sodium, and salmeterol over the ten years may be correlated with their lower effectiveness or discovery of adverse side effects. With regards to combination drugs that may be more effective than the individual drugs alone, we observed increasing usage of Advair and Combivent. Given that many of the drugs have start dates in the RCTs preceding 1994, we are unable to make conclusions about lag time and future work includes processing discharge summaries prior to the studied 10-year time period.
While we focused our review on trends in the patient record, we also made some initial observations about trends generated from disease and drug information in the Medline RCTs. We noticed a continuous up and down (oscillating) frequency of reports across the 40+ years. While further analysis is needed, we hypothesized that the ongoing reporting of certain drugs is due to their use as controls in RCTs against which newer medications are compared, or as part of combination agents.
DISCUSSION
In this work, we demonstrated the use of biomedical and clinical text to detect trends for specific diseases. While this study investigated disease-drug trends at the population-level, we believe that the approach is generalizable to other aspects of diseases (e.g., procedures and findings) as well as for detecting trends for providers. This approach involved using NLP to extract information from Medline RCTs and discharge summaries; however, this methodology does not preclude the use of other forms and sources of patient data for detecting practice patterns such as coded medical orders and diagnoses or outpatient data. Future investigations could include studying additional diseases, applying the methods for other types of trends, and exploring the use of other types of data (e.g., from ambulatory order entry systems or outpatient notes) and how they compare with or complement trend detection in discharge summaries. The initial interpretation of trends primarily focused on studying the direction (increasing or decreasing) and possible causes. Subsequent interpretations may include taking into consideration various factors such as disease incidence and prevalence data, drug pricing and insurance, and comorbidities.
Some of the challenges encountered in this study were related to the granularity of disease and drug concepts. These issues were encountered in our previous study and we have been exploring methods for resolving them. In reviewing the output from the NLP systems for the discharge summaries and RCTs, we found a range of UMLS codes for each disease in this study. For example, in the case of asthma, a series of related concepts are extracted from the documents including asthma, extrinsic asthma, intrinsic asthma, acute asthma, and chronic asthma. Similarly, for HIV/AIDS, we find various UMLS codes for concepts such as AIDS, HIV seropositivity, and HIV. For the two diseases of interest in this study, we used a single UMLS code (for the most general and frequent concept) to identify the disease-specific documents, but further investigations are needed to identify which codes to consider for a particular disease. The creation of these disease classes may be valuable for generating more accurate drug lists and trends for diseases at different levels of granularity (e.g., the general concept of asthma or more specifically, intrinsic or extrinsic asthma).
Other challenges encountered include determining disease-specific drugs, characterizing these drugs, and mapping of new or combination agents. Here, we leveraged results from our disease-drug association study that identified stronger associations from RCTs and discharge summaries; however, we may be interested in exploring the other associations identified or other sources of disease-drug knowledge. Next steps also include exploring semiautomated or automated approaches for grouping or categorizing drugs by purpose or function (e.g., determining if the drug is for the primary condition or for common co-morbidities).
CONCLUSION
Given the constantly evolving nature of healthcare, the ability to detect changes in an automated or semiautomated fashion may assist with performance measurement and quality improvement efforts. Clinical narrative data (e.g., discharge summaries) offer a rich source of information about patients such as diseases and medications while biomedical literature (e.g., RCTs) provides information regarding the testing and effectiveness of drug therapies. Analysis and comparison of knowledge within these documents over time could reveal emerging and disappearing trends in the drugs that are associated with particular diseases. The methods we have developed will continue to be refined and extended to a range of disease related information (e.g., drug therapy, diagnostic and therapeutic procedures, and symptoms) as they were shown to be promising for gaining insight into the evolution of disease management, studying adoption of therapies into practice, and assisting in performance evaluation.
Acknowledgments
The authors thank Hui Cao, Hua Xu, and Lyudmila Shagina for their contributions. This work is supported in part by grants LM007659, LM008635, LM006910, and LM008805 from the National Library of Medicine.
References
- 1.Chen E, Hripcsak G, Xu H, Markatou M, Friedman C.Automated acquisition of disease-drug associations from biomedical and clinical documents. 2007. (submitted) [DOI] [PMC free article] [PubMed]
- 2.Feldman R, Dagan I. Knowledge discovery in textual databases (KDT). First International Conference on Knowledge Discovery and Data Mining; 1995. pp. 112–7. [Google Scholar]
- 3.Lent B, Agrawal R, Srikant R. Discovering trends in text databases. Third International Conference on Knowledge Discovery and Data Mining; 1997. pp. 227–30. [Google Scholar]
- 4.Haimowitz I, Kohane I. Automated trend detection with alternate temporal hypotheses. Proceedings of the 13th International Joint Conference on Artificial Intelligence; 1993. pp. 146–51. [Google Scholar]
- 5.Kontostathis A, Galitsky L, Pottenger W, Roy S, Phelps D. A survey of emerging trend detection in textual data mining. Survey of Text Mining. 2003:185–224. [Google Scholar]
- 6.Swan R, Jensen D. TimeMines: Constructing timelines with statistical models of word usage. KDD-2000 Workshop on Text Mining. 2000:73–80. [Google Scholar]
- 7.Bordons M, Bravo C, Barrigon S. Time-tracking of the research profile of a drug using bibliometric tools. J Am Soc Inf Sci. 2004;55(5):445–461. [Google Scholar]
- 8.Morris S, Yen G, Wu Z, Asnake B. Time line visualization of research fronts. J Am Soc Inf Sci. 2003;54(5):413–422. [Google Scholar]
- 9.Chen C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci. 2006;57(3):359–377. [Google Scholar]
- 10.Srinivasan P, Wedenmeyer M. Mining concept profiles with the vector model or where on earth are diseases being studied. Third SIAM International Conference on Data Mining; 2003. [Google Scholar]
- 11.Tsay MY, Yang YH. Bibliometric analysis of the literature of randomized controlled trials. J Med Libr Assoc. 2005;93(4):450–8. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Chen Y. Searching for clinical evidence in CiteSpace. Proc AMIA Symp. 2005:121–5. [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. Proc AMIA Symp. 2002:722–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Fiszman M, Rindflesch TC, Kilicoglu H. Summarizing drug information in medline citations. Proc AMIA Symp. 2006:254–8. [PMC free article] [PubMed] [Google Scholar]
- 15.Rindflesch TC, Pakhomov SV, Fiszman M, Kilicoglu H, Sanchez VR. Medical facts to support inferencing in natural language processing. Proc AMIA Symp. 2005:634–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc. 1999;6(1):76–87. doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Friedman C.Extracting phenotypic information from the literature via natural language processing Medinfo 200411(Pt 2):758–62. [PubMed] [Google Scholar]
- 20.Lussier Y, Borlawsky T, Rappaport D, Liu Y, Friedman C. PhenoGO: Assigning phenotypic context to gene ontology annotation with natural language processing. Pacific Symposium on Biocomputing; 2006. pp. 64–75. [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H, Hripcsak G, Markatou M. A statistical methodology for analyzing cooccurrence data from a large sample. J Biomed Inform. 2007 Jun;40(3):343–52. doi: 10.1016/j.jbi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao H, Markatou M, Melton GB, Chiang MF, Hripcsak G. Mining a clinical data warehouse to discover disease-finding associations using co-occurrence statistics. Proc AMIA Symp. 2005:106–10. [PMC free article] [PubMed] [Google Scholar]
- 23.Chen E, Hiripcsak G, Friedman C. Disseminating natural language processed clinical narratives. Proc AMIA Annu Fall Symp. 2006:126–30. [PMC free article] [PubMed] [Google Scholar]