Abstract
Prostate cancer removal surgeries that result in tumor found at the surgical margin, otherwise known as a positive surgical margin, have a significantly higher chance of biochemical recurrence and clinical progression. To support clinical outcomes assessment a system was designed to automatically identify, extract, and classify key phrases from pathology reports describing this outcome. Heuristics and boundary detection were used to extract phrases. Phrases were then classified using support vector machines into one of three classes: ‘positive (involved) margins,’ ‘negative (uninvolved) margins,’ and ‘not-applicable or definitive.’ A total of 851 key phrases were extracted from a sample of 782 reports produced between 1996 and 2006 from two major hospitals. Despite differences in reporting style, at least 1 sentence containing a diagnosis was extracted from 780 of the 782 reports (99.74%). Of the 851 sentences extracted, 97.3% contained diagnoses. Overall accuracy of automated classification of extracted sentences into the three categories was 97.18%.
Introduction
According to the American Cancer Society, there were an estimated 234,460 new cases of prostate cancer in the United States in 2006 [1]. A prevalent treatment for prostate cancer is removal of the prostate and the surrounding lymph nodes in a procedure known as a radical retropubic prostatectomy (RRP). One unintended consequence of RRP is a positive surgical margin, which results from incising inadvertently into the prostate or incising into the extraprostatic tumor [2]. Positive margins are correlated with decreased cancer-specific and overall survival and a 2 to 4 times greater chance of biochemical cancer recurrence [3].
Identifying the number of patients with positive surgical margin is a requisite step toward improving RRP outcomes. Unfortunately, surgical margin status is largely reported in non-standardized, free text form within the associated pathology reports. As a result, margin status must be manually abstracted from these documents, a timely process.
The Clinical Outcomes Assessment Tool (COAT) is being designed as both an interface and Java API to facilitate medical records-based clinical outcomes assessment. As part of an ongoing study in the urological domain, COAT is being used to automatically identify phrases in pathology reports that reference the margin status of a prostate specimen. Once identified, these phrases (herein referred to as margin sentences) are automatically classified into one of three classes: 1) positive (involved) margin; 2) negative (uninvolved) margin; and 3) not applicable or definitive enough to diagnose margin status. We report an early evaluation of its ability to automatically identify and to classify margin sentences from a large sample of 782 free-text pathology reports taken from two hospitals.
Background
Reporting of margin status appears in the pathology report, along with other key prognostic parameters as set forth by the College of American Pathologists’ (CAP) prostate cancer protocol, including pathologic TNM stage (tumor staging) and Gleason score (histological analysis of tumor). Unlike the other CAP measures, which typically appear in a standard format (e.g., T2bN0Mx for TNM stage and 3+3=6 for Gleason score), margin status has no standard format for representation and often appears in sentence or phrase form. Examples of typical margin sentences extracted from pathology reports in the sample are provided in Table 1.
Table 1.
Examples of margin status references
| Positive surgical margin |
| “surgical margins involved at right apex” |
| “base margin positive focal left” |
| “tumor is present focally at the margin of resection” |
| Negative surgical margin |
| “no tumor present at the soft tissue resection margin” |
| “no carcinoma is present at the inked margin” |
The challenge of extracting information of interest from the clinical record has been approached with a variety of techniques. Robust natural language processing (NLP) technologies such as MedLEE [4] and MetaMap/MMTx [5] employ comprehensive medical dictionaries and part-of-speech taggers to structure clinical data. A few examples of successful clinical applications of such systems include formatting the contents of radiology reports [6], identifying candidates for clinical trials [7], and detection public health outbreaks [8]. Full NLP systems are not always necessary for identifying a fewer number of predetermined lexical targets. In many cases, fast and lightweight information extraction systems have been used to capitalize on the relative consistency of the appearance of specific clinical values in medical reports. These approaches typically employ patternrecognition techniques such as regular expression matching [9]. Several systems have combined extracted text features with machine learning algorithms to classify clinical documents or clinical values of interest within the documents [10]. One particular machine learning technique that has proven successful in classification of text features is support vector machines (SVMs) [11]. SVMs are linear classifiers that attempt to find a hyperplane that maximizes the margin between two different classes of instances. SVMs have been used in the clinical domain for several NLP-related tasks including document classification [12] and complex concept identification in radiology reports [13].
The availability of downloadable application programming interfaces (APIs) for various components required for medical language processing have made it possible for researchers to assemble pipelines of functionalities to accomplish specific information extraction tasks. For example, the Cancer Biomedical Informatics Grid’s (caBIG) text extraction tool ca- TIES [14] and Zeng et al.’s HITEx [15] feature language tools from the General Architecture for Text Engineering (GATE) [16]. While caTIES uses the NLM’s MetaMap/MMTx for medical concept mapping, HITEx implements only portions of the Unified Medical Language System [17] relevant to its tasks. The Clinical Outcomes Assessment Tool (COAT) is similarly designed, assembling a suite of functionalities specific to the task of facilitating records-based clinical outcomes assessment. It features a Java API of generalizable text processing facilities (record import, tokenization, regular expression matching, etc.), integration with MetaMap/MMTx for concept mapping, and Weka’s API for data and text mining [18]. These features are combined in an interface created for outcomes researchers to manipulate, manage, visualize, and analyze clinical data for outcomes assessment. COAT is being developed at UCLA in cooperation with the Center for Surgery and Public Health at Brigham and Women’s Hospital (BWH). In this current study, COAT was extended to identify and classify margin sentences in cooperation with urologists at both UCLA and BWH.
Methods
The goal of this study was to facilitate RRP outcomes assessment by identifying key phrases which describe the margin status of patients from pathology reports. Once extracted, a classification algorithm was applied to automatically classify the extracted sentences. A consideration in this study was how best to balance the level of customization needed to achieve an acceptable level of performance in identifying and classifying margin sentences, relative to the extensibility of this technique to encompass different institutions. To explore this issue, a sample of pathology reports from two major teaching hospitals, UCLA Medical Center and Brigham and Women’s Hospital, was collected for analysis and testing.
Data
A corpus of 787 pathology reports for patients having undergone RRPs was randomly selected from BWH (n = 456) and the UCLA Medical Center (n = 331). The pathology reports selected from BWH were drawn from RRP surgeries conducted at BWH between Jan. 1, 1996 and June 1, 2006. The UCLA pathology reports were drawn from surgeries performed between Jan. 1, 1998 and June 1, 2006. The population of patients having RRP procedures at both institutions during these time periods was approximately 3480. A total of 5 reports (.6% of sample) were excluded from the sample for being inaccurately coded as prostate cancer or for not featuring some reference to the surgical margin status as required by the American College of Surgeons Commission on Cancer [19]. As a result, the final number of reports comprising the sample was 453 from BWH and 329 from UCLA for a total sample of 782 reports. To conduct this study, IRB approvals were obtained from both institutions.
Heuristics Design
The first step in designing a method to automatically identify and extract margin sentences was an exploratory pilot analysis of the pathology reports. A random sample of 30 pathology reports was taken from each institution (60 total) to manually identify potential consistencies and differences in the description of margin status.
Reports from both BWH and UCLA feature a summary section in which margin status is described in narrative sentence form, along with other CAP prognostic parameters (tumor stage, Gleason score, etc.). Most UCLA reports also included a “Microscopic Examination” section that featured semi-structured text describing margin status and other analytic measures. For example:
Surgical Margin: Less than 1 mm from margin
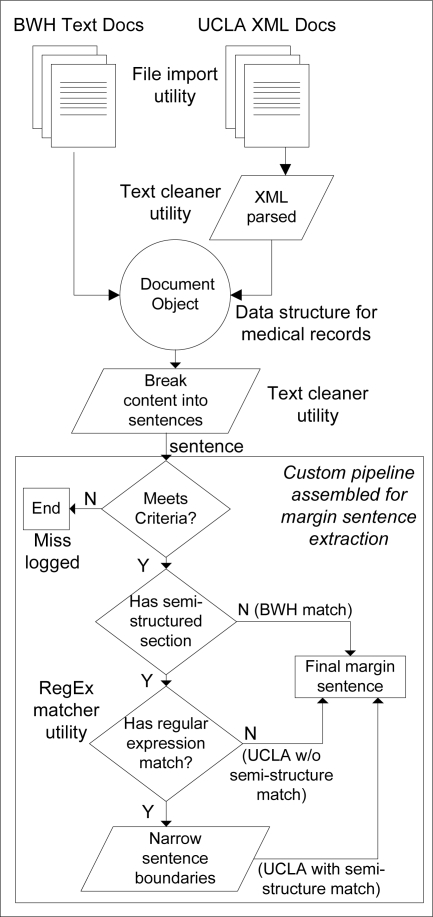
These features were incorporated in the design of an identification and extraction algorithm. A graphical overview of the pipeline used is provided in Figure 1.
Figure 1.
Workflow and classes for identifying and extracting margin sentences
Identification & Extraction of Margin Sentences
Sentence boundary detection was used to break up the reports and all text converted to lowercase. Based on the pilot study results a pair of two, two-word combinations appearing consistently in the corpus was used to flag potential sentences of interest. This set of trigger rules was supplemented with three additional combinations after iterating through the sample and logging misses, for a total of five trigger rules. Table 2 shows the rules used for extracting sentences describing margin status that were applied. To capitalize on the consistency of the semi-structured Microscopic Exam section in UCLA reports, the extraction algorithm was designed to make an initial pass on UCLA reports to identify margin status in this section using regular expression matching. The expression matched case-insensitive appearances of the word “margin” or “margins” followed by a colon and the word or words that followed.
Table 2.
Rules used for extracting margin sentences
| First Run Margin Sentence Rules |
| resection and (margin or margins) |
| surgical and (margin or margins) |
| Margin Sentence Rules Added |
| apical and (margin or margins) |
| tumor and (margin or margins) |
| carcinoma and (margin or margins) |
Regular expression: (?i)\\w+\\s*\\W*((margin)s*(:)\\s*\\W*\\w+)
In the case of a match, a modified boundary detection algorithm was used to accommodate the bulleted-style of text featured in the Microscopic Exam section. It captured all text between double line breaks when the semi-structured margin sentence regular expression was matched. This initial pass to capture semi-structured margin status in UCLA reports was the only variation in handling data from one hospital versus the other. The IDs of reports for which no match was found were logged and these reports were manually reviewed for causes of failures.
Classification of Margin Sentences
The sentences extracted from the sample of 782 reports were manually classified into one of three classes by the author (LWD) based a review of the literature and consultation with urologists to create a training set and gold standard for classification. All sentences were classified into one of three categories: 1) ‘positive (involved) surgical margin,’ 2) ‘negative (uninvolved) surgical margin,’ and 3) ‘not applicable or no explicit diagnosis.’ This third category was used to classify sentences extracted with no relevance to margin status, as well as sentences in which the diagnosis could not be definitively determined (i.e., false positives). Examples of category three sentences from the sample are provided in Table 3.
Table 3.
Examples of category 3 (false positive) sentences
| “the apical and basal margins are amputated and fixed separately” |
| “note benign prostate glands are focally present at an inked resection margin” |
Once classified, the sentences extracted from the reports were tokenized into vectors of lowercase words. A feature vector was created from all unique token appearances. Vectors of tokens appearing in classified sentences, as well as their assigned category were also created. COAT was integrated with Weka’s Java API and the vectors were passed to an implementation of an SVM classifier that used a polynomial kernel function and sequential minimal optimization (SMO) for training [20]. The performance of the classifier was evaluated using a 10-fold cross-validation.
Results
Identification of Margin Sentences
At least one potential margin sentence was extracted for 780 of the 782 documents. Extrapolated to the collection of approximately 3480 pathology reports, at least one sentence describing margin status should be extracted in 99.74% of reports (C.I. 99.56%, 99.92% with 95% confidence). The sentences describing margin status that were missed are listed below in Table 4.
Table 4.
Missed sentences describing margin status
| “Tumor is within 0.1 cm of the ink on both sides.” |
| “Margins, negative.” |
For the 780 documents from which sentences were extracted, 851 potential sentences were identified. Of the 851 sentences extracted, there were 23 false positives (category 3) for a sentence extraction precision of 97.3%. Of the 23 false positive sentences, 22 were from BWH, and 1 from UCLA. Examples of sentences falsely considered to be describing surgical margin status are featured in the previous Table 3.
Each document yielding a false positive sentence also produced at least 1 true positive sentence. In other words, all of the 780 documents from which sentences were identified produced at least one true positive sentence (category 1 or 2) from which a diagnosis of margin status could be made. The results of margin sentence extraction are provided in Table 5.
Table 5.
Margin sentence extraction results
| Reports | |
| RRP path reports with diagnosis of carcinoma of the prostate | 782 |
| Reports from which true positive (category 1 or 2) sentences were extracted | 780 (99.74%) |
| Sentences Extracted | |
| Number of sentences extracted | 851 |
| Positive margin sentences (category 1) | 131 |
| Negative margin sentences (category 2) | 697 |
| Not applicable or definitive (category 3) | 23 |
| Number of true positives (precision) | 828 (97.3%) |
| Distribution of False Positives | |
| BWH false positive sentences | 22 |
| UCLA false positive | 1 |
Classification of Margin Sentences
The SVM classifier correctly classified 827 of 851 sentences for an overall accuracy of 97.18%. Sensitivity and specificity for the three categories was as follows; category 1 (positive margin) = 96.95%, 98.77%, category 2 (negative margin) = 98.71%, 91.67, and category 3 (not applicable to or definitive of margin status) = 52.17%, 99.88%. Summaries of the overall classification results, sensitivity and specificity, area under receiver operator curves (ROC), and a confusion matrix are provided in the tables below.
Discussion
The overall performance of the system in identifying, extracting, and classifying sentences was promising. The only customization included in handling reports from the two different hospitals was a preliminary iteration in the algorithm to capitalize on the existence of any semi-structured reporting of margin status.
The preliminary pilot analysis of a small subset of reports was a useful starting point for designing heuristics for capturing potential margin sentences. It led to the five simple rules based on keyword appearance to capture sentences from 780 of 782 reports (99.74%) with only 23 (2.7%) extracted sentences not containing a margin status diagnosis. It also informed the decision to modify the extraction algorithm to capitalize the appearance of semi-structured reporting.
The larger collection of BWH documents explains some of the differential in false positives, but not the ratio of 22 to 1. In addition, only 3 of the 22 BWH false positive reports was created in 1996 or 1997, ruling out some significant temporal factor in producing false positives. Instead, the majority of BWH false positives might be attributed to the lack of consistency in the way margin status and related concepts are referenced in BWH RRP pathology reports. If this is the case, then the results offer support for the standardization of key analytic measures to facilitate automated outcomes assessment research.
The strong performance of the SVM classifier (97.19% accuracy) was partially an indication of the success of the boundary detection techniques in extracting representative feature vectors (margin sentences). It was also additional evidence of the power of SVMs in classifying text using distance-based techniques. We were also encouraged by the classifier’s performance in light of no modifications made to accommodate classification of margin sentences from one hospital versus the other.
Performance was predictably poor (12/23 for 53.3%) for classifying category three (not applicable) sentences due to a training set of only 23 sentences in the collection of 851 total sentences extracted. However, only one of the 11 misses for category 3 was falsely classified as a positive surgical margin (category 1) sentence while the rest were classified as negative margin sentences. This observation is important as it implies that, using this technique, incorrectly classified category 3 sentences are unlikely to inflate the number of cases resulting in positive surgical margin. While only 1.1% (8 sentences) of all negative margin sentences was incorrectly classified as positive margin, the number of positive margin cases was falsely inflated by 6.3%. The effect of the large ratio of negative to positive margin cases must therefore be considered in efforts to extend this technique for discovering the total number of positive margin cases in a collection.
The conclusions drawn from this study are limited by the sample used. First, both UCLA and BWH are American College of Surgeons Commission on Cancer-approved hospitals which indicates that both have achieved a baseline of “quality” in regards to their oncology services [19]. The format and inclusion of margin status in non-approved facilities may differ systematically. Second, this study did not account for the effects of poor data quality introduced by incorrectly assigned administrative codes or missing values.
Conclusion
We have demonstrated the ability to automatically identify and classify sentences describing a key outcome of prostate cancer surgery from pathology reports with high accuracy. This is a fundamental first step in supporting automated prostate cancer surgery outcomes assessment. Currently, COAT is being extended to capture other key quality measures including Gleason score and tumor stage. An evaluation of the effects of incorrect administrative code assignments and missing values on produced results is also in progress.
Table 6.
Overall classification accuracy
| Total Number of Sentences | 851 |
| Correctly Classified Sentences | 827 (97.19%) |
| Incorrectly Classified Sentences | 24 (2.82%) |
Table 7.
Sensitivity and specificity of classification
| Sensitivity | Specificity | |
|---|---|---|
| Category 1 (positive margin) | 96.95% | 98.77% |
| Category 2 (negative margin) | 98.71% | 91.67% |
| Category 3 (not applicable to margin status) | 52.17% | 99.88% |
Table 8.
Area under ROC for classification results
| Area under ROC | |
|---|---|
| Category 1 (positive margin) | .9789 |
| Category 2 (negative margin) | .9482 |
| Category 3 (not applicable) | .8457 |
Table 9.
Confusion matrix of classification results
| classified as → | Category 1 (positive) | Category 2 (negative) | Category 3 (NA) |
|---|---|---|---|
| Category 1 | 127 (97%) | 4 (3%) | 0 (0%) |
| Category 2 | 8 (1.1%) | 688 (98.7%) | 1 (.1%) |
| Category 3 | 1 (4.3%) | 10 (43.5%) | 12 (52.2%) |
Acknowledgments
This work was supported in part by the NLM Medical Informatics Training Grant # LM07356. This paper benefited from suggestions from Dr. Jim Sayre, Dr. David Miller, Dr. Jim Hu, and Vijay Bashyam.
References
- [1].American Cancer Society Overview: Prostate Cancer. [Web site] 2006 [cited 2006; Available: http://www.cancer.org/docroot/CRI/CRI_2_1x.asp?dt=36.
- [2].Richie J. Management of patients with positive surgical margins following radical prostatectomy. The Urological Clinics of North America. 1994;21:717. [PubMed] [Google Scholar]
- [3].Hull G, Rabbani F, et al. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. The Journal of Urology. 2002;167:528. doi: 10.1016/S0022-5347(01)69079-7. [DOI] [PubMed] [Google Scholar]
- [4].Friedman C, Alderson P, et al. A general natural language text processor for clinical radiology. Journal of American Medical Informatics Association. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Aronson A. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. In: Belfus H, editor. Vol. 2001. AMIA Symposium; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- [6].Hripsak G, Austin J, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224(1):157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- [7].Xu H, Anderson K, et al. Facilitating Cancer Research using Natural Language Processing of Pathology Reports. MedInfo. 2004;2004 [PubMed] [Google Scholar]
- [8].Chapman W, Fiszman M, et al. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap Medinfo2004 2004///11(Pt 1)487–91. [PubMed] [Google Scholar]
- [9].Turchin A, Kolatkar N, et al. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. Journal of the American Medical Informatics Association. 2006;13(6):691–8. doi: 10.1197/jamia.M2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chapman WW, Fizman M, et al. A comparison of classification algorithms to automatically identify chest X-ray reports that support pneumonia. J Biomed Inform. 2001;34(1):14. doi: 10.1006/jbin.2001.1000. [DOI] [PubMed] [Google Scholar]
- [11].Joachims T. Kluwer Academic Publishers; 2002. Learning to Classify Text Using Support Vector Machines: Methods, Theory and Algorithms. [Google Scholar]
- [12].Yetisgen-Yildiz M, Pratt W.The effect of feature representation on MEDLINE document classification Proceedings of the American Medical Informatics Association2005Washington, DC2005. [PMC free article] [PubMed] [Google Scholar]
- [13].Bashyam V, Taira R.Identifying Anatomical Phrases in Clinical Reports by Shallow Semantic Parsing Methods Proceedings of the IEEE Symposium on Computational Intelligence and Data Mining2007Honolulu, Hawaii2007. [Google Scholar]
- [14].Cancer Biomedical Informatics Grid. About caTIES 2007. [cited 2007 March 10, 2007]; Available: http://caties.cabig.upmc.edu/overview.html.
- [15].Zeng Q, Goryachev S, et al. Extracting principle diagnosis, co-morbidity, and smoking status for asthma research: evaluation of a natural language processing system. BMC Medical Informatics and Decision Making. 2006;6(30) doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cunningham H. GATE, A General Architecure for Text Engineering. Computers and the Humanities. 2004;36(2):223–54. [Google Scholar]
- [17].National Library of Medicine. UMLS Fact Sheet. 2006 [cited 2006 August 10]; Available from:
- [18].Witten I, Frank E. Data Mining: Practical machine learning tools and techniques. 2nd Edition ed. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- [19].American College of Surgeons Commission on Cancer Categories of Approval. [cited Feb. 24, 2007];Available: http://www.facs.org/cancer/coc/categories.html#3.
- [20].Platt J.Machines using sequential minimal optimizationIn: B S, C B, A S, eds. Advances in Kernel Methods - Support Vector Learning 1998.