Abstract
OBJECTIVES:
To develop and evaluate an information processing method based on terminologies, in order to index medical documents in any given documentary context.
METHODS:
We designed a model using both symbolic general knowledge extracted from the Unified Medical Language System (UMLS) and statistical knowledge extracted from a domain of application. Using statistical knowledge allowed us to contextualize the general knowledge for every particular situation. For each document studied, the extracted terms are ranked to highlight the most significant ones. The model was tested on a set of 17,079 French standardized discharge summaries (SDSs).
RESULTS:
The most important ICD-10 term of each SDS was ranked 1st or 2nd by the method in nearly 90% of the cases. CONCLUSIONS: The use of several terminologies leads to more precise indexing. The improvement achieved in the model’s implementation performances as a result of using semantic relationships is encouraging.
Introduction
The manual indexing of medical documents needs qualified professionals with specialized knowledge of the terminology used for coding and is heavily time-consuming. Moreover its performances depend on the regularity and consistency of the indexers. Automated and semi-automated indexing are less likely to be affected by such limitations and thus their use could thus improve medical document indexing.
Several automated indexing methods are available such as the one described by Salton1 using the vector model and adopted in the SMART system. Another example of automated indexing is “latent semantic indexing” introduced by Chute et al. in 19912. Lastly, the OKAPI system uses a probabilistic model described by Sparck-Jones et al. in 20003. Other studies have addressed the selection of candidate describers retrieved in lexicons as opposed to full text analysis: either by reducing the text to be indexed in order to retain only the sections in which the relevant describers are likely to be found (e.g. MeSHMap4, MTI5); or by revising the list of extracted potential terms. On the other hand, the SAPHIRE6 indexing system uses canonical concepts. This system has been used by several authors including Huang et al.7 who used the concepts underpinning the Unified Medical Language System (UMLS)8 but who encountered major variations in precision and recall according to the terminologies used.
Medical Subject Headings (MeSH) is the most frequently used thesaurus for the indexing of medical publications9. Nevertheless, coding and indexing medical documents in other contexts require other terminologies. In France, the VUMeF project (Vocabulaire Unifié Médical Français)10 focuses on the indexing of medical documents using standard nomenclatures: MeSH for documentation, ICD-10 for disease coding, and SNOMED for the clinical coding of pathologies and medical procedures. In order to achieve this objective, several teams have created automated tools for the extraction of MeSH11 and ICD-10 terms12. In the framework of the present study, we propose a heuristic method designed to classify terms that have been automatically or manually extracted by order of significance in order to best convey the content of the documents13. Some terms can be considered “major” or more significant than others to describe content such as the major MesH terms in the Medline database.
Our research aims at developing and evaluating an information processing method based on terminologies in order to index medical documents in different documentary contexts.
Methods
We developed a model based on the following heuristic reasoning: the importance of a term is function of the number of relationships it has with other terms. Those relationships could use general symbolic knowledge issued from the UMLS and statistical data relative to an indexing domain in any given documentary context
Information Processing Model:
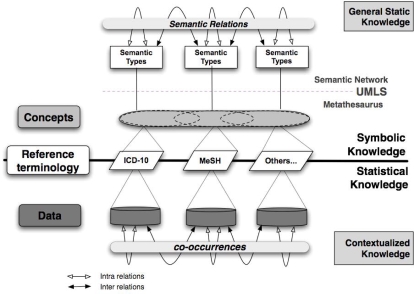
The model we designed (Figure 1) uses three levels of representation: data, terminologies and concepts. The data are connected to codes or terms in corresponding terminologies which are themselves connected to the concepts in the UMLS Metathesaurus when the terminologies are integrated in the UMLS. Each concept is linked to one or several semantic types which share semantic connections thus establishing possible links between concepts, and therefore between data. Statistical knowledge is mainly given by the frequencies of co-occurrences of codes from reference documents that have been manually indexed and validated. These co-occurrences can relate to a single terminology (e.g. diagnoses coded under ICD-10) or to different terminologies (e.g. ICD-10 and MeSH).
Figure 1.
Intra and inter referential information processing model.
The model contains two knowledge components: an invariable part (symbolic knowledge) and a contextual part (statistical knowledge).
Knowledge contextualization:
The statistical data allows contextualization of the knowledge according to a specific domain of use. Indeed, it is necessary to take into account the indexing context for two reasons:
The rules can vary according to the aim of the created indexing system: e.g. computerized medical files and medical publications each have their own indexing rules.
The terminologies used for indexing can differ (e.g. MeSH, ICD-10 or SNOMED).
Application of the model:
The model is designed to exploit terms extracted from medical documents and these could come from several terminologies.
A first version of the method has already been tested on Medline records and on medical documents indexed on the internet using MeSH13. To test our model with a large number of documents that integrate terms from several terminologies we exploited in this study standardized discharge summaries (SDSs). Diagnoses were coded using ICD-10 and technical medical procedures using the French Joint Classification of Technical Medical Procedures (or Classification Commune des Actes Médicaux: CCAM)14.
The aim was to retrieve, from the ICD-10 terms recorded in an SDS, the code considered the most significant, i.e the principal diagnosis (PD) at the time of coding.
We used two knowledge sources from the 2005AC version of the UMLS as semantic repositories15: the Metathesarus and the Semantic Network. The method was applied to anonymous SDSs from a University hospital in Marseilles, France. Two extractions were performed: 1) the whole 29,000 SDSs recorded in 2005, as a training set to create the statistical knowledge; 2) the 17,079 SDSs recorded during the first half of 2006, as a test set to evaluate the method.
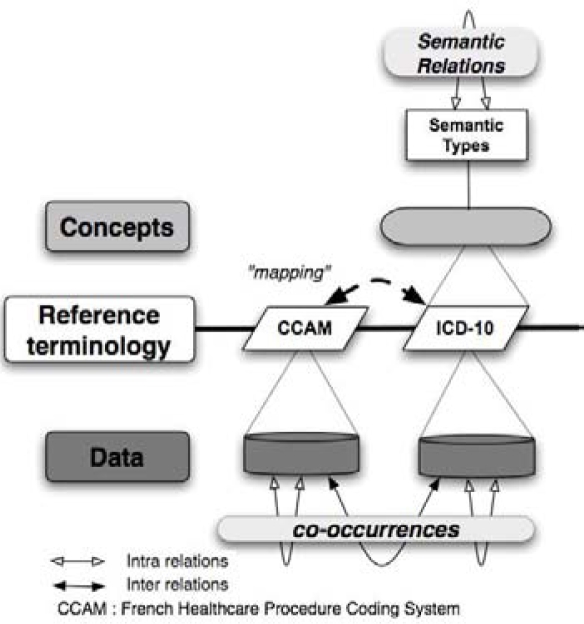
The French Healthcare Procedure Coding System (CCAM) was set up in France to encode technical medical procedures. This coding system uses a 7-character semi-structured code. The classification is not integrated into the UMLS. Thus, we had to perform a “mapping” between CCAM and ICD-10 (Fig 2). It was not a mapping of the CCAM into the UMLS. This component connects sub-chapters of ICD-10 with sub-chapters of CCAM using semantic links inspired by the semantic relationships of the UMLS.
Figure 2.
Intra and inter-referential information processing model adapted to our situation of test.
Semantic relationships:
All the possible associations between codes do not necessarily lend themselves to medical interpretation. Semantic relationships are used to determine whether the co-occurrence of two terms has a medical meaning. The co-occurrences which do not have sense are discarded. Among the 54 semantic relationships from the UMLS, we selected, on the basis of previous studies16, 17, 15 semantic relationships18 relevant to our study. This allowes us to considerably reduce the number of semantic links between concepts.
The table of frequencies of co-occurrences codes:
Our model requires the creation of a table of frequencies of co-occurrences codes, in the chosen nomenclatures. This co-occurrences table is used for knowledge contextualization. The frequencies attached to co-occurrence codes indicate the strength of a link between two codes in the area under investigation. In our study, we created a ICD-10 code co-occurrences table using codes taken from the SDSs in the training set.
Implementation of the method:
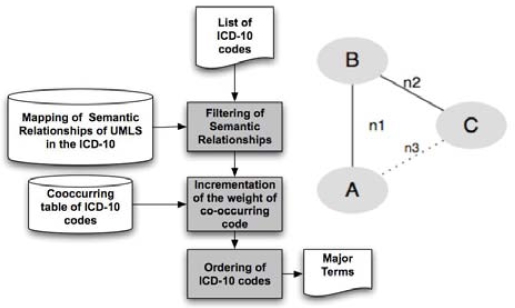
(Figure 3) A score is calculated for each index term extracted from a document in order to rank the terms according to their estimated relative importance in the document.
Figure 3.
Simplified architecture flow-chart illustrating the implementation of the model and the score calculation method.
For each SDS in the test set, each pair of ICD-10 codes composing the SDS was analyzed in turn:
Only those pairs of codes which display a semantic relationship between their components are retained. (e.g. A-B and B-C, Figure 3). If no semantic relationship is found between the components of the pair, the pair is discarded (e.g. A-C, Figure 3).
The co-occurrences table attributes a weight to the retained pairs. Each of the codes of the couple is incremented with a weight equal to their frequency of co-occurrences. (e.g. n1 and n2, but not n3 because there is no semantic relations, Figure 3)
When the whole set of combinations has been explored, the sum of the weights attributed by the different relationships by which a code is linked is affected to it.
The contribution of the CCAM:
When there are one or several CCAM codes in a SDS, all the ICD-10 and CCAM code pairs are analyzed. The mapping performed between ICD-10 and CCAM permits a filtering process which selects the pairs of codes which satisfy a semantic relationship of the type treats or diagnoses. Thus, a ICD-10 code semantically and statistically related to a CCAM procedure will be attributed an additional weight which will impact the ranking.
Evaluation:
We tested the method on the 2006 test set by retrieving the codes ranked 1st or 2nd and checking whether either of these was the principal diagnosis (PD), considered the most important term. We compared the results of the ranking depending on the number of ICD-10 codes in a given SDS. We proceeded in three stages firstly by using only the ICD-10 co-ccurrences without using the semantic network nor CCAM co-ccurrences, then using only the ICD-10 co-ccurrences and the semantic network and finally using all three knowledge sources: ICD-10 co-ccurrences, the CCAM co-ccurrences and the semantic network. We were then able to analyze their respective contributions to improving the ranking. The various results were compared depending on the number of ICD-10 codes in the SDS by performing Chi-square test on paired data using McNemar tests.
Results
Description of the population:
The test set contained 17,079 SDSs and fifty-one per cent of these contained only two ICD-10 codes (Table 1).
Table 1.
Description of the population of the test set
| Number of ICD-10 codes in a SDS* | Number of SDS | % | Cumulated % |
|---|---|---|---|
| 2 | 8,710 | 51.0 | 51.0 |
| 3 | 3,616 | 21.2 | 72.2 |
| 4 | 2,310 | 13.5 | 85.7 |
| 5 | 1,286 | 7.5 | 93.2 |
| 6 and more
|
1,157
|
6.8
|
100.0
|
| Total | 17,079 | 100.0 |
We discard 24 102 SDSs with only a single code as determining ranking was irrelevant.
Standardized discharge summaries with two ICD-10 codes:
In 91% of cases, we found the PD ranked 1st for SDSs with only two ICD-10 codes.
Standardized discharge summaries with three ICD-10 codes or more:
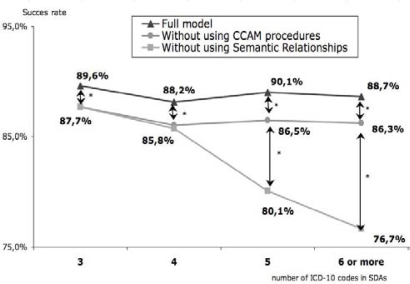
Figure 4 shows the percentage of PDs in 1st or 2nd rank as a function of the number of ICD-10 codes in the SDS. The top curve shows the complete model using ICD-10 co-occurrences, semantic relationships and the CCAM co-oocurences. For 3-code SDSs, the PD is ranked 1st or 2nd in 89.6% of cases.
Figure 4.
Success rate in finding the principal diagnosis in 1st or 2nd position according to the number of ICD-10 codes in the standardized discharge summaries. *p < 10–4
The results obtained for SDSs without using the CCAM co-oocurences are 3% lower whatever the number of codes in the SDSs (p<10-4).
The bottom curve represents the application of the method excluding semantic relationships and thus using only the statistical data regarding co-occurrences. The results with no semantics are much lower (p<10–5) than the other methods for SDSs with 5, 6 codes or more.
Discussion
Using all available knowledge, the method we describe here succeeds in top-ranking the most important ICD-code of SDSs in 90% of cases. These results are consistent with those obtained in previous studies performed with MeSH using Medline records13.Thus this method could participate in indexing SDSs or more importantly any other medical document. Nevertheless, to improve the performances of this system, further studies should try to characterize misindexing, which could help to increase the model accuracy. It could also lead to identify documents for which automated indexation has to be reviewed by professionals.
The contribution of semantic relationships:
The UMLS semantic network partially improves results as compared to those obtained using a purely statistical method. Nonetheless, semantic relationships make a significant (p<10–5) contribution to classification of SDSs with more than 4 diagnostic codes. This limited contribution is probably due to the small number of different semantic types within ICD-10 codes19. It is true that ‘Disease or Syndrome’ constitute half of these. Selection of the most appropriate semantic relationships20 can be made according to the application context, as we have done in retaining only some of them.
The contribution of a second specific terminology, the CCAM:
The use of multiple terminologies for indexing documents improves the ranking process, as shown by our results when we integrate CCAM co-oocurences in our calculation. The benefit obtained using CCAM procedures would probably be even greater if it was integrated into the UMLS. Indeed, the manual character of the mapping we performed on the top level terms limited the advantages to be drawn from this second terminology.
Integration of new terminologies into the UMLS increases the number of possible uses, as noted by Lindberg et al.8 and McCray & Nelson15. A study of the types of concept and a more specific use of semantic relationships are other means of further improvement17..
The method developed in this study was applied to ICD-10 codes extracted from SDSs. It seems also valid for other medical documents such as e-mails, examination assessments, computerized medical files, etc., provided that terms belonging to a standard nomenclature have previously been extracted.
Conclusion
The proposed model is general in character and could be applied to all nomenclatures integrated into the UMLS. This study demonstrates the benefits to be derived from using several semantic and statistical knowledge sources in order to enhance indexing quality.
Acknowledgments
We wish to thank Dr Christian Trapé who helped us perform the mapping between CCAM and ICD-10. Mrs Marthe-Aline Jutand and Dr Roch Giorgi for their well-informed advice on statistics. Mr George Morgan and Dr Philip Robinson for help with manuscript preparation. The US. NLM which kindly provided the authors with the UMLS knowledge sources.
References
- 1.Salton G. Automatic text analysis. Science. 1970 Apr 17;168(929):335–43. doi: 10.1126/science.168.3929.335. [DOI] [PubMed] [Google Scholar]
- 2.Chute CG, Yang Y, Evans DA. Latent semantic indexing of medical diagnoses using UMLS semantic structures. Proc Annu Symp Comput Appl Med Care. 1991:185–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Sparck Jones K, Walker S, Robertson SE.A probabilistic model of information retrieval: development and comparative experiments Information Processing and Management 200036Part 1779–808.Part 2 9–40. [Google Scholar]
- 4.Srinivasan P. MeSHmap: a text mining tool for MEDLINE. Proc AMIA Symp. 2001:642–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ.The NLM indexing initiative's medical text indexer Medinfo 200411(Pt 1)268–72. [PubMed] [Google Scholar]
- 6.Hersh WR, Greenes RA. SAPHIRE--an information retrieval system featuring concept matching, automatic indexing, probabilistic retrieval, and hierarchical relationships. Comput Biomed Res. 1990 Oct;23(5):410–25. doi: 10.1016/0010-4809(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Lowe HJ, Hersh WR. A pilot study of contextual UMLS indexing to improve the precision of concept-based representation in XML-structured clinical radiology reports. J Am Med Inform Assoc. 2003 Nov-Dec;10(6):580–7. doi: 10.1197/jamia.M1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilbur WJ, Kim W. The dimensions of indexing. AMIA Annu Symp Proc. 2003:714–0. [PMC free article] [PubMed] [Google Scholar]
- 10.Darmoni SJ, Jarrousse E, Zweigenbaum P, Le Beux P, Namer F, Baud R, Joubert M, Vallee H, Cote RA, Buemi A, Bourigault D, Recource G, Jeanneau S, Rodrigues JM. VUMeF: extending the french involvement in the UMLS Metathesaurus. AMIA Annu Symp Proc. 2003;824 [PMC free article] [PubMed] [Google Scholar]
- 11.Neveol A, Mork JG, Aronson AR, Darmoni SJ. Evaluation of french and english MeSH indexing systems with a parallel corpus. AMIA Annu Symp Proc. 2005:565–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira S, Neveol A, Massari P, Joubert M, Darmoni S. Construction of a semi-automated ICD-10 coding help system to optimize medical and economic coding. Stud Health Technol Inform. 2006;124:845–50. [PubMed] [Google Scholar]
- 13.Joubert M, Peretti A, Darmoni S, Dahamna B, Fieschi M. Contribution to an automated indexing of french-language health web sites. Proc AMIA Symp. 2006:409–13. [PMC free article] [PubMed] [Google Scholar]
- 14.Classification Commune des Actes Médicaux. [cited 2007 march 9th]; Available from: http://www.ccam.sante.fr/
- 15.McCray AT, Nelson SJ. The representation of meaning in the UMLS. Methods Inf Med. 1995 Mar;:34, 1–2, 193–201. [PubMed] [Google Scholar]
- 16.Fung KW, Bodenreider O. Utilizing the UMLS for semantic mapping between terminologies. AMIA Annu Symp Proc. 2005:266–70. [PMC free article] [PubMed] [Google Scholar]
- 17.McCray AT, Burgun A, Bodenreider O.Aggregating UMLS semantic types for reducing conceptual complexity Medinfo 200110(Pt 1)216–20. [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. J Biomed Inform. 2003 Dec;36(6):414–32. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimino JJ. Auditing the Unified Medical Language System with semantic methods. J Am Med Inform Assoc. 1998 Jan-Feb;5(1):41–51. doi: 10.1136/jamia.1998.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgun A, Bodenreider O.Methods for exploring the semantics of the relationships between co-occurring UMLS concepts Medinfo 200110(Pt 1)171–5. [PMC free article] [PubMed] [Google Scholar]