Abstract
caGrid is the core Grid architecture of the NCI-sponsored cancer Biomedical Informatics Grid (caBIGTM) program. The current release, caGrid version 1.0, is developed as the production Grid software infrastructure of caBIGTM. Based on feedback from adopters of the previous version (caGrid 0.5), it has been significantly enhanced with new features and improvements to existing components. This paper presents an overview of caGrid 1.0, its main components, and enhancements over caGrid 0.5.
Introduction
The National Cancer Institute (NCI) initiated in 2004 a program, called cancer Biomedical Informatics Grid (caBIG™), to address the increasingly complex information management, integration, and analysis requirements of cancer research. The primary objectives of this program are to provide solutions for more effective sharing of data and tools in an open, voluntary network of cancer centers and research institutions and to enable researchers to leverage combined expertise, knowledge, and resources at multiple organizations. To achieve these goals, the caBIG™ community has been developing standards, applications, data and analytical resources, and a common middleware infrastructure, called caGrid1.
caGrid is a services oriented Grid software infrastructure, building on the Grid Services architecture2. It is designed to provide services, tools, and runtime support that will link applications and resources within the guidelines and policies accepted by the caBIGTM community and allow researchers to easily contribute to and leverage the resources in a multi-institutional environment. The first public release of caGrid was version 0.5, which provided the design and a reference implementation of the basic Grid architecture of the caBIG™ program. It was intended as a test bed infrastructure for the caBIG™ community to evaluate the architecture and provide feedback through implementation of several reference applications. Additional information about the caGrid 0.5 effort, and a good overview of the motivation of the Grid approach of caBIG™, can be found in an earlier paper1. The current release of caGrid is version 1.0 (caGrid 1.0), which was released to the caBIG™ participants and the research community at large in December 2006. It is a significant improvement over caGrid 0.5 with new features and enhancements to existing components. It is being used as the production Grid environment in caBIGTM. caGrid is not only built on open source, it is itself made publicly available under a liberal open source license for use both in and outside of caBIGTM. Additional information and downloads of the software can be found at the project web site*. This paper presents an overview of caGrid 1.0 and its enhancements over caGrid 0.5.
caGrid Framework
Objectives of caGrid
The design of caGrid is mainly driven by the requirements and use cases from the cancer research community. These use cases can be grouped into three main categories: 1) discovery, 2) integrated and large-scale data analysis, and 3) coordinated research. The first use case represents the need to support precisely targeted searches that return precisely defined attributes and data values from heterogeneous information sources. Using caGrid, a researcher can, for example, discover “all data sources in the environment that host Gene or Protein data” and submit queries to retrieve data subsets corresponding to his/her search criteria. The second use case reflects the need to support integration of different data types and analysis of large volumes of data. caGrid allows institutions or laboratories to expose their local molecular, specimen, and image datasets, collected from high throughput and high resolution instruments, to their collaborators, without having to deposit them in a centralized system. Collaborating researchers can remotely access these datasets and carry out operations to correlate these data types with clinical information and pathology annotations. The third use case is recognition of the fact that many types of basic and clinical cancer research involve coordinated access to information across multiple institutions. Examples are cooperative groups and multi-institutional clinical trials.
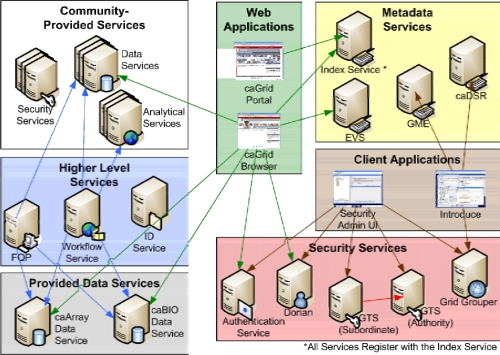
In order to support the use cases, caGrid aims to create an infrastructure wherein the structure and semantics of data can be programmatically determined and to provide a powerful means by which distributed data and analytical resources can be programmatically discovered and accessed. caGrid leverages Grid Services technologies and makes innovative use of several Grid systems, including the Globus Toolkit (http://www.globus.org) and Mobius3, and tools developed by the NCI such as the caCORE infrastructure4 to accomplish its goals. As a primary principle of caBIG™ is open standards, caGrid is built upon the Grid Services standards5 as a services oriented architecture. Each data and analytical resource in caGrid is implemented as a Grid Service, which interacts with other resources and clients using Grid Service protocols. The caGrid infrastructure also consists of coordination services, runtime environment to support the deployment, execution, and invocation of data and analytical services, and tools for easier development of services, management of security, and composition of services into workflows. The coordination services provide support for common Grid-wide operations required by clients and other services. These operations include metadata management; advertisement and discovery; federated query; workflow management; and security. The coordination services can be replicated and distributed to achieve better performance and scalability to large numbers of clients. Figure 1 shows the production deployment of the caGrid infrastructure with coordination services (e.g., metadata services, security services, workflow service) and data and analytical services provided by the community. Users can access these services via web portals or application specific client programs.
Figure 1.
caGrid infrastructure and environment.
caGrid 1.0 is built upon Globus Toolkit 4.0 (GT4), which is the most commonly deployed reference implementation of the Web Services Resource Framework (WSRF)5. However, it aims to be programming language and toolkit agnostic. Specifically, caGrid services are standard WSRF v1.2 services and can be accessed by any specification-compliant client.
caGrid implements several important functions on top of the basic Grid Services architecture to better address the informatics needs of cancer research. In the rest of this section we present an overview of these key features of the caGrid framework.
Interoperability and Model Driven Architecture
A primary distinction of the requirements implemented in caGrid is the attention given to syntactic and semantic interoperability. A major complication in the medical domain arises from the fact that there are a variety of representations of data sets and semantics associated with data elements and values. Controlled vocabularies, common data elements (CDEs), information models (or domain models), and well-defined application programming interfaces (APIs) are critical for interoperability and for correct interpretation of information in such an environment. To enable syntactic and semantic interoperability, the caBIG community has developed guidelines and a set of requirements to represent the interoperability level of an application in terms of vocabularies, data elements, information/domain models, and APIs. These levels and guidelines are outlined in the caBIG compatibility guidelines document*. For a resource to be caBIG compliant, it should have well defined APIs, which provide object-oriented access to backend resources, employ approved terminologies and common data elements based on these terminologies for its data models, and expose a published information model. caGrid adds to these requirements 1) service interfaces in the form of Grid services and XML for data exchange and 2) a common framework across the caBIG federation for the representation, advertisement, discovery, and invocation of distributed resources.
caGrid adopts a model-driven architecture best practice. Client and service APIs in caGrid represent an object-oriented view of data and analytical resources. These APIs operate on registered data models, expressed as object classes and relationships between the classes in UML. caGrid leverages existing NCI data modeling infrastructure to manage, curate, and employ the data models. Data models are defined in UML and converted into common data elements, which are in turn registered in the Cancer Data Standards Repository4 (caDSR). The definitions of these data elements draw from vocabulary registered in the Enterprise Vocabulary Services4 (EVS). The concepts and relationships of data elements thus are semantically described. Clients and services communicate through the Grid using messages encoded in XML. In caGrid, when an objects is transferred over the Grid between clients and services, it is serialized into a XML document that adheres to a XML schema registered in the Mobius Global Model Exchange3 (GME) service. As the caDSR and EVS define the properties, relationships, and semantics of caBIG™ data types, the GME defines the syntax of their XML materialization.
Semantic Discovery of Resources
A critical requirement of the caGrid framework is that it supports the ability of researchers to discover distributed resources. This ability is enabled by taking advantage of rich structural and semantic descriptions of data models and services. Each caGrid service is required to describe itself using service metadata. caGrid provides support for rich service metadata. At the base is the common service metadata standard to which every service is required to adhere. This metadata contains information about the service-providing cancer center, such as the point of contact and the institution’s name providing the service. It also describes the objects used as input and output of the service’s operations. The definitions of the objects themselves are described in terms of their underlying concepts, attributes, attribute value domains, and associations to other objects. In addition, the service metadata specifies the operations or methods the service provides, and allows semantic concepts, extracted from the EVS, to be applied to them. This base metadata is extended for different types of services. Data Services, for example, provide an additional metadata standard, which details the domain model, including associations and inheritance information, from which the objects being exposed by the service are drawn. In this way, all services fully define their object types by referencing the corresponding data models registered in caDSR, and identify their underlying semantic concepts from EVS.
When a service is deployed, its service metadata is registered with an indexing registry service, called the Index Service (IS) in the caGrid infrastructure. This service can be thought of the repository of information about all advertised and available services in the environment. caGrid 1.0 provides a series of high-level APIs for performing searches on these metadata standards, thus facilitating discovery of resources based on data models and semantic information associated with them. For instance, using the API, a researcher can submit a query to the IS to search for data services that “serve Radiology images from Institutions X and Y”.
Security
Security is a required component both for protecting intellectual property and to ensure protection and privacy of patient related information. The underlying system should enable data owners to set and enforce their access control policies in collaboration with others or autonomously. caGrid provides a comprehensive set of services for security. These services enable Grid-wide management of user credentials, support for grouping of users into virtual organizations for role based access control, and management of trust fabric in the Grid.
caGrid Version 1.0 Components and Enhancements
Based on lessons learned from caGrid 0.5 and feedback from the community, caGrid 1.0 has been enhanced in the areas of coordination services, runtime environment, and tooling support to satisfy the additional requirements and comply with current standards. In this section, we describe the improvements and additional features of caGrid 1.0.
Enhanced Metadata Support
In caGrid 0.5, caDSR and EVS were accessible only through their own APIs and proprietary communication protocols. This limited their use by client applications, other caGrid tools, and caGrid services. caGrid 1.0 has implemented Grid service access to both the EVS and caDSR. Also, the new service metadata standards include additions of information extracted from the caDSR and EVS, providing richer metadata and better support for semantic discovery.
Enhanced Support for Service Development and Deployment
One of the barriers to adoption of Grid technologies in application domains is that Grid middleware toolkits like GT4 require a good understanding of the Grid and low-level details of the toolkit to develop and deploy Grid services. Since caGrid is envisioned to be used by developers with different levels of knowledge of the Grid, one of the priorities has been to provide high-level tools to facilitate easier development of services. To this end, we have developed the Introduce toolkit6. Initially implemented as a tool for analytical services, Introduce has become a unified Grid service authoring toolkit in caGrid 1.0, implemented as an extensible framework and graphical workbench. The toolkit reduces the service developer’s efforts by abstracting away the need to manage the low level details of the WSRF specification and integration with the GT4. Developers with existing caBIG™ Silver compatible systems need only follow simple a wizard-like process for creating the “adapter” between the Grid and their system.
Support for Grid Workflows
One significant feature provided by caGrid 1.0, which lacked in caGrid 0.5, is the addition of service support for orchestration of Grid services using the industry standard Business Process Execution Language7 (BPEL). The caBIGTM environment is expected to provide an increasing number of analytical and data services developed and deployed by different institutions. Powerful applications can be created by composing workflows that access multiple services, thus harnessing data and analytical methods exposed as services more effectively. In such applications, information can be queried and extracted from one or more data sources and processed through a network of analytical services. caGrid 1.0 provides a workflow management service, enabling the execution and monitoring of BPEL-defined workflows in a secure Grid environment.
Federated Query Support
Another higher-level support service made available in caGrid 1.0 is the Federated Query Infrastructure. It provides a mechanism to perform basic distributed aggregations and joins of queries over multiple data services. An extension to the standard Data Service query language, implemented in caGrid 0.5, has been developed to describe distributed query scenarios, as well as various enhancements to the Data Service query language itself. The Federated Query Infrastructure contains three main client-facing components: an API implementing the business logic of federated query support, a Grid service providing remote access to that engine, and a Grid service for managing status and results of queries that were invoked asynchronously using the query service.
Support for Large Dataset Retrieval
Numerous improvements to the handling of large data sets and distributed information processing have been made. Support for WS-Enumeration has been implemented and added to the Globus Toolkit. This provides the capability for a Grid client to enumerate over results provided by a Grid service (much like a Grid-enabled cursor). This support is integrated into the caGrid Data Service tools, providing a mechanism for iterating on large query results from a service. Additionally, the initial effort to standardize a “bulk data transport” interface for large data has been started in caGrid 1.0, which is intended to provide uniform mechanism by which clients may access data sets form arbitrary services. This work currently supports access via WS-Enumeration, WS-Transfer, and GridFTP8. The bulk data transport effort was mainly motivated by the caBIG in-vivo imaging middleware effort9, which builds on caGrid, and is employed in that middleware system.
Security
caGrid 1.0 provides a complete overhaul of federated security infrastructure of caGrid 0.5 to satisfy caBIG™ security needs, culminating in the creation of the Grid Authentication and Authorization with Reliably Distributed Services (GAARDS) infrastructure10*. GAARDS provides services and tools for the administration and enforcement of security policy in an enterprise Grid. These services and tools provide support for Grid user management, identity federation, trust management, group/VO management, access control policy management and enforcement, and integration between existing security domains and the Grid security domain. GAARDS consists of three main components. Dorian11 provides support for provisioning and federation of Grid user identities and credentials. GAARDS provides flexible mechanism for creating and managing user accounts on the Grid. System administrators at a user’s institution can create new Grid user accounts directly with Dorian or link existing user accounts to Dorian. Part of the authentication process in the Grid is to verify that the client credentials were issued by a trusted Grid credential provider (e.g., Dorian). To support this requirement, the Grid Trust Service (GTS) of GAARDS maintains a federated trust fabric of all the trusted credential providers in the Grid. Authorization is another important requirement. The Grid Grouper service of GAARDS provides a group-based authorization solution for the Grid, wherein Grid services and applications enforce local authorization policy based on membership to groups defined and managed at the Grid level. Services can use Grid Grouper to enforce their internal access control policies.
Conclusions
caGrid 1.0 release represents a major milestone in caBIG™ towards achieving the program goals. It is developed to be used as a production infrastructure. It provides a comprehensive set of core services, toolkits for the development and deployment of community provided services, and APIs for building client applications. It already has been employed in application development; information on the applications and services, developed using caGrid 1.0, is available at the caGrid project web site.
Acknowledgments
The authors would like to acknowledge the contributions of the entire caGrid technical team, the members of the Architecture Workspace, the NCICB caCORE team, and the caBIG™ community. The authors also would like to thank Ken Buetow for his vision of a Cancer Grid and his tireless efforts in making caBIG™ a reality, and Patrick McConnell for his contributions on caGrid requirements and being a liaison between the caGrid developer team and the caBIG application community. This work was supported in part by the NCI caGrid developer grant 79077CBS10, the State of Ohio BRTT Program grants ODOD AGMT-TECH 04-049 and BRTT02-0003, and the NCI grant U54 CA113001.
Footnotes
https://cabig.nci.nih.gov/workspaces/Architecture/caGrid
https://cabig.nci.nih.gov/guidelines_documentation/
More detailed information about the security infrastructure is also available at the caGrid project site and wiki site (http://www.cagrid.org).
References
- 1.Saltz J, Oster S, Hastings S, et al. caGrid: Design and Implementation of the Core Architecture of the Cancer Biomedical Informatics Grid. Bioinformatics. 2006;22(15):1910–1916. doi: 10.1093/bioinformatics/btl272. [DOI] [PubMed] [Google Scholar]
- 2.Foster I, Kesselman C, Nick J, Tuecke S. Grid Services for Distributed System Integration. Computer. 2002;35(6):37–46. [Google Scholar]
- 3.Hastings S, Langella S, Oster S, Saltz J. Distributed Data Management and Integration: The Mobius Project. Proceedings of GGF11 Semantic Grid Applications Workshop; Honolulu, Hawaii, USA. 2004. pp. 20–38. [Google Scholar]
- 4.Phillips J, Chilukuri R, Fragoso G, Warzel D, Covitz PA. The caCORE Software Development Kit: Streamlining construction of interoperable biomedical information services. BMC Medical Informatics and Decision Making. 2006;6(2) doi: 10.1186/1472-6947-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czajkowski K, Ferguson DF, Foster I, et al. The WS-Resource Framework version 1.0 http://www.globus.org/wsrf/specs/ws-wsrf.pdf, 2004
- 6.Hastings S, Oster S, Langella S, Ervin D, Kurc T, Saltz J. Introduce: An Open Source Toolkit for Rapid Development of Strongly Typed Grid Services. Journal of Grid Computing (in press, available online) 2007 [Google Scholar]
- 7.Bussiness Process Execution Language for Web Services version 1.1. 2006. http://www-128.ibm.com/developerworks/library/specification/ws-bpel/
- 8.Allcock WE, Foster I, Madduri R. Reliable Data Transport: A Critical Service for the Grid. Proceedings of Building Service Based Grids Workshop; Honolulu, Hawaii, USA. 2004. [Google Scholar]
- 9.Gurcan M, Pan T, Sharma A, et al. GridImage: A Novel Use of Grid Computing to Support Interactive Human and Computer-Assisted Detection Decision Support. Journal of Digital Imaging (in press, available online) 2007 doi: 10.1007/s10278-007-9020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langella S, Oster S, Hastings S, et al. The Cancer Biomedical Informatics Grid (caBIG) Security Infrastructure. AMIA 2007 Annual Symposium; Chicago, IL. 2007. [PMC free article] [PubMed] [Google Scholar]
- 11.Langella S, Oster S, Hastings S, Siebenlist F, Kurc T, Saltz J. Dorian: Grid Service Infrastructure for Identity Management and Federation; The 19th, IEEE, Symposium on Computer-Based Medical Systems; Salt Lake City, Utah. 2006. [Google Scholar]